Project
PRJNA195909:zebrafish embryo and larva development
Navigation
Downloads
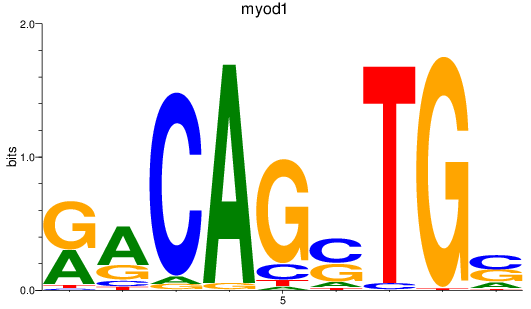
Results for myod1
Z-value: 1.10
Transcription factors associated with myod1
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
myod1
|
ENSDARG00000030110 | myogenic differentiation 1 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| myod1 | dr11_v1_chr25_-_31423493_31423493 | 0.30 | 4.3e-01 | Click! |
Activity profile of myod1 motif
Sorted Z-values of myod1 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr7_+_49715750 | 1.11 |
ENSDART00000019446
|
ascl1b
|
achaete-scute family bHLH transcription factor 1b |
| chr7_+_39402864 | 1.07 |
ENSDART00000025852
|
tnni2b.1
|
troponin I type 2b (skeletal, fast), tandem duplicate 1 |
| chr6_+_40629066 | 0.85 |
ENSDART00000103757
|
slc6a11a
|
solute carrier family 6 (neurotransmitter transporter), member 11a |
| chr3_-_32817274 | 0.76 |
ENSDART00000142582
|
mylpfa
|
myosin light chain, phosphorylatable, fast skeletal muscle a |
| chr10_+_31244619 | 0.75 |
ENSDART00000145562
ENSDART00000184412 |
robo4
|
roundabout, axon guidance receptor, homolog 4 (Drosophila) |
| chr6_-_42003780 | 0.72 |
ENSDART00000032527
|
cav3
|
caveolin 3 |
| chr3_-_31804481 | 0.72 |
ENSDART00000028270
|
gfap
|
glial fibrillary acidic protein |
| chr6_+_3680651 | 0.69 |
ENSDART00000013588
|
klhl41b
|
kelch-like family member 41b |
| chr21_-_25741096 | 0.58 |
ENSDART00000181756
|
cldnh
|
claudin h |
| chr8_-_18582922 | 0.58 |
ENSDART00000123917
|
tmem47
|
transmembrane protein 47 |
| chr11_-_5865744 | 0.57 |
ENSDART00000104360
|
gamt
|
guanidinoacetate N-methyltransferase |
| chr23_+_23658474 | 0.57 |
ENSDART00000162838
|
agrn
|
agrin |
| chr20_+_34913069 | 0.56 |
ENSDART00000007584
|
snap25a
|
synaptosomal-associated protein, 25a |
| chr21_-_23331619 | 0.56 |
ENSDART00000007806
|
zbtb16a
|
zinc finger and BTB domain containing 16a |
| chr17_+_27434626 | 0.55 |
ENSDART00000052446
|
vgll2b
|
vestigial-like family member 2b |
| chr21_-_25741411 | 0.55 |
ENSDART00000101211
|
cldnh
|
claudin h |
| chr5_-_28679135 | 0.54 |
ENSDART00000193585
|
tnc
|
tenascin C |
| chr24_+_7861373 | 0.53 |
ENSDART00000019705
|
bmp6
|
bone morphogenetic protein 6 |
| chr13_+_42124566 | 0.52 |
ENSDART00000074707
|
cdc42ep3
|
CDC42 effector protein (Rho GTPase binding) 3 |
| chr12_-_4070058 | 0.50 |
ENSDART00000042200
|
aldoab
|
aldolase a, fructose-bisphosphate, b |
| chr14_+_22113331 | 0.50 |
ENSDART00000109759
|
tmx2a
|
thioredoxin-related transmembrane protein 2a |
| chr20_-_32446406 | 0.50 |
ENSDART00000026635
|
nr2e1
|
nuclear receptor subfamily 2, group E, member 1 |
| chr14_-_17563773 | 0.49 |
ENSDART00000082667
|
fgfrl1a
|
fibroblast growth factor receptor like 1a |
| chr21_+_27382893 | 0.49 |
ENSDART00000005682
|
actn3a
|
actinin alpha 3a |
| chr19_+_19767567 | 0.49 |
ENSDART00000169074
|
hoxa3a
|
homeobox A3a |
| chr4_+_12031958 | 0.49 |
ENSDART00000044154
|
tnnt2c
|
troponin T2c, cardiac |
| chr2_-_21335131 | 0.49 |
ENSDART00000057022
|
klhl40a
|
kelch-like family member 40a |
| chr18_-_6634424 | 0.48 |
ENSDART00000062423
ENSDART00000179955 |
tnni1c
|
troponin I, skeletal, slow c |
| chr6_+_47843760 | 0.47 |
ENSDART00000140943
|
padi2
|
peptidyl arginine deiminase, type II |
| chr14_-_30704075 | 0.47 |
ENSDART00000134098
|
efemp2a
|
EGF containing fibulin extracellular matrix protein 2a |
| chr22_-_651719 | 0.47 |
ENSDART00000148692
|
apobec2a
|
apolipoprotein B mRNA editing enzyme, catalytic polypeptide-like 2a |
| chr5_+_36693859 | 0.46 |
ENSDART00000019259
|
dlb
|
deltaB |
| chr3_+_32526263 | 0.45 |
ENSDART00000150897
|
si:ch73-367p23.2
|
si:ch73-367p23.2 |
| chr9_-_7539297 | 0.45 |
ENSDART00000081550
ENSDART00000081553 |
desma
|
desmin a |
| chr5_-_63515210 | 0.45 |
ENSDART00000022348
|
prdm12b
|
PR domain containing 12b |
| chr23_+_36063599 | 0.45 |
ENSDART00000103147
|
hoxc12a
|
homeobox C12a |
| chr2_-_44255537 | 0.44 |
ENSDART00000011188
ENSDART00000093298 |
atp1a2a
|
ATPase Na+/K+ transporting subunit alpha 2 |
| chr24_+_20575259 | 0.44 |
ENSDART00000010488
|
klhl40b
|
kelch-like family member 40b |
| chr18_+_38288877 | 0.44 |
ENSDART00000134247
|
lmo2
|
LIM domain only 2 (rhombotin-like 1) |
| chr19_+_1184878 | 0.44 |
ENSDART00000163539
|
scrt1a
|
scratch family zinc finger 1a |
| chr19_-_2317558 | 0.43 |
ENSDART00000190300
|
sp8a
|
sp8 transcription factor a |
| chr21_-_7265219 | 0.43 |
ENSDART00000158852
|
egfl7
|
EGF-like-domain, multiple 7 |
| chr16_-_14074594 | 0.42 |
ENSDART00000090234
|
trim109
|
tripartite motif containing 109 |
| chr19_-_7450796 | 0.42 |
ENSDART00000104750
|
mllt11
|
MLLT11, transcription factor 7 cofactor |
| chr17_+_52822831 | 0.42 |
ENSDART00000193368
|
meis2a
|
Meis homeobox 2a |
| chr23_-_5683147 | 0.42 |
ENSDART00000102766
ENSDART00000067351 |
tnnt2a
|
troponin T type 2a (cardiac) |
| chr3_+_37574885 | 0.42 |
ENSDART00000055225
|
wnt9b
|
wingless-type MMTV integration site family, member 9B |
| chr23_-_3409140 | 0.42 |
ENSDART00000002309
|
mafba
|
v-maf avian musculoaponeurotic fibrosarcoma oncogene homolog Ba |
| chr2_+_55982940 | 0.42 |
ENSDART00000097753
ENSDART00000097751 |
nmrk2
|
nicotinamide riboside kinase 2 |
| chr1_+_39553040 | 0.42 |
ENSDART00000137676
|
tenm3
|
teneurin transmembrane protein 3 |
| chr14_+_6159356 | 0.42 |
ENSDART00000157730
|
bscl2l
|
Bernardinelli-Seip congenital lipodystrophy 2, like |
| chr5_-_40734045 | 0.41 |
ENSDART00000010896
|
isl1
|
ISL LIM homeobox 1 |
| chr11_+_14622379 | 0.41 |
ENSDART00000112589
|
efna2b
|
ephrin-A2b |
| chr14_-_33044955 | 0.41 |
ENSDART00000170626
|
kdrl
|
kinase insert domain receptor like |
| chr11_-_41966854 | 0.41 |
ENSDART00000055709
|
her2
|
hairy-related 2 |
| chr7_-_18881358 | 0.41 |
ENSDART00000021502
|
mllt3
|
MLLT3, super elongation complex subunit |
| chr9_-_98982 | 0.40 |
ENSDART00000147882
|
lims2
|
LIM and senescent cell antigen-like domains 2 |
| chr15_+_43906043 | 0.40 |
ENSDART00000010881
|
naalad2
|
N-acetylated alpha-linked acidic dipeptidase 2 |
| chr23_-_29003864 | 0.40 |
ENSDART00000148257
|
casz1
|
castor zinc finger 1 |
| chr16_+_46294337 | 0.40 |
ENSDART00000040769
|
nr2f5
|
nuclear receptor subfamily 2, group F, member 5 |
| chr19_-_8877469 | 0.40 |
ENSDART00000193951
|
celf3a
|
cugbp, Elav-like family member 3a |
| chr7_+_31891110 | 0.39 |
ENSDART00000173883
|
mybpc3
|
myosin binding protein C, cardiac |
| chr9_-_22831836 | 0.39 |
ENSDART00000142585
|
neb
|
nebulin |
| chr14_-_25956804 | 0.38 |
ENSDART00000135627
ENSDART00000146022 ENSDART00000039660 |
sparc
|
secreted protein, acidic, cysteine-rich (osteonectin) |
| chr22_-_26595027 | 0.37 |
ENSDART00000184162
|
CABZ01072309.1
|
|
| chr13_-_15994419 | 0.37 |
ENSDART00000079724
ENSDART00000042377 ENSDART00000046079 ENSDART00000050481 ENSDART00000016430 |
ikzf1
|
IKAROS family zinc finger 1 (Ikaros) |
| chr4_-_16354292 | 0.36 |
ENSDART00000139919
|
lum
|
lumican |
| chr4_-_16412084 | 0.36 |
ENSDART00000188460
|
dcn
|
decorin |
| chr19_-_6385594 | 0.35 |
ENSDART00000104950
|
atp1a3a
|
ATPase Na+/K+ transporting subunit alpha 3a |
| chr16_-_42894628 | 0.35 |
ENSDART00000045600
|
hfe2
|
hemochromatosis type 2 |
| chr1_+_4101741 | 0.35 |
ENSDART00000163793
|
slitrk6
|
SLIT and NTRK-like family, member 6 |
| chr8_-_26609259 | 0.34 |
ENSDART00000027301
|
sema3b
|
sema domain, immunoglobulin domain (Ig), short basic domain, secreted, (semaphorin) 3B |
| chr24_-_33703504 | 0.34 |
ENSDART00000079292
|
cavin4b
|
caveolae associated protein 4b |
| chr17_+_52822422 | 0.34 |
ENSDART00000158273
ENSDART00000161414 |
meis2a
|
Meis homeobox 2a |
| chr8_+_1065458 | 0.33 |
ENSDART00000081432
|
sprb
|
sepiapterin reductase b |
| chr9_-_48281941 | 0.33 |
ENSDART00000099787
|
klhl41a
|
kelch-like family member 41a |
| chr14_-_29905962 | 0.33 |
ENSDART00000142605
|
sorbs2b
|
sorbin and SH3 domain containing 2b |
| chr2_+_55982300 | 0.33 |
ENSDART00000183903
|
nmrk2
|
nicotinamide riboside kinase 2 |
| chr23_-_31512496 | 0.33 |
ENSDART00000158755
ENSDART00000143425 |
eya4
|
EYA transcriptional coactivator and phosphatase 4 |
| chr10_-_33621739 | 0.32 |
ENSDART00000142655
ENSDART00000128049 |
hunk
|
hormonally up-regulated Neu-associated kinase |
| chr8_-_1051438 | 0.32 |
ENSDART00000067093
ENSDART00000170737 |
smyd1b
|
SET and MYND domain containing 1b |
| chr13_+_25449681 | 0.32 |
ENSDART00000101328
|
atoh7
|
atonal bHLH transcription factor 7 |
| chr17_+_38573471 | 0.32 |
ENSDART00000040627
|
sptb
|
spectrin, beta, erythrocytic |
| chr13_+_42011287 | 0.31 |
ENSDART00000131147
|
cyp1b1
|
cytochrome P450, family 1, subfamily B, polypeptide 1 |
| chr13_-_31296358 | 0.31 |
ENSDART00000030946
|
prdm8
|
PR domain containing 8 |
| chr13_+_23157053 | 0.31 |
ENSDART00000162359
|
sorbs1
|
sorbin and SH3 domain containing 1 |
| chr24_-_26283359 | 0.31 |
ENSDART00000128618
|
and1
|
actinodin1 |
| chr3_+_32526799 | 0.30 |
ENSDART00000185755
|
si:ch73-367p23.2
|
si:ch73-367p23.2 |
| chr18_+_402048 | 0.30 |
ENSDART00000166345
|
gpib
|
glucose-6-phosphate isomerase b |
| chr23_-_21453614 | 0.30 |
ENSDART00000079274
|
her4.1
|
hairy-related 4, tandem duplicate 1 |
| chr10_+_15777258 | 0.30 |
ENSDART00000140511
|
apba1b
|
amyloid beta (A4) precursor protein-binding, family A, member 1b |
| chr15_+_1397811 | 0.30 |
ENSDART00000102125
|
schip1
|
schwannomin interacting protein 1 |
| chr19_-_2421793 | 0.30 |
ENSDART00000180238
|
TMEM196 (1 of many)
|
transmembrane protein 196 |
| chr12_+_16967715 | 0.30 |
ENSDART00000138174
|
slc16a12b
|
solute carrier family 16, member 12b |
| chr7_+_6652967 | 0.30 |
ENSDART00000102681
|
pnp5a
|
purine nucleoside phosphorylase 5a |
| chr20_+_26683933 | 0.30 |
ENSDART00000139852
ENSDART00000077751 |
foxq1b
|
forkhead box Q1b |
| chr4_-_24019711 | 0.29 |
ENSDART00000077926
|
celf2
|
cugbp, Elav-like family member 2 |
| chr17_+_52823015 | 0.29 |
ENSDART00000160507
ENSDART00000186979 |
meis2a
|
Meis homeobox 2a |
| chr20_+_35382482 | 0.29 |
ENSDART00000135284
|
vsnl1a
|
visinin-like 1a |
| chr4_+_5798223 | 0.29 |
ENSDART00000059440
|
si:ch73-352p4.8
|
si:ch73-352p4.8 |
| chr7_-_69857692 | 0.29 |
ENSDART00000124764
|
myoz2a
|
myozenin 2a |
| chr15_-_15357178 | 0.29 |
ENSDART00000106120
|
ywhag2
|
3-monooxygenase/tryptophan 5-monooxygenase activation protein, gamma polypeptide 2 |
| chr15_+_7187228 | 0.29 |
ENSDART00000109394
|
her13
|
hairy-related 13 |
| chr6_-_21189295 | 0.29 |
ENSDART00000137136
|
obsl1a
|
obscurin-like 1a |
| chr16_+_5156420 | 0.29 |
ENSDART00000012053
|
elovl4a
|
ELOVL fatty acid elongase 4a |
| chr20_-_49681850 | 0.28 |
ENSDART00000025926
|
col12a1b
|
collagen, type XII, alpha 1b |
| chr16_-_50229193 | 0.28 |
ENSDART00000161782
ENSDART00000010081 |
etfb
|
electron-transfer-flavoprotein, beta polypeptide |
| chr24_+_10414028 | 0.28 |
ENSDART00000193257
|
myca
|
MYC proto-oncogene, bHLH transcription factor a |
| chr5_+_37854685 | 0.28 |
ENSDART00000051222
ENSDART00000185283 |
ins
|
preproinsulin |
| chr23_+_21459263 | 0.28 |
ENSDART00000104209
|
her4.3
|
hairy-related 4, tandem duplicate 3 |
| chr10_+_15777064 | 0.27 |
ENSDART00000114483
|
apba1b
|
amyloid beta (A4) precursor protein-binding, family A, member 1b |
| chr3_-_28075756 | 0.27 |
ENSDART00000122037
|
rbfox1
|
RNA binding fox-1 homolog 1 |
| chr6_-_49673476 | 0.27 |
ENSDART00000112226
|
apcdd1l
|
adenomatosis polyposis coli down-regulated 1-like |
| chr11_-_32723851 | 0.27 |
ENSDART00000155592
|
pcdh17
|
protocadherin 17 |
| chr13_-_31452516 | 0.27 |
ENSDART00000193268
|
rtn1a
|
reticulon 1a |
| chr23_-_3408777 | 0.26 |
ENSDART00000193245
|
mafba
|
v-maf avian musculoaponeurotic fibrosarcoma oncogene homolog Ba |
| chr8_-_7093507 | 0.26 |
ENSDART00000045669
|
si:dkey-222n6.2
|
si:dkey-222n6.2 |
| chr13_+_1575276 | 0.26 |
ENSDART00000165987
|
DST
|
dystonin |
| chr6_-_40058686 | 0.26 |
ENSDART00000103240
|
uroc1
|
urocanate hydratase 1 |
| chr23_+_19790962 | 0.25 |
ENSDART00000142228
|
flna
|
filamin A, alpha (actin binding protein 280) |
| chr7_-_35708450 | 0.25 |
ENSDART00000193886
|
irx5a
|
iroquois homeobox 5a |
| chr14_-_35672890 | 0.25 |
ENSDART00000074710
|
pdgfc
|
platelet derived growth factor c |
| chr5_+_23630384 | 0.25 |
ENSDART00000013745
|
cx39.9
|
connexin 39.9 |
| chr15_+_7057050 | 0.25 |
ENSDART00000061828
|
foxl2a
|
forkhead box L2a |
| chr12_-_28570989 | 0.25 |
ENSDART00000008010
|
pdk2a
|
pyruvate dehydrogenase kinase, isozyme 2a |
| chr2_-_16254782 | 0.24 |
ENSDART00000133708
|
arhgef4
|
Rho guanine nucleotide exchange factor (GEF) 4 |
| chr13_-_16226312 | 0.24 |
ENSDART00000163952
|
zgc:110045
|
zgc:110045 |
| chr5_+_62611400 | 0.24 |
ENSDART00000132054
|
abr
|
active BCR-related |
| chr18_-_14734678 | 0.24 |
ENSDART00000142462
|
tshz3a
|
teashirt zinc finger homeobox 3a |
| chr24_-_24163201 | 0.24 |
ENSDART00000140170
|
map7d2b
|
MAP7 domain containing 2b |
| chr10_+_6496185 | 0.24 |
ENSDART00000164770
|
reep5
|
receptor accessory protein 5 |
| chr1_-_51734524 | 0.24 |
ENSDART00000109640
ENSDART00000122628 |
junba
|
JunB proto-oncogene, AP-1 transcription factor subunit a |
| chr18_+_17418254 | 0.24 |
ENSDART00000140191
|
ces3
|
carboxylesterase 3 |
| chr12_+_18524953 | 0.23 |
ENSDART00000090332
|
neurl2
|
neuralized E3 ubiquitin protein ligase 2 |
| chr14_-_9281232 | 0.23 |
ENSDART00000054693
|
asb12b
|
ankyrin repeat and SOCS box-containing 12b |
| chr5_+_64900223 | 0.23 |
ENSDART00000191677
|
ptgs1
|
prostaglandin-endoperoxide synthase 1 |
| chr3_-_54544612 | 0.23 |
ENSDART00000018044
|
angptl6
|
angiopoietin-like 6 |
| chr13_+_22264914 | 0.23 |
ENSDART00000060576
|
myoz1a
|
myozenin 1a |
| chr21_-_39639954 | 0.23 |
ENSDART00000026766
|
aldocb
|
aldolase C, fructose-bisphosphate, b |
| chr12_-_6063328 | 0.23 |
ENSDART00000002583
|
aarsd1
|
alanyl-tRNA synthetase domain containing 1 |
| chr1_-_14233815 | 0.23 |
ENSDART00000044896
|
camk2d2
|
calcium/calmodulin-dependent protein kinase (CaM kinase) II delta 2 |
| chr24_-_24162930 | 0.22 |
ENSDART00000080602
|
map7d2b
|
MAP7 domain containing 2b |
| chr8_+_44926946 | 0.22 |
ENSDART00000098567
|
zgc:154046
|
zgc:154046 |
| chr14_+_31651533 | 0.22 |
ENSDART00000172835
|
fhl1a
|
four and a half LIM domains 1a |
| chr1_+_41609676 | 0.22 |
ENSDART00000183675
|
mogs
|
mannosyl-oligosaccharide glucosidase |
| chr14_-_21219659 | 0.22 |
ENSDART00000089867
|
ppp2r2cb
|
protein phosphatase 2, regulatory subunit B, gamma b |
| chr8_+_23521974 | 0.22 |
ENSDART00000188130
ENSDART00000129378 |
sema3gb
|
sema domain, immunoglobulin domain (Ig), short basic domain, secreted, (semaphorin) 3Gb |
| chr5_+_44846280 | 0.22 |
ENSDART00000084370
|
kank1a
|
KN motif and ankyrin repeat domains 1a |
| chr6_-_7052408 | 0.22 |
ENSDART00000150033
ENSDART00000149232 |
bin1b
|
bridging integrator 1b |
| chr24_+_7884880 | 0.22 |
ENSDART00000139467
|
bmp6
|
bone morphogenetic protein 6 |
| chr17_+_38262408 | 0.22 |
ENSDART00000017493
|
nkx2.1
|
NK2 homeobox 1 |
| chr23_+_37458602 | 0.21 |
ENSDART00000181686
|
cdaa
|
cytidine deaminase a |
| chr5_-_13818833 | 0.21 |
ENSDART00000192198
|
add2
|
adducin 2 (beta) |
| chr6_+_28752943 | 0.21 |
ENSDART00000078447
ENSDART00000146574 |
tprg1
|
tumor protein p63 regulated 1 |
| chr3_-_35554809 | 0.21 |
ENSDART00000010944
|
dctn5
|
dynactin 5 |
| chr12_+_23866368 | 0.21 |
ENSDART00000188652
ENSDART00000192478 |
svila
|
supervillin a |
| chr8_-_27687095 | 0.21 |
ENSDART00000086946
|
mov10b.1
|
Moloney leukemia virus 10b, tandem duplicate 1 |
| chr16_-_29146624 | 0.21 |
ENSDART00000159814
ENSDART00000009826 |
mef2d
|
myocyte enhancer factor 2d |
| chr11_+_31285127 | 0.21 |
ENSDART00000160154
|
si:dkey-238i5.2
|
si:dkey-238i5.2 |
| chr10_-_13178853 | 0.20 |
ENSDART00000163740
ENSDART00000166327 ENSDART00000160265 ENSDART00000164299 |
musk
|
muscle, skeletal, receptor tyrosine kinase |
| chr14_-_41678357 | 0.20 |
ENSDART00000185925
|
fgfrl1b
|
fibroblast growth factor receptor like 1b |
| chr14_+_31657412 | 0.20 |
ENSDART00000105767
|
fhl1a
|
four and a half LIM domains 1a |
| chr15_-_8517376 | 0.20 |
ENSDART00000186289
|
npas1
|
neuronal PAS domain protein 1 |
| chr23_-_9925568 | 0.20 |
ENSDART00000081268
|
si:ch211-220i18.4
|
si:ch211-220i18.4 |
| chr6_+_29305190 | 0.20 |
ENSDART00000078647
|
si:ch211-201h21.5
|
si:ch211-201h21.5 |
| chr17_+_53311618 | 0.20 |
ENSDART00000166517
|
asb2b
|
ankyrin repeat and SOCS box containing 2b |
| chr12_+_18718065 | 0.20 |
ENSDART00000152935
|
mkl1b
|
megakaryoblastic leukemia (translocation) 1b |
| chr13_-_27916439 | 0.20 |
ENSDART00000139081
ENSDART00000087097 |
ogfrl1
|
opioid growth factor receptor-like 1 |
| chr19_-_15281996 | 0.20 |
ENSDART00000103784
|
edn2
|
endothelin 2 |
| chr7_+_31879649 | 0.19 |
ENSDART00000099789
|
mybpc3
|
myosin binding protein C, cardiac |
| chr20_+_34320635 | 0.19 |
ENSDART00000153207
|
ivns1abpa
|
influenza virus NS1A binding protein a |
| chr13_+_4205724 | 0.19 |
ENSDART00000134105
|
dlk2
|
delta-like 2 homolog (Drosophila) |
| chr10_+_19026988 | 0.19 |
ENSDART00000137456
|
igsf9a
|
immunoglobulin superfamily, member 9a |
| chr8_+_21146262 | 0.19 |
ENSDART00000045684
|
porcn
|
porcupine O-acyltransferase |
| chr22_+_29091545 | 0.19 |
ENSDART00000044559
|
pla2g6
|
phospholipase A2, group VI (cytosolic, calcium-independent) |
| chr1_-_17650223 | 0.19 |
ENSDART00000043484
|
si:dkey-256e7.5
|
si:dkey-256e7.5 |
| chr19_-_2420990 | 0.19 |
ENSDART00000181498
|
TMEM196 (1 of many)
|
transmembrane protein 196 |
| chr13_-_8692860 | 0.19 |
ENSDART00000058107
|
mcfd2
|
multiple coagulation factor deficiency 2 |
| chr11_-_13107106 | 0.19 |
ENSDART00000184477
|
elovl1b
|
ELOVL fatty acid elongase 1b |
| chr20_+_48413712 | 0.19 |
ENSDART00000159983
|
CABZ01059119.1
|
|
| chr3_+_14388010 | 0.19 |
ENSDART00000171726
ENSDART00000165452 |
tmem56b
|
transmembrane protein 56b |
| chr25_+_5249513 | 0.18 |
ENSDART00000126814
|
CABZ01039863.1
|
|
| chr18_+_44768829 | 0.18 |
ENSDART00000016271
|
ilvbl
|
ilvB (bacterial acetolactate synthase)-like |
| chr13_+_51579851 | 0.18 |
ENSDART00000163847
|
nkx6.2
|
NK6 homeobox 2 |
| chr25_-_20258508 | 0.18 |
ENSDART00000133860
ENSDART00000006840 ENSDART00000173434 |
dnm1l
|
dynamin 1-like |
| chr18_-_50151979 | 0.18 |
ENSDART00000127496
|
loxl1
|
lysyl oxidase-like 1 |
| chr21_+_29077509 | 0.18 |
ENSDART00000128561
|
ecscr
|
endothelial cell surface expressed chemotaxis and apoptosis regulator |
| chr21_-_4250682 | 0.18 |
ENSDART00000099389
|
dnlz
|
DNL-type zinc finger |
| chr9_+_28232522 | 0.18 |
ENSDART00000031761
|
fzd5
|
frizzled class receptor 5 |
| chr11_+_27364338 | 0.18 |
ENSDART00000186759
|
fbln2
|
fibulin 2 |
| chr4_-_75175407 | 0.18 |
ENSDART00000180125
|
CABZ01043953.1
|
|
| chr19_-_30904590 | 0.18 |
ENSDART00000137633
|
si:ch211-194e15.5
|
si:ch211-194e15.5 |
| chr23_+_43950674 | 0.18 |
ENSDART00000167813
|
corin
|
corin, serine peptidase |
| chr13_-_15986871 | 0.17 |
ENSDART00000189394
|
ikzf1
|
IKAROS family zinc finger 1 (Ikaros) |
| chr7_+_27253063 | 0.17 |
ENSDART00000191138
|
sox6
|
SRY (sex determining region Y)-box 6 |
| chr19_-_22478888 | 0.17 |
ENSDART00000090679
|
pleca
|
plectin a |
| chr19_-_42588510 | 0.17 |
ENSDART00000102583
|
sytl1
|
synaptotagmin-like 1 |
| chr6_-_39764995 | 0.17 |
ENSDART00000085277
|
pfkmb
|
phosphofructokinase, muscle b |
| chr24_-_24146875 | 0.17 |
ENSDART00000173052
|
map7d2b
|
MAP7 domain containing 2b |
| chr12_+_17504559 | 0.17 |
ENSDART00000020628
|
cyth3a
|
cytohesin 3a |
Network of associatons between targets according to the STRING database.
First level regulatory network of myod1
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 0.9 | GO:0098528 | skeletal muscle fiber differentiation(GO:0098528) |
| 0.3 | 0.8 | GO:0001954 | positive regulation of cell-matrix adhesion(GO:0001954) |
| 0.2 | 0.6 | GO:0006600 | creatine metabolic process(GO:0006600) creatine biosynthetic process(GO:0006601) |
| 0.2 | 0.6 | GO:1902571 | regulation of serine-type peptidase activity(GO:1902571) |
| 0.2 | 0.5 | GO:0014814 | regulation of synaptic growth at neuromuscular junction(GO:0008582) axon regeneration at neuromuscular junction(GO:0014814) positive regulation of synaptic growth at neuromuscular junction(GO:0045887) |
| 0.2 | 0.9 | GO:0003210 | cardiac atrium formation(GO:0003210) |
| 0.2 | 0.7 | GO:0021742 | abducens nucleus development(GO:0021742) |
| 0.2 | 0.5 | GO:0043388 | positive regulation of DNA binding(GO:0043388) |
| 0.2 | 0.5 | GO:0051574 | positive regulation of histone H3-K9 methylation(GO:0051574) |
| 0.1 | 0.4 | GO:0065001 | specification of axis polarity(GO:0065001) |
| 0.1 | 0.4 | GO:0003228 | atrial cardiac muscle tissue development(GO:0003228) |
| 0.1 | 0.5 | GO:0010882 | regulation of cardiac muscle contraction by calcium ion signaling(GO:0010882) |
| 0.1 | 0.4 | GO:0070983 | dendrite guidance(GO:0070983) regulation of homophilic cell adhesion(GO:1903385) |
| 0.1 | 0.5 | GO:0019240 | citrulline biosynthetic process(GO:0019240) |
| 0.1 | 0.3 | GO:0019557 | formate metabolic process(GO:0015942) histidine catabolic process to glutamate and formamide(GO:0019556) histidine catabolic process to glutamate and formate(GO:0019557) formamide metabolic process(GO:0043606) |
| 0.1 | 0.4 | GO:0090199 | regulation of mitochondrial membrane potential(GO:0051881) regulation of release of cytochrome c from mitochondria(GO:0090199) positive regulation of release of cytochrome c from mitochondria(GO:0090200) |
| 0.1 | 0.2 | GO:0048496 | maintenance of organ identity(GO:0048496) |
| 0.1 | 0.6 | GO:0034389 | lipid particle organization(GO:0034389) |
| 0.1 | 0.7 | GO:0046716 | muscle cell cellular homeostasis(GO:0046716) |
| 0.1 | 1.2 | GO:0035914 | skeletal muscle cell differentiation(GO:0035914) |
| 0.1 | 0.2 | GO:0006216 | cytidine catabolic process(GO:0006216) cytidine deamination(GO:0009972) cytidine metabolic process(GO:0046087) |
| 0.1 | 0.5 | GO:0060017 | parathyroid gland development(GO:0060017) |
| 0.1 | 0.3 | GO:0048521 | positive regulation of epithelial cell differentiation(GO:0030858) negative regulation of behavior(GO:0048521) negative regulation of feeding behavior(GO:2000252) |
| 0.1 | 0.4 | GO:0007405 | neuroblast proliferation(GO:0007405) |
| 0.1 | 0.4 | GO:0071691 | cardiac muscle thin filament assembly(GO:0071691) |
| 0.1 | 0.2 | GO:0061355 | Wnt protein secretion(GO:0061355) |
| 0.1 | 0.6 | GO:2001239 | histone dephosphorylation(GO:0016576) negative regulation of signal transduction in absence of ligand(GO:1901099) regulation of extrinsic apoptotic signaling pathway in absence of ligand(GO:2001239) negative regulation of extrinsic apoptotic signaling pathway in absence of ligand(GO:2001240) |
| 0.1 | 0.4 | GO:0061298 | retina vasculature development in camera-type eye(GO:0061298) |
| 0.1 | 0.2 | GO:0009097 | isoleucine metabolic process(GO:0006549) isoleucine biosynthetic process(GO:0009097) |
| 0.1 | 0.2 | GO:0019371 | cyclooxygenase pathway(GO:0019371) |
| 0.1 | 0.1 | GO:1903779 | regulation of cardiac conduction(GO:1903779) |
| 0.1 | 0.2 | GO:0099625 | regulation of membrane repolarization(GO:0060306) regulation of ventricular cardiac muscle cell membrane repolarization(GO:0060307) regulation of cardiac muscle cell membrane repolarization(GO:0099623) ventricular cardiac muscle cell membrane repolarization(GO:0099625) |
| 0.1 | 0.5 | GO:0031268 | pseudopodium organization(GO:0031268) pseudopodium assembly(GO:0031269) regulation of pseudopodium assembly(GO:0031272) positive regulation of pseudopodium assembly(GO:0031274) |
| 0.1 | 0.2 | GO:0042364 | water-soluble vitamin biosynthetic process(GO:0042364) |
| 0.1 | 0.4 | GO:1900026 | positive regulation of substrate adhesion-dependent cell spreading(GO:1900026) |
| 0.1 | 0.3 | GO:0003100 | regulation of systemic arterial blood pressure by endothelin(GO:0003100) vein smooth muscle contraction(GO:0014826) regulation of prostaglandin-endoperoxide synthase activity(GO:0060584) positive regulation of prostaglandin-endoperoxide synthase activity(GO:0060585) |
| 0.0 | 0.3 | GO:0035332 | positive regulation of hippo signaling(GO:0035332) |
| 0.0 | 0.6 | GO:0030241 | skeletal muscle myosin thick filament assembly(GO:0030241) |
| 0.0 | 0.3 | GO:0033292 | T-tubule organization(GO:0033292) |
| 0.0 | 0.6 | GO:0032481 | positive regulation of type I interferon production(GO:0032481) |
| 0.0 | 0.2 | GO:0019254 | carnitine metabolic process, CoA-linked(GO:0019254) |
| 0.0 | 1.0 | GO:0030388 | fructose 1,6-bisphosphate metabolic process(GO:0030388) |
| 0.0 | 0.5 | GO:0021592 | fourth ventricle development(GO:0021592) |
| 0.0 | 0.2 | GO:0051823 | regulation of synapse structural plasticity(GO:0051823) |
| 0.0 | 2.3 | GO:0060048 | cardiac muscle contraction(GO:0060048) |
| 0.0 | 0.2 | GO:0010259 | multicellular organism aging(GO:0010259) |
| 0.0 | 0.2 | GO:1903146 | regulation of mitophagy(GO:1903146) |
| 0.0 | 0.3 | GO:0072498 | embryonic skeletal joint development(GO:0072498) |
| 0.0 | 0.2 | GO:0006309 | apoptotic DNA fragmentation(GO:0006309) cellular component disassembly involved in execution phase of apoptosis(GO:0006921) apoptotic nuclear changes(GO:0030262) |
| 0.0 | 0.3 | GO:0006729 | tetrahydrobiopterin biosynthetic process(GO:0006729) tetrahydrobiopterin metabolic process(GO:0046146) |
| 0.0 | 0.7 | GO:0048251 | elastic fiber assembly(GO:0048251) |
| 0.0 | 0.2 | GO:0006107 | oxaloacetate metabolic process(GO:0006107) |
| 0.0 | 0.2 | GO:2000096 | positive regulation of Wnt signaling pathway, planar cell polarity pathway(GO:2000096) |
| 0.0 | 0.2 | GO:0038083 | peptidyl-tyrosine autophosphorylation(GO:0038083) |
| 0.0 | 0.2 | GO:2001256 | regulation of store-operated calcium entry(GO:2001256) |
| 0.0 | 0.1 | GO:0051037 | histone displacement(GO:0001207) regulation of transcription involved in meiotic cell cycle(GO:0051037) positive regulation of transcription involved in meiotic cell cycle(GO:0051039) |
| 0.0 | 0.1 | GO:0042543 | protein N-linked glycosylation via arginine(GO:0042543) |
| 0.0 | 0.1 | GO:0035279 | mRNA cleavage involved in gene silencing by miRNA(GO:0035279) mRNA cleavage involved in gene silencing(GO:0098795) |
| 0.0 | 0.1 | GO:2001244 | positive regulation of intrinsic apoptotic signaling pathway(GO:2001244) |
| 0.0 | 0.2 | GO:0031954 | positive regulation of protein autophosphorylation(GO:0031954) |
| 0.0 | 0.6 | GO:0048936 | peripheral nervous system neuron axonogenesis(GO:0048936) |
| 0.0 | 0.4 | GO:0051764 | actin crosslink formation(GO:0051764) |
| 0.0 | 0.5 | GO:0071594 | T cell differentiation in thymus(GO:0033077) thymocyte aggregation(GO:0071594) |
| 0.0 | 0.1 | GO:0033152 | immunoglobulin V(D)J recombination(GO:0033152) |
| 0.0 | 0.1 | GO:1903964 | monounsaturated fatty acid metabolic process(GO:1903964) monounsaturated fatty acid biosynthetic process(GO:1903966) |
| 0.0 | 0.3 | GO:0051224 | negative regulation of protein transport(GO:0051224) negative regulation of establishment of protein localization(GO:1904950) |
| 0.0 | 0.1 | GO:0099612 | protein localization to juxtaparanode region of axon(GO:0071205) protein localization to axon(GO:0099612) |
| 0.0 | 0.0 | GO:0031293 | membrane protein intracellular domain proteolysis(GO:0031293) |
| 0.0 | 0.1 | GO:0015862 | uridine transport(GO:0015862) pyrimidine nucleoside transport(GO:0015864) |
| 0.0 | 0.2 | GO:0051124 | synaptic growth at neuromuscular junction(GO:0051124) |
| 0.0 | 0.2 | GO:0050975 | sensory perception of touch(GO:0050975) |
| 0.0 | 0.3 | GO:0042572 | retinol metabolic process(GO:0042572) |
| 0.0 | 0.1 | GO:0045737 | positive regulation of cyclin-dependent protein serine/threonine kinase activity(GO:0045737) positive regulation of cyclin-dependent protein kinase activity(GO:1904031) |
| 0.0 | 0.4 | GO:0006883 | cellular sodium ion homeostasis(GO:0006883) |
| 0.0 | 0.2 | GO:0021982 | pineal gland development(GO:0021982) |
| 0.0 | 0.4 | GO:0030878 | thyroid gland development(GO:0030878) |
| 0.0 | 0.2 | GO:0006450 | regulation of translational fidelity(GO:0006450) |
| 0.0 | 0.4 | GO:0048026 | positive regulation of mRNA splicing, via spliceosome(GO:0048026) |
| 0.0 | 0.1 | GO:2000480 | negative regulation of cAMP-dependent protein kinase activity(GO:2000480) |
| 0.0 | 0.0 | GO:0042755 | eating behavior(GO:0042755) |
| 0.0 | 0.1 | GO:0071470 | cellular response to osmotic stress(GO:0071470) |
| 0.0 | 0.1 | GO:0019264 | glycine biosynthetic process from serine(GO:0019264) |
| 0.0 | 0.6 | GO:0016082 | synaptic vesicle priming(GO:0016082) |
| 0.0 | 0.1 | GO:0044211 | CTP salvage(GO:0044211) |
| 0.0 | 0.2 | GO:0070262 | peptidyl-serine dephosphorylation(GO:0070262) |
| 0.0 | 0.2 | GO:0035479 | angioblast cell migration from lateral mesoderm to midline(GO:0035479) |
| 0.0 | 0.2 | GO:0060612 | adipose tissue development(GO:0060612) |
| 0.0 | 0.1 | GO:0003207 | cardiac chamber formation(GO:0003207) |
| 0.0 | 0.2 | GO:0055003 | cardiac myofibril assembly(GO:0055003) |
| 0.0 | 0.1 | GO:2001223 | negative regulation of neuron migration(GO:2001223) |
| 0.0 | 0.3 | GO:0070570 | regulation of axon regeneration(GO:0048679) regulation of neuron projection regeneration(GO:0070570) |
| 0.0 | 0.3 | GO:0030316 | osteoclast differentiation(GO:0030316) |
| 0.0 | 0.4 | GO:0032474 | otolith morphogenesis(GO:0032474) |
| 0.0 | 0.1 | GO:0071376 | response to corticotropin-releasing hormone(GO:0043435) cellular response to corticotropin-releasing hormone stimulus(GO:0071376) |
| 0.0 | 0.1 | GO:0030205 | dermatan sulfate metabolic process(GO:0030205) |
| 0.0 | 0.9 | GO:0050919 | negative chemotaxis(GO:0050919) |
| 0.0 | 0.5 | GO:0030199 | collagen fibril organization(GO:0030199) |
| 0.0 | 0.1 | GO:0006388 | tRNA splicing, via endonucleolytic cleavage and ligation(GO:0006388) |
| 0.0 | 0.1 | GO:0010887 | negative regulation of cholesterol storage(GO:0010887) |
| 0.0 | 0.4 | GO:0010508 | positive regulation of autophagy(GO:0010508) |
| 0.0 | 0.1 | GO:0006797 | polyphosphate metabolic process(GO:0006797) polyphosphate catabolic process(GO:0006798) |
| 0.0 | 0.3 | GO:0051156 | glucose 6-phosphate metabolic process(GO:0051156) |
| 0.0 | 0.1 | GO:1902765 | L-arginine import(GO:0043091) amino acid import across plasma membrane(GO:0089718) arginine import(GO:0090467) L-arginine import across plasma membrane(GO:0097638) L-arginine transport(GO:1902023) L-arginine import into cell(GO:1902765) amino acid import into cell(GO:1902837) L-arginine transmembrane transport(GO:1903400) arginine transmembrane transport(GO:1903826) |
| 0.0 | 0.3 | GO:0034625 | fatty acid elongation, saturated fatty acid(GO:0019367) fatty acid elongation, unsaturated fatty acid(GO:0019368) fatty acid elongation, monounsaturated fatty acid(GO:0034625) fatty acid elongation, polyunsaturated fatty acid(GO:0034626) |
| 0.0 | 0.4 | GO:0032526 | response to retinoic acid(GO:0032526) |
| 0.0 | 0.5 | GO:0021575 | hindbrain morphogenesis(GO:0021575) |
| 0.0 | 0.2 | GO:0031581 | hemidesmosome assembly(GO:0031581) |
| 0.0 | 0.3 | GO:0055013 | cardiac muscle cell development(GO:0055013) |
| 0.0 | 0.2 | GO:0003310 | pancreatic A cell differentiation(GO:0003310) |
| 0.0 | 0.1 | GO:0034427 | nuclear-transcribed mRNA catabolic process, exonucleolytic, 3'-5'(GO:0034427) |
| 0.0 | 0.1 | GO:0030183 | B cell differentiation(GO:0030183) |
| 0.0 | 0.0 | GO:0003347 | epicardial cell to mesenchymal cell transition(GO:0003347) negative regulation of adherens junction organization(GO:1903392) |
| 0.0 | 0.1 | GO:0060754 | mast cell chemotaxis(GO:0002551) regulation of mast cell chemotaxis(GO:0060753) positive regulation of mast cell chemotaxis(GO:0060754) mast cell migration(GO:0097531) |
| 0.0 | 0.5 | GO:0010862 | positive regulation of pathway-restricted SMAD protein phosphorylation(GO:0010862) |
| 0.0 | 0.2 | GO:0045851 | vacuolar acidification(GO:0007035) pH reduction(GO:0045851) intracellular pH reduction(GO:0051452) |
| 0.0 | 0.1 | GO:0032889 | regulation of vacuole fusion, non-autophagic(GO:0032889) |
| 0.0 | 0.6 | GO:0010842 | retina layer formation(GO:0010842) |
| 0.0 | 0.3 | GO:0035622 | intrahepatic bile duct development(GO:0035622) |
| 0.0 | 0.4 | GO:0045746 | negative regulation of Notch signaling pathway(GO:0045746) |
| 0.0 | 0.1 | GO:0060337 | response to type I interferon(GO:0034340) type I interferon signaling pathway(GO:0060337) cellular response to type I interferon(GO:0071357) |
| 0.0 | 0.5 | GO:0000245 | spliceosomal complex assembly(GO:0000245) |
| 0.0 | 0.1 | GO:0010996 | response to auditory stimulus(GO:0010996) |
| 0.0 | 0.2 | GO:0030150 | protein import into mitochondrial matrix(GO:0030150) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.6 | GO:0070032 | synaptobrevin 2-SNAP-25-syntaxin-1a-complexin I complex(GO:0070032) |
| 0.1 | 0.2 | GO:0005948 | acetolactate synthase complex(GO:0005948) |
| 0.1 | 0.1 | GO:0005790 | smooth endoplasmic reticulum(GO:0005790) |
| 0.1 | 2.4 | GO:0005861 | troponin complex(GO:0005861) |
| 0.0 | 1.2 | GO:0031672 | A band(GO:0031672) |
| 0.0 | 0.2 | GO:0033263 | CORVET complex(GO:0033263) |
| 0.0 | 0.1 | GO:0042709 | succinate-CoA ligase complex(GO:0042709) |
| 0.0 | 1.0 | GO:0033017 | sarcoplasmic reticulum membrane(GO:0033017) |
| 0.0 | 0.1 | GO:0005958 | DNA-dependent protein kinase-DNA ligase 4 complex(GO:0005958) |
| 0.0 | 0.1 | GO:1990745 | EARP complex(GO:1990745) |
| 0.0 | 0.3 | GO:0005945 | 6-phosphofructokinase complex(GO:0005945) |
| 0.0 | 0.1 | GO:0055087 | Ski complex(GO:0055087) |
| 0.0 | 0.1 | GO:0008091 | spectrin(GO:0008091) |
| 0.0 | 1.9 | GO:0030018 | Z disc(GO:0030018) |
| 0.0 | 0.1 | GO:0005797 | Golgi medial cisterna(GO:0005797) |
| 0.0 | 0.1 | GO:0072669 | tRNA-splicing ligase complex(GO:0072669) |
| 0.0 | 0.2 | GO:0045095 | keratin filament(GO:0045095) |
| 0.0 | 0.2 | GO:0031314 | extrinsic component of mitochondrial inner membrane(GO:0031314) |
| 0.0 | 0.1 | GO:0008275 | gamma-tubulin small complex(GO:0008275) |
| 0.0 | 0.1 | GO:0005853 | eukaryotic translation elongation factor 1 complex(GO:0005853) |
| 0.0 | 0.8 | GO:0044853 | caveola(GO:0005901) plasma membrane raft(GO:0044853) |
| 0.0 | 0.1 | GO:0097224 | sperm connecting piece(GO:0097224) |
| 0.0 | 0.2 | GO:0030056 | hemidesmosome(GO:0030056) |
| 0.0 | 0.1 | GO:0016586 | RSC complex(GO:0016586) |
| 0.0 | 0.2 | GO:0000220 | vacuolar proton-transporting V-type ATPase, V0 domain(GO:0000220) |
| 0.0 | 0.1 | GO:0033010 | paranodal junction(GO:0033010) |
| 0.0 | 0.1 | GO:0070876 | SOSS complex(GO:0070876) |
| 0.0 | 0.1 | GO:0001669 | acrosomal vesicle(GO:0001669) |
| 0.0 | 0.1 | GO:0070062 | extracellular exosome(GO:0070062) |
| 0.0 | 0.0 | GO:0071556 | integral component of cytoplasmic side of endoplasmic reticulum membrane(GO:0071458) integral component of lumenal side of endoplasmic reticulum membrane(GO:0071556) lumenal side of endoplasmic reticulum membrane(GO:0098553) |
| 0.0 | 0.1 | GO:0001650 | fibrillar center(GO:0001650) |
| 0.0 | 0.0 | GO:1990879 | CST complex(GO:1990879) |
| 0.0 | 0.1 | GO:0098894 | presynaptic endocytic zone(GO:0098833) presynaptic endocytic zone membrane(GO:0098835) extrinsic component of presynaptic membrane(GO:0098888) extrinsic component of presynaptic endocytic zone(GO:0098894) |
| 0.0 | 0.1 | GO:0000015 | phosphopyruvate hydratase complex(GO:0000015) |
| 0.0 | 0.1 | GO:0072546 | ER membrane protein complex(GO:0072546) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.7 | GO:0071253 | connexin binding(GO:0071253) |
| 0.2 | 0.5 | GO:1990226 | histone methyltransferase binding(GO:1990226) |
| 0.1 | 0.7 | GO:0050262 | ribosylnicotinamide kinase activity(GO:0050262) |
| 0.1 | 0.7 | GO:0004126 | cytidine deaminase activity(GO:0004126) |
| 0.1 | 0.7 | GO:0004332 | fructose-bisphosphate aldolase activity(GO:0004332) |
| 0.1 | 0.9 | GO:0030172 | troponin C binding(GO:0030172) troponin I binding(GO:0031013) |
| 0.1 | 0.2 | GO:0003984 | acetolactate synthase activity(GO:0003984) |
| 0.1 | 0.8 | GO:0005391 | sodium:potassium-exchanging ATPase activity(GO:0005391) |
| 0.1 | 0.9 | GO:0032036 | myosin heavy chain binding(GO:0032036) |
| 0.1 | 0.2 | GO:0004771 | sterol esterase activity(GO:0004771) |
| 0.1 | 0.2 | GO:0032574 | 5'-3' RNA helicase activity(GO:0032574) |
| 0.1 | 0.7 | GO:0070700 | BMP receptor binding(GO:0070700) |
| 0.1 | 0.9 | GO:0005007 | fibroblast growth factor-activated receptor activity(GO:0005007) |
| 0.1 | 0.2 | GO:0004666 | prostaglandin-endoperoxide synthase activity(GO:0004666) |
| 0.1 | 0.7 | GO:0005332 | gamma-aminobutyric acid:sodium symporter activity(GO:0005332) |
| 0.1 | 0.2 | GO:0004092 | carnitine O-acetyltransferase activity(GO:0004092) |
| 0.1 | 0.5 | GO:0016813 | hydrolase activity, acting on carbon-nitrogen (but not peptide) bonds, in linear amidines(GO:0016813) |
| 0.1 | 0.5 | GO:0031433 | telethonin binding(GO:0031433) FATZ binding(GO:0051373) |
| 0.1 | 0.3 | GO:0070330 | aromatase activity(GO:0070330) |
| 0.1 | 0.3 | GO:0031708 | endothelin B receptor binding(GO:0031708) |
| 0.0 | 0.1 | GO:0017050 | D-erythro-sphingosine kinase activity(GO:0017050) |
| 0.0 | 0.1 | GO:0004466 | long-chain-acyl-CoA dehydrogenase activity(GO:0004466) |
| 0.0 | 0.2 | GO:0035256 | G-protein coupled glutamate receptor binding(GO:0035256) |
| 0.0 | 0.1 | GO:0004774 | succinate-CoA ligase activity(GO:0004774) succinate-CoA ligase (ADP-forming) activity(GO:0004775) succinate-CoA ligase (GDP-forming) activity(GO:0004776) |
| 0.0 | 0.2 | GO:0048156 | tau protein binding(GO:0048156) |
| 0.0 | 0.3 | GO:0004731 | purine-nucleoside phosphorylase activity(GO:0004731) |
| 0.0 | 0.2 | GO:0030060 | L-malate dehydrogenase activity(GO:0030060) |
| 0.0 | 0.8 | GO:0005112 | Notch binding(GO:0005112) |
| 0.0 | 0.2 | GO:0004740 | pyruvate dehydrogenase (acetyl-transferring) kinase activity(GO:0004740) |
| 0.0 | 0.7 | GO:0043236 | laminin binding(GO:0043236) |
| 0.0 | 0.3 | GO:0072349 | modified amino acid transmembrane transporter activity(GO:0072349) |
| 0.0 | 0.4 | GO:0038191 | neuropilin binding(GO:0038191) |
| 0.0 | 0.1 | GO:0043183 | vascular endothelial growth factor receptor 1 binding(GO:0043183) |
| 0.0 | 0.2 | GO:0000014 | single-stranded DNA endodeoxyribonuclease activity(GO:0000014) |
| 0.0 | 0.1 | GO:0016886 | ligase activity, forming phosphoric ester bonds(GO:0016886) |
| 0.0 | 0.8 | GO:0005518 | collagen binding(GO:0005518) |
| 0.0 | 0.1 | GO:0033857 | inositol heptakisphosphate kinase activity(GO:0000829) diphosphoinositol-pentakisphosphate kinase activity(GO:0033857) |
| 0.0 | 0.2 | GO:0005161 | platelet-derived growth factor receptor binding(GO:0005161) |
| 0.0 | 0.3 | GO:0003872 | 6-phosphofructokinase activity(GO:0003872) |
| 0.0 | 0.1 | GO:0008179 | adenylate cyclase binding(GO:0008179) |
| 0.0 | 0.1 | GO:0004768 | stearoyl-CoA 9-desaturase activity(GO:0004768) acyl-CoA desaturase activity(GO:0016215) palmitoyl-CoA 9-desaturase activity(GO:0032896) |
| 0.0 | 0.6 | GO:0001540 | beta-amyloid binding(GO:0001540) |
| 0.0 | 0.2 | GO:0002161 | aminoacyl-tRNA editing activity(GO:0002161) |
| 0.0 | 0.2 | GO:0004985 | opioid receptor activity(GO:0004985) |
| 0.0 | 0.2 | GO:0019136 | deoxynucleoside kinase activity(GO:0019136) |
| 0.0 | 0.7 | GO:0098632 | protein binding involved in cell adhesion(GO:0098631) protein binding involved in cell-cell adhesion(GO:0098632) |
| 0.0 | 0.3 | GO:0016861 | intramolecular oxidoreductase activity, interconverting aldoses and ketoses(GO:0016861) |
| 0.0 | 0.1 | GO:0004862 | cAMP-dependent protein kinase inhibitor activity(GO:0004862) |
| 0.0 | 0.4 | GO:0046875 | ephrin receptor binding(GO:0046875) |
| 0.0 | 0.2 | GO:0070008 | serine-type exopeptidase activity(GO:0070008) |
| 0.0 | 0.1 | GO:0070905 | glycine hydroxymethyltransferase activity(GO:0004372) serine binding(GO:0070905) |
| 0.0 | 0.1 | GO:0004849 | uridine kinase activity(GO:0004849) |
| 0.0 | 0.2 | GO:0008467 | [heparan sulfate]-glucosamine 3-sulfotransferase 1 activity(GO:0008467) |
| 0.0 | 0.2 | GO:0030506 | ankyrin binding(GO:0030506) |
| 0.0 | 0.1 | GO:0004032 | alditol:NADP+ 1-oxidoreductase activity(GO:0004032) |
| 0.0 | 0.1 | GO:0047757 | chondroitin-glucuronate 5-epimerase activity(GO:0047757) |
| 0.0 | 0.1 | GO:0008420 | CTD phosphatase activity(GO:0008420) |
| 0.0 | 0.5 | GO:0017147 | Wnt-protein binding(GO:0017147) |
| 0.0 | 0.1 | GO:0008526 | phosphatidylinositol transporter activity(GO:0008526) |
| 0.0 | 0.1 | GO:0001642 | group III metabotropic glutamate receptor activity(GO:0001642) |
| 0.0 | 0.1 | GO:0004309 | exopolyphosphatase activity(GO:0004309) |
| 0.0 | 0.1 | GO:0016933 | extracellular-glycine-gated ion channel activity(GO:0016933) extracellular-glycine-gated chloride channel activity(GO:0016934) |
| 0.0 | 0.3 | GO:0009922 | fatty acid elongase activity(GO:0009922) 3-oxo-arachidoyl-CoA synthase activity(GO:0102336) 3-oxo-cerotoyl-CoA synthase activity(GO:0102337) 3-oxo-lignoceronyl-CoA synthase activity(GO:0102338) |
| 0.0 | 0.2 | GO:0048495 | Roundabout binding(GO:0048495) |
| 0.0 | 0.2 | GO:0008327 | methyl-CpG binding(GO:0008327) |
| 0.0 | 0.6 | GO:0030215 | semaphorin receptor binding(GO:0030215) |
| 0.0 | 0.5 | GO:0017075 | syntaxin-1 binding(GO:0017075) |
| 0.0 | 0.1 | GO:0035259 | glucocorticoid receptor binding(GO:0035259) |
| 0.0 | 0.1 | GO:0015189 | L-lysine transmembrane transporter activity(GO:0015189) |
| 0.0 | 0.1 | GO:0004634 | phosphopyruvate hydratase activity(GO:0004634) |
| 0.0 | 0.2 | GO:0004720 | protein-lysine 6-oxidase activity(GO:0004720) |
| 0.0 | 1.3 | GO:0004879 | RNA polymerase II transcription factor activity, ligand-activated sequence-specific DNA binding(GO:0004879) |
| 0.0 | 0.2 | GO:0015271 | outward rectifier potassium channel activity(GO:0015271) |
| 0.0 | 0.2 | GO:0051117 | ATPase binding(GO:0051117) |
| 0.0 | 0.2 | GO:0042043 | neurexin family protein binding(GO:0042043) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.8 | ST MYOCYTE AD PATHWAY | Myocyte Adrenergic Pathway is a specific case of the generalized Adrenergic Pathway. |
| 0.0 | 1.3 | PID BMP PATHWAY | BMP receptor signaling |
| 0.0 | 0.5 | PID INTEGRIN A9B1 PATHWAY | Alpha9 beta1 integrin signaling events |
| 0.0 | 0.8 | PID NFAT TFPATHWAY | Calcineurin-regulated NFAT-dependent transcription in lymphocytes |
| 0.0 | 0.7 | NABA PROTEOGLYCANS | Genes encoding proteoglycans |
| 0.0 | 0.4 | PID INSULIN GLUCOSE PATHWAY | Insulin-mediated glucose transport |
| 0.0 | 0.5 | PID ILK PATHWAY | Integrin-linked kinase signaling |
| 0.0 | 0.3 | PID WNT NONCANONICAL PATHWAY | Noncanonical Wnt signaling pathway |
| 0.0 | 0.7 | NABA BASEMENT MEMBRANES | Genes encoding structural components of basement membranes |
| 0.0 | 0.2 | PID RAS PATHWAY | Regulation of Ras family activation |
| 0.0 | 0.4 | PID NFAT 3PATHWAY | Role of Calcineurin-dependent NFAT signaling in lymphocytes |
| 0.0 | 0.1 | PID S1P META PATHWAY | Sphingosine 1-phosphate (S1P) pathway |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.3 | REACTOME SYNTHESIS SECRETION AND DEACYLATION OF GHRELIN | Genes involved in Synthesis, Secretion, and Deacylation of Ghrelin |
| 0.0 | 0.9 | REACTOME A TETRASACCHARIDE LINKER SEQUENCE IS REQUIRED FOR GAG SYNTHESIS | Genes involved in A tetrasaccharide linker sequence is required for GAG synthesis |
| 0.0 | 0.2 | REACTOME VEGF LIGAND RECEPTOR INTERACTIONS | Genes involved in VEGF ligand-receptor interactions |
| 0.0 | 0.3 | REACTOME ENDOGENOUS STEROLS | Genes involved in Endogenous sterols |
| 0.0 | 0.4 | REACTOME KERATAN SULFATE DEGRADATION | Genes involved in Keratan sulfate degradation |
| 0.0 | 0.4 | REACTOME SYNTHESIS SECRETION AND INACTIVATION OF GIP | Genes involved in Synthesis, Secretion, and Inactivation of Glucose-dependent Insulinotropic Polypeptide (GIP) |
| 0.0 | 0.1 | REACTOME NCAM SIGNALING FOR NEURITE OUT GROWTH | Genes involved in NCAM signaling for neurite out-growth |
| 0.0 | 0.1 | REACTOME INTEGRATION OF PROVIRUS | Genes involved in Integration of provirus |
| 0.0 | 0.2 | REACTOME ACYL CHAIN REMODELLING OF PE | Genes involved in Acyl chain remodelling of PE |
| 0.0 | 0.0 | REACTOME PROSTACYCLIN SIGNALLING THROUGH PROSTACYCLIN RECEPTOR | Genes involved in Prostacyclin signalling through prostacyclin receptor |
| 0.0 | 0.6 | REACTOME NUCLEAR SIGNALING BY ERBB4 | Genes involved in Nuclear signaling by ERBB4 |
| 0.0 | 0.6 | REACTOME NUCLEAR RECEPTOR TRANSCRIPTION PATHWAY | Genes involved in Nuclear Receptor transcription pathway |
| 0.0 | 0.2 | REACTOME PURINE SALVAGE | Genes involved in Purine salvage |
| 0.0 | 0.3 | REACTOME SMOOTH MUSCLE CONTRACTION | Genes involved in Smooth Muscle Contraction |
| 0.0 | 0.3 | REACTOME N GLYCAN TRIMMING IN THE ER AND CALNEXIN CALRETICULIN CYCLE | Genes involved in N-glycan trimming in the ER and Calnexin/Calreticulin cycle |
| 0.0 | 0.0 | REACTOME ADVANCED GLYCOSYLATION ENDPRODUCT RECEPTOR SIGNALING | Genes involved in Advanced glycosylation endproduct receptor signaling |
| 0.0 | 0.6 | REACTOME CLASS B 2 SECRETIN FAMILY RECEPTORS | Genes involved in Class B/2 (Secretin family receptors) |
| 0.0 | 0.1 | REACTOME CREATION OF C4 AND C2 ACTIVATORS | Genes involved in Creation of C4 and C2 activators |
| 0.0 | 0.4 | REACTOME NETRIN1 SIGNALING | Genes involved in Netrin-1 signaling |