Project
PRJNA195909:zebrafish embryo and larva development
Navigation
Downloads
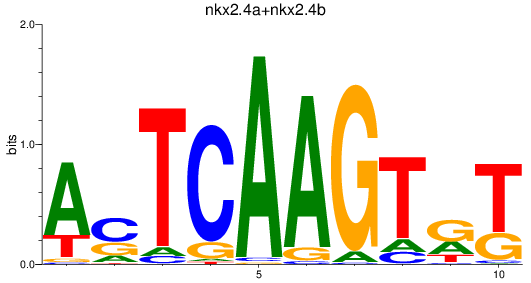
Results for nkx2.4a+nkx2.4b
Z-value: 0.33
Transcription factors associated with nkx2.4a+nkx2.4b
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
nkx2.4a
|
ENSDARG00000075107 | NK2 homeobox 4a |
|
nkx2.4b
|
ENSDARG00000104107 | NK2 homeobox 4b |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| nkx2.4a | dr11_v1_chr17_-_42097267_42097267 | 0.82 | 7.2e-03 | Click! |
| nkx2.4b | dr11_v1_chr20_+_48803248_48803248 | 0.80 | 8.9e-03 | Click! |
Activity profile of nkx2.4a+nkx2.4b motif
Sorted Z-values of nkx2.4a+nkx2.4b motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr1_+_25696798 | 0.41 |
ENSDART00000054228
|
lrata
|
lecithin retinol acyltransferase a |
| chr2_+_42724404 | 0.38 |
ENSDART00000075392
|
basp1
|
brain abundant, membrane attached signal protein 1 |
| chr7_-_35708450 | 0.36 |
ENSDART00000193886
|
irx5a
|
iroquois homeobox 5a |
| chr14_-_26536504 | 0.34 |
ENSDART00000105933
|
tgfbi
|
transforming growth factor, beta-induced |
| chr15_+_9072821 | 0.33 |
ENSDART00000154463
|
si:dkey-202g17.3
|
si:dkey-202g17.3 |
| chr12_-_16898140 | 0.32 |
ENSDART00000152656
|
MGC174155
|
Cathepsin L1-like |
| chr7_+_25323742 | 0.32 |
ENSDART00000110347
|
cyp26b1
|
cytochrome P450, family 26, subfamily b, polypeptide 1 |
| chr9_-_23033818 | 0.31 |
ENSDART00000022392
|
rnd3b
|
Rho family GTPase 3b |
| chr22_+_12770877 | 0.31 |
ENSDART00000044683
|
ftcd
|
formimidoyltransferase cyclodeaminase |
| chr17_+_27434626 | 0.31 |
ENSDART00000052446
|
vgll2b
|
vestigial-like family member 2b |
| chr10_+_21807497 | 0.28 |
ENSDART00000164634
|
pcdh1g32
|
protocadherin 1 gamma 32 |
| chr8_+_39607466 | 0.27 |
ENSDART00000097427
|
msi1
|
musashi RNA-binding protein 1 |
| chr10_+_31244619 | 0.27 |
ENSDART00000145562
ENSDART00000184412 |
robo4
|
roundabout, axon guidance receptor, homolog 4 (Drosophila) |
| chr20_+_48782068 | 0.25 |
ENSDART00000159275
|
nkx2.2b
|
NK2 homeobox 2b |
| chr1_-_21599219 | 0.25 |
ENSDART00000148327
|
adamtsl7
|
ADAMTS-like 7 |
| chr12_-_16720432 | 0.24 |
ENSDART00000152261
ENSDART00000152154 |
si:dkey-26g8.4
|
si:dkey-26g8.4 |
| chr19_+_10847118 | 0.23 |
ENSDART00000189784
|
apoa4a
|
apolipoprotein A-IV a |
| chr15_-_26552393 | 0.23 |
ENSDART00000150152
|
serpinf2b
|
serpin peptidase inhibitor, clade F (alpha-2 antiplasmin, pigment epithelium derived factor), member 2b |
| chr6_-_13114821 | 0.21 |
ENSDART00000164579
ENSDART00000165375 |
zgc:194469
|
zgc:194469 |
| chr5_-_33460959 | 0.21 |
ENSDART00000085636
|
si:ch211-182d3.1
|
si:ch211-182d3.1 |
| chr17_-_42213285 | 0.20 |
ENSDART00000140549
|
nkx2.2a
|
NK2 homeobox 2a |
| chr8_-_49345388 | 0.18 |
ENSDART00000053203
|
plp2
|
proteolipid protein 2 |
| chr9_+_7724152 | 0.17 |
ENSDART00000061716
|
mnx2a
|
motor neuron and pancreas homeobox 2a |
| chr13_-_13294847 | 0.17 |
ENSDART00000125883
ENSDART00000013534 |
fgfr3
|
fibroblast growth factor receptor 3 |
| chr4_+_15942075 | 0.17 |
ENSDART00000147964
|
si:dkey-117n7.2
|
si:dkey-117n7.2 |
| chr24_+_3307857 | 0.17 |
ENSDART00000106527
|
gyg1b
|
glycogenin 1b |
| chr1_-_14234076 | 0.16 |
ENSDART00000040049
|
camk2d2
|
calcium/calmodulin-dependent protein kinase (CaM kinase) II delta 2 |
| chr15_-_26552652 | 0.16 |
ENSDART00000152336
|
serpinf2b
|
serpin peptidase inhibitor, clade F (alpha-2 antiplasmin, pigment epithelium derived factor), member 2b |
| chr15_-_8517555 | 0.16 |
ENSDART00000140213
|
npas1
|
neuronal PAS domain protein 1 |
| chr15_-_12011390 | 0.15 |
ENSDART00000187403
|
si:dkey-202l22.6
|
si:dkey-202l22.6 |
| chr1_-_14233815 | 0.14 |
ENSDART00000044896
|
camk2d2
|
calcium/calmodulin-dependent protein kinase (CaM kinase) II delta 2 |
| chr8_+_23485079 | 0.14 |
ENSDART00000026316
|
sema3gb
|
sema domain, immunoglobulin domain (Ig), short basic domain, secreted, (semaphorin) 3Gb |
| chr21_-_41147818 | 0.13 |
ENSDART00000167339
ENSDART00000192730 |
msx2b
|
muscle segment homeobox 2b |
| chr4_+_22365061 | 0.12 |
ENSDART00000039277
|
lhfpl3
|
LHFPL tetraspan subfamily member 3 |
| chr3_+_33340939 | 0.12 |
ENSDART00000128786
|
pyya
|
peptide YYa |
| chr19_+_23982466 | 0.12 |
ENSDART00000080673
|
syt11a
|
synaptotagmin XIa |
| chr21_+_17110598 | 0.12 |
ENSDART00000101282
ENSDART00000191864 |
bcr
|
breakpoint cluster region |
| chr23_+_6795709 | 0.12 |
ENSDART00000149136
|
si:ch211-117c9.5
|
si:ch211-117c9.5 |
| chr7_-_35126374 | 0.11 |
ENSDART00000141211
|
hsd11b2
|
hydroxysteroid (11-beta) dehydrogenase 2 |
| chr1_-_38816685 | 0.11 |
ENSDART00000075230
|
asb5b
|
ankyrin repeat and SOCS box containing 5b |
| chr9_-_35155089 | 0.11 |
ENSDART00000077901
|
appb
|
amyloid beta (A4) precursor protein b |
| chr11_+_36158134 | 0.11 |
ENSDART00000189827
ENSDART00000163330 |
grm2b
|
glutamate receptor, metabotropic 2b |
| chr19_-_27776649 | 0.11 |
ENSDART00000135348
|
adcy2b
|
adenylate cyclase 2b (brain) |
| chr21_-_14310159 | 0.10 |
ENSDART00000155097
|
si:ch211-196i2.1
|
si:ch211-196i2.1 |
| chr6_+_58832155 | 0.10 |
ENSDART00000144842
|
dctn2
|
dynactin 2 (p50) |
| chr6_-_13114406 | 0.10 |
ENSDART00000188015
|
zgc:194469
|
zgc:194469 |
| chr17_+_43032529 | 0.10 |
ENSDART00000055611
ENSDART00000154863 |
isca2
|
iron-sulfur cluster assembly 2 |
| chr10_-_9346360 | 0.10 |
ENSDART00000039536
|
mrrf
|
mitochondrial ribosome recycling factor |
| chr14_+_14225048 | 0.10 |
ENSDART00000168749
|
nlgn3a
|
neuroligin 3a |
| chr8_+_48480529 | 0.10 |
ENSDART00000186631
|
PRDM16
|
si:ch211-263k4.2 |
| chr9_+_25832329 | 0.10 |
ENSDART00000130059
|
zeb2a
|
zinc finger E-box binding homeobox 2a |
| chr17_-_42213822 | 0.09 |
ENSDART00000187904
ENSDART00000180029 |
nkx2.2a
|
NK2 homeobox 2a |
| chr4_-_13580348 | 0.09 |
ENSDART00000067160
|
opn1sw1
|
opsin 1 (cone pigments), short-wave-sensitive 1 |
| chr22_-_36856405 | 0.09 |
ENSDART00000029588
|
kng1
|
kininogen 1 |
| chr10_-_32524035 | 0.09 |
ENSDART00000137608
|
dgat2
|
diacylglycerol O-acyltransferase 2 |
| chr8_-_23776399 | 0.09 |
ENSDART00000114800
|
INAVA
|
si:ch211-163l21.4 |
| chr8_-_17516448 | 0.09 |
ENSDART00000100667
|
skia
|
v-ski avian sarcoma viral oncogene homolog a |
| chr20_-_38449903 | 0.09 |
ENSDART00000136303
|
itpkb
|
inositol-trisphosphate 3-kinase B |
| chr20_+_54738210 | 0.08 |
ENSDART00000151399
|
pak7
|
p21 protein (Cdc42/Rac)-activated kinase 7 |
| chr25_-_27842654 | 0.08 |
ENSDART00000154852
ENSDART00000156906 |
asb15a
|
ankyrin repeat and SOCS box containing 15a |
| chr25_+_19485198 | 0.08 |
ENSDART00000156730
|
glsl
|
glutaminase like |
| chr20_+_27020201 | 0.08 |
ENSDART00000126919
ENSDART00000016014 |
chga
|
chromogranin A |
| chr7_+_59649399 | 0.08 |
ENSDART00000123520
ENSDART00000040771 |
rpl34
|
ribosomal protein L34 |
| chr14_+_21106444 | 0.07 |
ENSDART00000075744
ENSDART00000132363 |
aldob
|
aldolase b, fructose-bisphosphate |
| chr16_+_24971717 | 0.07 |
ENSDART00000156958
|
fpr1
|
formyl peptide receptor 1 |
| chr9_+_18716485 | 0.07 |
ENSDART00000135125
|
serp2
|
stress-associated endoplasmic reticulum protein family member 2 |
| chr4_+_17245217 | 0.07 |
ENSDART00000184215
ENSDART00000110908 |
casc1
|
cancer susceptibility candidate 1 |
| chr5_+_34981584 | 0.06 |
ENSDART00000134795
|
ankra2
|
ankyrin repeat, family A (RFXANK-like), 2 |
| chr23_-_26532696 | 0.06 |
ENSDART00000124811
ENSDART00000180274 |
si:dkey-205h13.2
|
si:dkey-205h13.2 |
| chr22_+_3223489 | 0.06 |
ENSDART00000082011
|
lim2.2
|
lens intrinsic membrane protein 2.2 |
| chr8_-_25120231 | 0.06 |
ENSDART00000147308
|
amigo1
|
adhesion molecule with Ig-like domain 1 |
| chr21_+_30084823 | 0.06 |
ENSDART00000154573
|
prob1
|
proline-rich basic protein 1 |
| chr21_+_19062124 | 0.06 |
ENSDART00000134746
|
rpl17
|
ribosomal protein L17 |
| chr5_+_53824959 | 0.06 |
ENSDART00000169565
|
sat2b
|
spermidine/spermine N1-acetyltransferase family member 2b |
| chr14_+_8343498 | 0.06 |
ENSDART00000164551
|
nrg2b
|
neuregulin 2b |
| chr5_+_32882688 | 0.06 |
ENSDART00000008807
ENSDART00000185317 |
rpl12
|
ribosomal protein L12 |
| chr3_+_32557615 | 0.05 |
ENSDART00000151608
|
pax10
|
paired box 10 |
| chr6_-_42073367 | 0.05 |
ENSDART00000154304
|
grm2a
|
glutamate receptor, metabotropic 2a |
| chr7_-_18601206 | 0.05 |
ENSDART00000111636
|
DTX4
|
si:ch211-119e14.2 |
| chr14_+_23709134 | 0.05 |
ENSDART00000191162
ENSDART00000179754 ENSDART00000054266 |
gnpda1
|
glucosamine-6-phosphate deaminase 1 |
| chr12_+_10599604 | 0.05 |
ENSDART00000152538
|
lrrc3ca
|
leucine rich repeat containing 3Ca |
| chr17_+_8184649 | 0.05 |
ENSDART00000091818
|
tulp4b
|
tubby like protein 4b |
| chr14_+_14224730 | 0.05 |
ENSDART00000180112
ENSDART00000184891 ENSDART00000174760 |
nlgn3a
|
neuroligin 3a |
| chr17_-_22552678 | 0.04 |
ENSDART00000079401
|
si:ch211-125o16.4
|
si:ch211-125o16.4 |
| chr15_-_23485752 | 0.04 |
ENSDART00000152706
|
nlrx1
|
NLR family member X1 |
| chr4_+_65649267 | 0.04 |
ENSDART00000184068
|
si:dkey-205i10.1
|
si:dkey-205i10.1 |
| chr14_-_17622080 | 0.04 |
ENSDART00000112149
|
si:ch211-159i8.4
|
si:ch211-159i8.4 |
| chr7_-_61842593 | 0.04 |
ENSDART00000023770
|
HTRA3
|
HtrA serine peptidase 3 |
| chr9_-_12811936 | 0.04 |
ENSDART00000188490
|
myo10l3
|
myosin X-like 3 |
| chr16_-_34258931 | 0.04 |
ENSDART00000145485
|
phactr4b
|
phosphatase and actin regulator 4b |
| chr25_-_12730260 | 0.04 |
ENSDART00000171834
|
klhdc4
|
kelch domain containing 4 |
| chr7_-_5039562 | 0.04 |
ENSDART00000154438
|
ltb4r
|
leukotriene B4 receptor |
| chr13_-_2112008 | 0.04 |
ENSDART00000110814
|
fam83b
|
family with sequence similarity 83, member B |
| chr14_-_8816933 | 0.04 |
ENSDART00000163383
|
si:dkeyp-115e12.7
|
si:dkeyp-115e12.7 |
| chr7_+_60551133 | 0.04 |
ENSDART00000148038
|
lrfn4b
|
leucine rich repeat and fibronectin type III domain containing 4b |
| chr24_-_205275 | 0.04 |
ENSDART00000108762
|
vopp1
|
VOPP1, WBP1/VOPP1 family member |
| chr2_+_16652238 | 0.04 |
ENSDART00000091351
|
gk5
|
glycerol kinase 5 (putative) |
| chr6_+_374875 | 0.03 |
ENSDART00000171698
|
si:zfos-169g10.3
|
si:zfos-169g10.3 |
| chr16_-_11766265 | 0.03 |
ENSDART00000141547
|
zgc:158701
|
zgc:158701 |
| chr10_+_8481660 | 0.03 |
ENSDART00000109559
|
BX682234.1
|
|
| chr22_-_28373698 | 0.03 |
ENSDART00000157592
|
si:ch211-213c4.5
|
si:ch211-213c4.5 |
| chr3_-_39305291 | 0.03 |
ENSDART00000102674
|
plcd3a
|
phospholipase C, delta 3a |
| chr20_+_27464721 | 0.03 |
ENSDART00000189552
|
kif26aa
|
kinesin family member 26Aa |
| chr3_+_58354852 | 0.03 |
ENSDART00000155715
|
vwa3a
|
von Willebrand factor A domain containing 3A |
| chr4_+_77038392 | 0.03 |
ENSDART00000186818
|
si:dkey-172k15.6
|
si:dkey-172k15.6 |
| chr1_-_11291324 | 0.03 |
ENSDART00000091205
|
sdk1b
|
sidekick cell adhesion molecule 1b |
| chr14_+_23158021 | 0.03 |
ENSDART00000084664
|
enox2
|
ecto-NOX disulfide-thiol exchanger 2 |
| chr13_+_25761279 | 0.03 |
ENSDART00000177818
ENSDART00000002863 |
neurog3
|
neurogenin 3 |
| chr22_+_19111444 | 0.03 |
ENSDART00000109655
|
fgf22
|
fibroblast growth factor 22 |
| chr12_+_19208338 | 0.03 |
ENSDART00000110415
|
sh3bp1
|
SH3-domain binding protein 1 |
| chr4_+_31405646 | 0.03 |
ENSDART00000128732
|
si:rp71-5o12.3
|
si:rp71-5o12.3 |
| chr10_-_10864331 | 0.03 |
ENSDART00000122657
|
nrarpa
|
NOTCH regulated ankyrin repeat protein a |
| chr3_-_11008532 | 0.03 |
ENSDART00000165086
|
CR382337.3
|
|
| chr14_+_23709543 | 0.03 |
ENSDART00000136909
|
gnpda1
|
glucosamine-6-phosphate deaminase 1 |
| chr1_-_34612613 | 0.03 |
ENSDART00000004361
|
klf5a
|
Kruppel-like factor 5a |
| chr4_+_50410452 | 0.02 |
ENSDART00000146006
|
si:dkey-156k2.8
|
si:dkey-156k2.8 |
| chr11_-_41130239 | 0.02 |
ENSDART00000173268
|
dnajc11b
|
DnaJ (Hsp40) homolog, subfamily C, member 11b |
| chr15_-_12011202 | 0.02 |
ENSDART00000160427
ENSDART00000168715 |
si:dkey-202l22.6
|
si:dkey-202l22.6 |
| chr10_-_10863936 | 0.02 |
ENSDART00000180568
|
nrarpa
|
NOTCH regulated ankyrin repeat protein a |
| chr7_+_20966434 | 0.02 |
ENSDART00000185570
|
efnb3b
|
ephrin-B3b |
| chr17_-_15201031 | 0.02 |
ENSDART00000124426
|
socs4
|
suppressor of cytokine signaling 4 |
| chr2_+_24203229 | 0.02 |
ENSDART00000138088
|
map4l
|
microtubule associated protein 4 like |
| chr2_+_47754419 | 0.02 |
ENSDART00000188221
|
mbnl1
|
muscleblind-like splicing regulator 1 |
| chr12_+_49100365 | 0.02 |
ENSDART00000171905
|
CABZ01075125.1
|
|
| chr10_-_15048781 | 0.02 |
ENSDART00000038401
ENSDART00000155674 |
si:ch211-95j8.2
|
si:ch211-95j8.2 |
| chr5_-_33825465 | 0.02 |
ENSDART00000110645
|
dab2ipb
|
DAB2 interacting protein b |
| chr17_-_31044803 | 0.02 |
ENSDART00000185941
ENSDART00000152016 |
eml1
|
echinoderm microtubule associated protein like 1 |
| chr10_+_9346383 | 0.02 |
ENSDART00000064975
ENSDART00000141869 |
rbm18
|
RNA binding motif protein 18 |
| chr22_+_24645325 | 0.02 |
ENSDART00000159531
|
lpar3
|
lysophosphatidic acid receptor 3 |
| chr13_-_2440622 | 0.02 |
ENSDART00000102767
ENSDART00000172616 |
fbxo9
|
F-box protein 9 |
| chr22_+_39096911 | 0.02 |
ENSDART00000157127
ENSDART00000153841 |
lmcd1
|
LIM and cysteine-rich domains 1 |
| chr23_+_7505943 | 0.01 |
ENSDART00000135787
|
hck
|
HCK proto-oncogene, Src family tyrosine kinase |
| chr10_-_17179415 | 0.01 |
ENSDART00000131751
|
depdc5
|
DEP domain containing 5 |
| chr8_+_36803415 | 0.01 |
ENSDART00000111680
|
iqsec2b
|
IQ motif and Sec7 domain 2b |
| chr4_-_65747476 | 0.01 |
ENSDART00000142421
|
si:dkey-28k24.2
|
si:dkey-28k24.2 |
| chr23_-_7826849 | 0.01 |
ENSDART00000157612
|
myt1b
|
myelin transcription factor 1b |
| chr16_-_31675669 | 0.01 |
ENSDART00000168848
ENSDART00000158331 |
c1r
|
complement component 1, r subcomponent |
| chr20_+_263056 | 0.01 |
ENSDART00000132669
|
tube1
|
tubulin, epsilon 1 |
| chr7_+_26049818 | 0.01 |
ENSDART00000173611
|
si:dkey-6n21.12
|
si:dkey-6n21.12 |
| chr17_-_8673567 | 0.01 |
ENSDART00000192714
ENSDART00000012546 |
ctbp2a
|
C-terminal binding protein 2a |
| chr12_-_19279103 | 0.01 |
ENSDART00000186669
|
si:ch211-141o9.10
|
si:ch211-141o9.10 |
| chr19_+_26424734 | 0.01 |
ENSDART00000187402
|
npr1a
|
natriuretic peptide receptor 1a |
| chr15_-_22139566 | 0.01 |
ENSDART00000149017
|
scn3b
|
sodium channel, voltage-gated, type III, beta |
| chr15_-_47937204 | 0.01 |
ENSDART00000154705
|
si:ch1073-111c8.3
|
si:ch1073-111c8.3 |
| chr9_+_31222026 | 0.01 |
ENSDART00000145573
|
clybl
|
citrate lyase beta like |
| chr7_+_42206847 | 0.01 |
ENSDART00000149250
|
phkb
|
phosphorylase kinase, beta |
| chr20_+_51478939 | 0.01 |
ENSDART00000149758
|
tlr5a
|
toll-like receptor 5a |
| chr24_+_33589667 | 0.01 |
ENSDART00000152097
|
dnah5l
|
dynein, axonemal, heavy chain 5 like |
| chr25_-_1079417 | 0.00 |
ENSDART00000163134
|
FO907089.1
|
|
| chr23_+_4324625 | 0.00 |
ENSDART00000146302
ENSDART00000136792 ENSDART00000135027 ENSDART00000179819 |
sgk2a
|
serum/glucocorticoid regulated kinase 2a |
| chr17_-_8673278 | 0.00 |
ENSDART00000171850
ENSDART00000017337 ENSDART00000148504 ENSDART00000148808 |
ctbp2a
|
C-terminal binding protein 2a |
| chr9_-_24970018 | 0.00 |
ENSDART00000026924
|
dnah7
|
dynein, axonemal, heavy chain 7 |
| chr2_-_24407732 | 0.00 |
ENSDART00000180612
ENSDART00000153688 ENSDART00000179913 |
si:dkey-208k22.6
|
si:dkey-208k22.6 |
| chr15_+_1137337 | 0.00 |
ENSDART00000191508
|
p2ry13
|
purinergic receptor P2Y13 |
| chr22_-_18241390 | 0.00 |
ENSDART00000083879
|
tmem161a
|
transmembrane protein 161A |
| chr5_-_28915130 | 0.00 |
ENSDART00000078592
|
npdc1b
|
neural proliferation, differentiation and control, 1b |
| chr16_+_41874001 | 0.00 |
ENSDART00000161278
|
scn1ba
|
sodium channel, voltage-gated, type I, beta a |
| chr19_-_23452345 | 0.00 |
ENSDART00000136875
|
pimr82
|
Pim proto-oncogene, serine/threonine kinase, related 82 |
Network of associatons between targets according to the STRING database.
First level regulatory network of nkx2.4a+nkx2.4b
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.3 | GO:0035992 | tendon formation(GO:0035992) |
| 0.1 | 0.3 | GO:0019557 | formate metabolic process(GO:0015942) histidine catabolic process to glutamate and formamide(GO:0019556) histidine catabolic process to glutamate and formate(GO:0019557) formamide metabolic process(GO:0043606) |
| 0.1 | 0.3 | GO:0021530 | spinal cord oligodendrocyte cell fate specification(GO:0021530) |
| 0.1 | 0.3 | GO:0001954 | positive regulation of cell-matrix adhesion(GO:0001954) |
| 0.0 | 0.4 | GO:0072498 | embryonic skeletal joint development(GO:0072498) |
| 0.0 | 0.4 | GO:0042572 | retinol metabolic process(GO:0042572) |
| 0.0 | 0.1 | GO:0006041 | glucosamine metabolic process(GO:0006041) glucosamine catabolic process(GO:0006043) |
| 0.0 | 0.1 | GO:0002792 | negative regulation of peptide secretion(GO:0002792) negative regulation of peptide hormone secretion(GO:0090278) |
| 0.0 | 0.1 | GO:0006651 | diacylglycerol biosynthetic process(GO:0006651) |
| 0.0 | 0.2 | GO:0021520 | spinal cord motor neuron cell fate specification(GO:0021520) |
| 0.0 | 0.1 | GO:0097428 | protein maturation by iron-sulfur cluster transfer(GO:0097428) |
| 0.0 | 0.1 | GO:0034650 | cortisol metabolic process(GO:0034650) |
| 0.0 | 0.1 | GO:0097105 | presynaptic membrane organization(GO:0097090) postsynaptic membrane assembly(GO:0097104) presynaptic membrane assembly(GO:0097105) |
| 0.0 | 0.3 | GO:0021508 | floor plate formation(GO:0021508) floor plate morphogenesis(GO:0033505) |
| 0.0 | 0.1 | GO:0090243 | fibroblast growth factor receptor signaling pathway involved in somitogenesis(GO:0090243) |
| 0.0 | 0.1 | GO:0030195 | negative regulation of blood coagulation(GO:0030195) negative regulation of hemostasis(GO:1900047) |
| 0.0 | 0.1 | GO:0002283 | neutrophil activation involved in immune response(GO:0002283) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.4 | GO:0047173 | phosphatidylcholine-retinol O-acyltransferase activity(GO:0047173) |
| 0.1 | 0.3 | GO:0008401 | retinoic acid 4-hydroxylase activity(GO:0008401) |
| 0.1 | 0.3 | GO:0016841 | folic acid binding(GO:0005542) ammonia-lyase activity(GO:0016841) |
| 0.0 | 0.1 | GO:0004342 | glucosamine-6-phosphate deaminase activity(GO:0004342) |
| 0.0 | 0.1 | GO:0031841 | neuropeptide Y receptor binding(GO:0031841) type 2 neuropeptide Y receptor binding(GO:0031843) |
| 0.0 | 0.2 | GO:0001641 | group II metabotropic glutamate receptor activity(GO:0001641) |
| 0.0 | 0.1 | GO:0043023 | ribosomal large subunit binding(GO:0043023) |
| 0.0 | 0.2 | GO:0005007 | fibroblast growth factor-activated receptor activity(GO:0005007) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.4 | PID INTEGRIN2 PATHWAY | Beta2 integrin cell surface interactions |
| 0.0 | 0.1 | ST PAC1 RECEPTOR PATHWAY | PAC1 Receptor Pathway |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.2 | REACTOME SIGNALING BY FGFR3 MUTANTS | Genes involved in Signaling by FGFR3 mutants |
| 0.0 | 0.0 | REACTOME FGFR1 LIGAND BINDING AND ACTIVATION | Genes involved in FGFR1 ligand binding and activation |
| 0.0 | 0.3 | REACTOME AMYLOIDS | Genes involved in Amyloids |
| 0.0 | 0.3 | REACTOME CYTOCHROME P450 ARRANGED BY SUBSTRATE TYPE | Genes involved in Cytochrome P450 - arranged by substrate type |