Project
PRJNA195909:zebrafish embryo and larva development
Navigation
Downloads
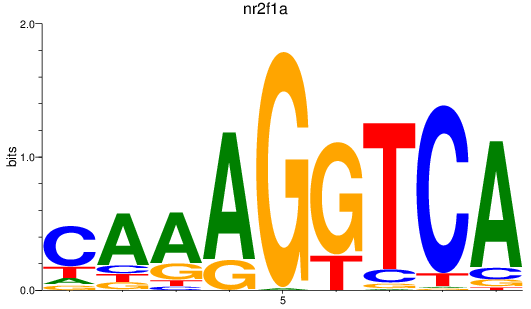
Results for nr2f1a
Z-value: 0.69
Transcription factors associated with nr2f1a
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
nr2f1a
|
ENSDARG00000052695 | nuclear receptor subfamily 2, group F, member 1a |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| nr2f1a | dr11_v1_chr5_+_49744713_49744713 | -0.18 | 6.4e-01 | Click! |
Activity profile of nr2f1a motif
Sorted Z-values of nr2f1a motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr1_+_24387659 | 0.64 |
ENSDART00000130356
|
qdprb2
|
quinoid dihydropteridine reductase b2 |
| chr3_-_5829501 | 0.64 |
ENSDART00000091017
|
pkn1b
|
protein kinase N1b |
| chr7_+_35068036 | 0.54 |
ENSDART00000022139
|
zgc:136461
|
zgc:136461 |
| chr17_-_2590222 | 0.49 |
ENSDART00000185711
|
CR759892.1
|
|
| chr3_+_7771420 | 0.48 |
ENSDART00000156809
ENSDART00000156309 |
hook2
|
hook microtubule-tethering protein 2 |
| chr9_+_29548195 | 0.47 |
ENSDART00000176057
|
rnf17
|
ring finger protein 17 |
| chr10_-_21542702 | 0.44 |
ENSDART00000146761
ENSDART00000134502 |
zgc:165539
|
zgc:165539 |
| chr6_+_154556 | 0.44 |
ENSDART00000193153
|
zglp1
|
zinc finger, GATA-like protein 1 |
| chr17_-_2573021 | 0.43 |
ENSDART00000074181
|
zp3.2
|
zona pellucida glycoprotein 3, tandem duplicate 2 |
| chr19_-_27564458 | 0.41 |
ENSDART00000123155
|
si:dkeyp-46h3.6
|
si:dkeyp-46h3.6 |
| chr3_-_3448095 | 0.41 |
ENSDART00000078886
|
A2ML1 (1 of many)
|
si:dkey-46g23.5 |
| chr17_-_6613458 | 0.39 |
ENSDART00000175024
|
si:ch211-189e2.3
|
si:ch211-189e2.3 |
| chr23_-_42810664 | 0.39 |
ENSDART00000102328
|
pfkfb2a
|
6-phosphofructo-2-kinase/fructose-2,6-biphosphatase 2a |
| chr22_-_24818066 | 0.38 |
ENSDART00000143443
|
vtg6
|
vitellogenin 6 |
| chr12_-_10508952 | 0.37 |
ENSDART00000152806
|
zgc:152977
|
zgc:152977 |
| chr22_+_29113796 | 0.35 |
ENSDART00000150264
|
pla2g6
|
phospholipase A2, group VI (cytosolic, calcium-independent) |
| chr17_-_2595736 | 0.35 |
ENSDART00000128797
|
zp3.2
|
zona pellucida glycoprotein 3, tandem duplicate 2 |
| chr24_-_10393969 | 0.35 |
ENSDART00000106260
|
ankha
|
ANKH inorganic pyrophosphate transport regulator a |
| chr7_-_51775688 | 0.34 |
ENSDART00000149793
|
bmp15
|
bone morphogenetic protein 15 |
| chr5_-_14509137 | 0.34 |
ENSDART00000180742
|
si:ch211-244o22.2
|
si:ch211-244o22.2 |
| chr23_+_39346930 | 0.34 |
ENSDART00000102843
|
src
|
v-src avian sarcoma (Schmidt-Ruppin A-2) viral oncogene homolog |
| chr8_-_23780334 | 0.34 |
ENSDART00000145179
ENSDART00000145894 |
zgc:195245
|
zgc:195245 |
| chr11_+_18037729 | 0.33 |
ENSDART00000111624
|
zgc:175135
|
zgc:175135 |
| chr16_+_26012569 | 0.33 |
ENSDART00000148846
|
prss59.1
|
protease, serine, 59, tandem duplicate 1 |
| chr12_+_21525496 | 0.33 |
ENSDART00000152974
|
ca10a
|
carbonic anhydrase Xa |
| chr17_+_6538733 | 0.32 |
ENSDART00000193712
|
slc5a6b
|
solute carrier family 5 (sodium/multivitamin and iodide cotransporter), member 6 |
| chr17_-_2578026 | 0.32 |
ENSDART00000065821
|
zp3.2
|
zona pellucida glycoprotein 3, tandem duplicate 2 |
| chr18_+_27515640 | 0.32 |
ENSDART00000181593
|
tp53i11b
|
tumor protein p53 inducible protein 11b |
| chr21_+_25226558 | 0.31 |
ENSDART00000168480
|
sycn.2
|
syncollin, tandem duplicate 2 |
| chr3_-_58455289 | 0.31 |
ENSDART00000052179
|
cdr2a
|
cerebellar degeneration-related protein 2a |
| chr23_+_39346774 | 0.30 |
ENSDART00000190985
|
src
|
v-src avian sarcoma (Schmidt-Ruppin A-2) viral oncogene homolog |
| chr11_+_45287541 | 0.30 |
ENSDART00000165321
ENSDART00000173116 |
pycr1b
|
pyrroline-5-carboxylate reductase 1b |
| chr18_-_12957451 | 0.30 |
ENSDART00000140403
|
srgap1a
|
SLIT-ROBO Rho GTPase activating protein 1a |
| chr10_+_19569052 | 0.30 |
ENSDART00000058425
|
CABZ01059627.1
|
|
| chr3_+_3454610 | 0.29 |
ENSDART00000024900
|
zgc:165453
|
zgc:165453 |
| chr18_+_44768829 | 0.29 |
ENSDART00000016271
|
ilvbl
|
ilvB (bacterial acetolactate synthase)-like |
| chr22_-_11833317 | 0.29 |
ENSDART00000125423
ENSDART00000000192 |
ptpn4b
|
protein tyrosine phosphatase, non-receptor type 4b |
| chr1_-_23110740 | 0.29 |
ENSDART00000171848
ENSDART00000086797 ENSDART00000189344 ENSDART00000190858 |
adgrl3.1
|
adhesion G protein-coupled receptor L3.1 |
| chr16_-_39131666 | 0.29 |
ENSDART00000075517
|
gdf6a
|
growth differentiation factor 6a |
| chr25_+_22571475 | 0.28 |
ENSDART00000161559
|
stra6
|
stimulated by retinoic acid 6 |
| chr18_+_44769027 | 0.28 |
ENSDART00000145190
|
ilvbl
|
ilvB (bacterial acetolactate synthase)-like |
| chr3_+_31662126 | 0.28 |
ENSDART00000113441
|
mylk5
|
myosin, light chain kinase 5 |
| chr19_-_10330778 | 0.27 |
ENSDART00000081465
ENSDART00000136653 ENSDART00000171232 |
ccdc106b
|
coiled-coil domain containing 106b |
| chr13_-_23956178 | 0.27 |
ENSDART00000133646
|
phactr2
|
phosphatase and actin regulator 2 |
| chr23_-_27050083 | 0.26 |
ENSDART00000142324
ENSDART00000133249 ENSDART00000138751 ENSDART00000128718 |
zgc:66440
|
zgc:66440 |
| chr14_-_7409364 | 0.26 |
ENSDART00000036463
|
dnd1
|
DND microRNA-mediated repression inhibitor 1 |
| chr19_-_27570333 | 0.26 |
ENSDART00000146562
ENSDART00000179060 |
si:dkeyp-46h3.5
si:dkeyp-46h3.8
|
si:dkeyp-46h3.5 si:dkeyp-46h3.8 |
| chr13_-_24880525 | 0.26 |
ENSDART00000136624
|
kat6b
|
K(lysine) acetyltransferase 6B |
| chr18_-_43866001 | 0.26 |
ENSDART00000150218
|
treh
|
trehalase (brush-border membrane glycoprotein) |
| chr20_+_35445462 | 0.26 |
ENSDART00000124497
|
tdrd6
|
tudor domain containing 6 |
| chr1_+_36772348 | 0.26 |
ENSDART00000109314
|
arhgap10
|
Rho GTPase activating protein 10 |
| chr24_-_24271629 | 0.26 |
ENSDART00000135060
|
rps6ka3b
|
ribosomal protein S6 kinase, polypeptide 3b |
| chr19_-_3056235 | 0.26 |
ENSDART00000137020
|
bop1
|
block of proliferation 1 |
| chr9_-_32912638 | 0.25 |
ENSDART00000110582
|
fam160a2
|
family with sequence similarity 160, member A2 |
| chr15_+_29393519 | 0.25 |
ENSDART00000193488
ENSDART00000112375 |
gdpd5b
|
glycerophosphodiester phosphodiesterase domain containing 5b |
| chr1_-_58868306 | 0.25 |
ENSDART00000166615
|
dnm2b
|
dynamin 2b |
| chr7_-_24520866 | 0.25 |
ENSDART00000077039
|
faah2b
|
fatty acid amide hydrolase 2b |
| chr18_-_26510545 | 0.25 |
ENSDART00000135133
|
FRMD5
|
si:ch211-69m14.1 |
| chr10_-_25561751 | 0.25 |
ENSDART00000147089
|
grik1a
|
glutamate receptor, ionotropic, kainate 1a |
| chr5_-_3839285 | 0.25 |
ENSDART00000122292
|
mlxipl
|
MLX interacting protein like |
| chr21_+_25221940 | 0.25 |
ENSDART00000108972
|
sycn.1
|
syncollin, tandem duplicate 1 |
| chr11_+_18130300 | 0.24 |
ENSDART00000169146
|
zgc:175135
|
zgc:175135 |
| chr5_-_37116265 | 0.24 |
ENSDART00000057613
|
il13ra2
|
interleukin 13 receptor, alpha 2 |
| chr23_+_24272421 | 0.24 |
ENSDART00000029974
|
clcnk
|
chloride channel K |
| chr11_+_18157260 | 0.24 |
ENSDART00000144659
|
zgc:173545
|
zgc:173545 |
| chr21_+_18405585 | 0.24 |
ENSDART00000139318
|
si:dkey-1d7.3
|
si:dkey-1d7.3 |
| chr6_+_49053319 | 0.24 |
ENSDART00000124524
|
sycp1
|
synaptonemal complex protein 1 |
| chr11_+_18053333 | 0.24 |
ENSDART00000075750
|
zgc:175135
|
zgc:175135 |
| chr16_-_16226180 | 0.24 |
ENSDART00000108965
|
gra
|
granulito |
| chr23_+_2740741 | 0.24 |
ENSDART00000134938
|
zgc:114123
|
zgc:114123 |
| chr12_+_1609563 | 0.23 |
ENSDART00000163559
|
SLC39A11
|
solute carrier family 39 member 11 |
| chr23_+_9522942 | 0.23 |
ENSDART00000137751
|
osbpl2b
|
oxysterol binding protein-like 2b |
| chr4_+_25917915 | 0.23 |
ENSDART00000138603
|
vezt
|
vezatin, adherens junctions transmembrane protein |
| chr2_+_52232630 | 0.23 |
ENSDART00000006216
|
plpp2a
|
phospholipid phosphatase 2a |
| chr10_+_10972795 | 0.23 |
ENSDART00000127331
|
cdc37l1
|
cell division cycle 37-like 1 |
| chr8_-_12468744 | 0.22 |
ENSDART00000135019
|
FIBCD1 (1 of many)
|
si:dkeyp-51b7.3 |
| chr11_+_45219558 | 0.22 |
ENSDART00000167828
|
tmc6b
|
transmembrane channel-like 6b |
| chr3_-_34561624 | 0.22 |
ENSDART00000129313
|
sept9a
|
septin 9a |
| chr21_-_28640316 | 0.22 |
ENSDART00000128237
|
nrg2a
|
neuregulin 2a |
| chr15_-_23482088 | 0.21 |
ENSDART00000185823
ENSDART00000185523 |
nlrx1
|
NLR family member X1 |
| chr8_+_36509885 | 0.21 |
ENSDART00000109530
|
slc7a4
|
solute carrier family 7, member 4 |
| chr19_-_867071 | 0.21 |
ENSDART00000122257
|
eomesa
|
eomesodermin homolog a |
| chr11_-_42230491 | 0.21 |
ENSDART00000164423
|
CABZ01030862.1
|
|
| chr5_-_67115872 | 0.21 |
ENSDART00000065262
|
rps6ka4
|
ribosomal protein S6 kinase, polypeptide 4 |
| chr21_+_1119046 | 0.20 |
ENSDART00000184678
|
CABZ01088049.1
|
|
| chr23_+_9522781 | 0.20 |
ENSDART00000136486
|
osbpl2b
|
oxysterol binding protein-like 2b |
| chr2_+_10642047 | 0.20 |
ENSDART00000091570
|
fam69aa
|
family with sequence similarity 69, member Aa |
| chr23_-_40194732 | 0.20 |
ENSDART00000164931
|
tgm1l2
|
transglutaminase 1 like 2 |
| chr8_-_33154677 | 0.20 |
ENSDART00000133300
|
zbtb34
|
zinc finger and BTB domain containing 34 |
| chr17_+_34206167 | 0.20 |
ENSDART00000136167
|
mpp5a
|
membrane protein, palmitoylated 5a (MAGUK p55 subfamily member 5) |
| chr12_+_23866368 | 0.20 |
ENSDART00000188652
ENSDART00000192478 |
svila
|
supervillin a |
| chr18_-_43866526 | 0.20 |
ENSDART00000111309
|
treh
|
trehalase (brush-border membrane glycoprotein) |
| chr14_-_14659023 | 0.20 |
ENSDART00000170355
ENSDART00000159888 ENSDART00000172241 |
nsdhl
|
NAD(P) dependent steroid dehydrogenase-like |
| chr25_-_37284370 | 0.20 |
ENSDART00000103222
|
nudt7
|
nudix (nucleoside diphosphate linked moiety X)-type motif 7 |
| chr8_+_25733872 | 0.20 |
ENSDART00000156340
|
si:ch211-167b20.8
|
si:ch211-167b20.8 |
| chr5_-_20678300 | 0.20 |
ENSDART00000088639
|
wscd2
|
WSC domain containing 2 |
| chr23_-_45705525 | 0.20 |
ENSDART00000148959
|
ednrab
|
endothelin receptor type Ab |
| chr24_+_39518774 | 0.20 |
ENSDART00000132939
|
dcun1d3
|
defective in cullin neddylation 1 domain containing 3 |
| chr21_-_9446747 | 0.19 |
ENSDART00000158790
|
ptpn13
|
protein tyrosine phosphatase, non-receptor type 13 |
| chr10_-_10969596 | 0.19 |
ENSDART00000092011
|
exd3
|
exonuclease 3'-5' domain containing 3 |
| chr19_+_31532043 | 0.19 |
ENSDART00000136289
|
tmem64
|
transmembrane protein 64 |
| chr23_-_14918276 | 0.19 |
ENSDART00000179831
|
ndrg3b
|
ndrg family member 3b |
| chr7_-_20611039 | 0.19 |
ENSDART00000170422
|
si:dkey-19b23.8
|
si:dkey-19b23.8 |
| chr13_-_23956361 | 0.19 |
ENSDART00000101150
|
phactr2
|
phosphatase and actin regulator 2 |
| chr19_-_27564980 | 0.19 |
ENSDART00000171967
|
si:dkeyp-46h3.8
|
si:dkeyp-46h3.8 |
| chr15_+_20799943 | 0.19 |
ENSDART00000154665
|
aldh3a1
|
aldehyde dehydrogenase 3 family, member A1 |
| chr13_+_23095228 | 0.19 |
ENSDART00000189068
ENSDART00000188624 |
pik3ap1
|
phosphoinositide-3-kinase adaptor protein 1 |
| chr13_+_10023256 | 0.19 |
ENSDART00000110035
|
srbd1
|
S1 RNA binding domain 1 |
| chr12_+_48681601 | 0.19 |
ENSDART00000187831
|
uros
|
uroporphyrinogen III synthase |
| chr15_+_35933094 | 0.19 |
ENSDART00000019976
|
rhbdd1
|
rhomboid domain containing 1 |
| chr19_-_42045372 | 0.19 |
ENSDART00000144275
|
trioa
|
trio Rho guanine nucleotide exchange factor a |
| chr5_-_69934558 | 0.18 |
ENSDART00000124954
|
ugt2a4
|
UDP glucuronosyltransferase 2 family, polypeptide A4 |
| chr4_-_858434 | 0.18 |
ENSDART00000006961
|
sobpb
|
sine oculis binding protein homolog (Drosophila) b |
| chr16_-_54919260 | 0.18 |
ENSDART00000156533
|
kdf1a
|
keratinocyte differentiation factor 1a |
| chr14_+_32926385 | 0.18 |
ENSDART00000139159
|
lnx2b
|
ligand of numb-protein X 2b |
| chr19_-_31707892 | 0.18 |
ENSDART00000088427
|
ripor2
|
RHO family interacting cell polarization regulator 2 |
| chr22_-_7050 | 0.18 |
ENSDART00000127829
|
atad3
|
ATPase family, AAA domain containing 3 |
| chr25_+_35889102 | 0.18 |
ENSDART00000023453
ENSDART00000125821 ENSDART00000135441 |
lsm14aa
|
LSM14A mRNA processing body assembly factor a |
| chr20_-_35508805 | 0.18 |
ENSDART00000169538
|
adgrf3b
|
adhesion G protein-coupled receptor F3b |
| chr5_+_63340637 | 0.18 |
ENSDART00000143742
|
si:ch73-376l24.4
|
si:ch73-376l24.4 |
| chr17_+_13031497 | 0.18 |
ENSDART00000115208
|
fbxo33
|
F-box protein 33 |
| chr6_-_41135215 | 0.17 |
ENSDART00000001861
|
slc6a22.1
|
solute carrier family 6 member 22, tandem duplicate 1 |
| chr6_-_49547680 | 0.17 |
ENSDART00000169678
|
ppp4r1l
|
protein phosphatase 4, regulatory subunit 1-like |
| chr12_-_3077395 | 0.17 |
ENSDART00000002867
ENSDART00000126315 |
rfng
|
RFNG O-fucosylpeptide 3-beta-N-acetylglucosaminyltransferase |
| chr4_-_4535189 | 0.17 |
ENSDART00000057519
|
zgc:194209
|
zgc:194209 |
| chr17_+_20569806 | 0.17 |
ENSDART00000113936
|
SAMD8
|
zgc:162183 |
| chr14_+_49683767 | 0.17 |
ENSDART00000192515
|
tspan17
|
tetraspanin 17 |
| chr2_+_23062085 | 0.17 |
ENSDART00000153745
|
csnk1g2a
|
casein kinase 1, gamma 2a |
| chr14_+_35428152 | 0.17 |
ENSDART00000172597
|
sytl4
|
synaptotagmin-like 4 |
| chr19_-_24136233 | 0.17 |
ENSDART00000143365
|
thap7
|
THAP domain containing 7 |
| chr18_-_127558 | 0.17 |
ENSDART00000149556
|
trpm7
|
transient receptor potential cation channel, subfamily M, member 7 |
| chr25_-_20049449 | 0.17 |
ENSDART00000104315
|
zgc:136858
|
zgc:136858 |
| chr5_+_9218318 | 0.17 |
ENSDART00000137774
|
si:ch211-12e13.1
|
si:ch211-12e13.1 |
| chr5_+_1109098 | 0.17 |
ENSDART00000166268
|
LO017790.1
|
|
| chr14_+_30413758 | 0.17 |
ENSDART00000092953
|
cnot7
|
CCR4-NOT transcription complex, subunit 7 |
| chr14_-_33478963 | 0.17 |
ENSDART00000132813
|
lamp2
|
lysosomal-associated membrane protein 2 |
| chr17_+_49484640 | 0.17 |
ENSDART00000179706
|
AREL1
|
apoptosis resistant E3 ubiquitin protein ligase 1 |
| chr22_-_16377960 | 0.17 |
ENSDART00000168170
|
ttc39c
|
tetratricopeptide repeat domain 39C |
| chr4_+_15942075 | 0.17 |
ENSDART00000147964
|
si:dkey-117n7.2
|
si:dkey-117n7.2 |
| chr12_-_48943467 | 0.16 |
ENSDART00000191829
|
CABZ01092907.1
|
|
| chr7_+_24520518 | 0.16 |
ENSDART00000173604
|
btr09
|
bloodthirsty-related gene family, member 9 |
| chr2_-_41562868 | 0.16 |
ENSDART00000084597
|
d2hgdh
|
D-2-hydroxyglutarate dehydrogenase |
| chr5_-_13076779 | 0.16 |
ENSDART00000192826
|
ypel1
|
yippee-like 1 |
| chr6_+_30533504 | 0.16 |
ENSDART00000155842
|
wwc3
|
WWC family member 3 |
| chr19_-_47587719 | 0.16 |
ENSDART00000111108
|
CABZ01071972.1
|
|
| chr20_+_6535176 | 0.16 |
ENSDART00000054652
|
si:ch211-191a24.4
|
si:ch211-191a24.4 |
| chr21_+_37513488 | 0.16 |
ENSDART00000185394
|
amot
|
angiomotin |
| chr24_+_16393302 | 0.16 |
ENSDART00000188670
ENSDART00000081759 ENSDART00000177790 |
sema5a
|
sema domain, seven thrombospondin repeats (type 1 and type 1-like), transmembrane domain (TM) and short cytoplasmic domain, (semaphorin) 5A |
| chr16_-_35532937 | 0.16 |
ENSDART00000193209
|
ctps1b
|
CTP synthase 1b |
| chr21_-_43428040 | 0.16 |
ENSDART00000148325
|
stk26
|
serine/threonine protein kinase 26 |
| chr19_-_8798178 | 0.16 |
ENSDART00000188232
|
cers2a
|
ceramide synthase 2a |
| chr4_-_8060962 | 0.16 |
ENSDART00000146622
|
wnk1b
|
WNK lysine deficient protein kinase 1b |
| chr14_+_26224541 | 0.16 |
ENSDART00000128971
|
gm2a
|
GM2 ganglioside activator |
| chr3_+_33745014 | 0.16 |
ENSDART00000159966
|
nacc1a
|
nucleus accumbens associated 1, BEN and BTB (POZ) domain containing a |
| chr24_+_39034090 | 0.16 |
ENSDART00000185763
|
capn15
|
calpain 15 |
| chr19_-_35439237 | 0.16 |
ENSDART00000145883
|
anln
|
anillin, actin binding protein |
| chr13_+_2357637 | 0.15 |
ENSDART00000017148
|
gclc
|
glutamate-cysteine ligase, catalytic subunit |
| chr15_+_47386939 | 0.15 |
ENSDART00000128224
|
FO904873.1
|
|
| chr18_+_45571378 | 0.15 |
ENSDART00000077251
|
kifc3
|
kinesin family member C3 |
| chr22_+_15960514 | 0.15 |
ENSDART00000181617
|
stil
|
scl/tal1 interrupting locus |
| chr6_-_1587291 | 0.15 |
ENSDART00000067592
ENSDART00000178877 |
zgc:123305
|
zgc:123305 |
| chr11_-_7410537 | 0.15 |
ENSDART00000009859
|
adgrl4
|
adhesion G protein-coupled receptor L4 |
| chr24_-_12770357 | 0.15 |
ENSDART00000060826
|
ipo4
|
importin 4 |
| chr19_+_3056450 | 0.15 |
ENSDART00000141324
ENSDART00000082353 |
hsf1
|
heat shock transcription factor 1 |
| chr1_-_9249943 | 0.15 |
ENSDART00000055011
|
zgc:136472
|
zgc:136472 |
| chr18_-_21746421 | 0.15 |
ENSDART00000188809
|
pskh1
|
protein serine kinase H1 |
| chr18_+_35229115 | 0.15 |
ENSDART00000129624
ENSDART00000184596 |
tbrg1
|
transforming growth factor beta regulator 1 |
| chr5_+_63375620 | 0.14 |
ENSDART00000185568
|
si:ch73-376l24.6
|
si:ch73-376l24.6 |
| chr3_-_54500354 | 0.14 |
ENSDART00000124215
|
trip10a
|
thyroid hormone receptor interactor 10a |
| chr3_+_1179601 | 0.14 |
ENSDART00000173378
|
triobpb
|
TRIO and F-actin binding protein b |
| chr2_-_57076687 | 0.14 |
ENSDART00000161523
|
slc25a42
|
solute carrier family 25, member 42 |
| chr2_+_44977889 | 0.14 |
ENSDART00000144024
|
alg3
|
asparagine-linked glycosylation 3 (alpha-1,3-mannosyltransferase) |
| chr16_-_33095161 | 0.14 |
ENSDART00000187648
|
dopey1
|
dopey family member 1 |
| chr13_-_21739142 | 0.14 |
ENSDART00000078460
|
si:dkey-191g9.5
|
si:dkey-191g9.5 |
| chr11_-_34219211 | 0.14 |
ENSDART00000098472
|
tmem44
|
transmembrane protein 44 |
| chr10_-_21362071 | 0.14 |
ENSDART00000125167
|
avd
|
avidin |
| chr10_+_24692076 | 0.14 |
ENSDART00000181600
|
tpte
|
transmembrane phosphatase with tensin homology |
| chr23_+_12545114 | 0.14 |
ENSDART00000105283
ENSDART00000166990 |
si:zfos-452g4.1
|
si:zfos-452g4.1 |
| chr6_-_47246948 | 0.14 |
ENSDART00000162435
|
grm4
|
glutamate receptor, metabotropic 4 |
| chr16_-_9423735 | 0.14 |
ENSDART00000185645
|
ccr8.1
|
chemokine (C-C motif) receptor 8.1 |
| chr5_-_69180587 | 0.14 |
ENSDART00000156681
ENSDART00000160753 |
zgc:171967
|
zgc:171967 |
| chr25_-_9805269 | 0.14 |
ENSDART00000192048
|
lrrc4c
|
leucine rich repeat containing 4C |
| chr9_+_38503233 | 0.14 |
ENSDART00000140331
|
lmln
|
leishmanolysin-like (metallopeptidase M8 family) |
| chr12_+_23991639 | 0.14 |
ENSDART00000003143
|
psme4b
|
proteasome activator subunit 4b |
| chr13_-_33114933 | 0.14 |
ENSDART00000140543
ENSDART00000075953 |
zfyve26
|
zinc finger, FYVE domain containing 26 |
| chr5_-_69180227 | 0.14 |
ENSDART00000154816
|
zgc:171967
|
zgc:171967 |
| chr15_-_6946286 | 0.14 |
ENSDART00000019330
|
ech1
|
enoyl CoA hydratase 1, peroxisomal |
| chr10_-_8197049 | 0.14 |
ENSDART00000129467
|
dhx29
|
DEAH (Asp-Glu-Ala-His) box polypeptide 29 |
| chr11_-_22997506 | 0.14 |
ENSDART00000167817
|
atp2b2
|
ATPase plasma membrane Ca2+ transporting 2 |
| chr24_-_10919588 | 0.14 |
ENSDART00000131204
|
asap1b
|
ArfGAP with SH3 domain, ankyrin repeat and PH domain 1b |
| chr9_+_38967998 | 0.14 |
ENSDART00000135581
|
map2
|
microtubule-associated protein 2 |
| chr2_-_20715094 | 0.14 |
ENSDART00000155439
|
dusp12
|
dual specificity phosphatase 12 |
| chr1_+_29741843 | 0.14 |
ENSDART00000136066
|
raph1b
|
Ras association (RalGDS/AF-6) and pleckstrin homology domains 1b |
| chr3_+_22905341 | 0.14 |
ENSDART00000111435
|
hdac5
|
histone deacetylase 5 |
| chr12_+_17603528 | 0.14 |
ENSDART00000111565
|
pms2
|
PMS1 homolog 2, mismatch repair system component |
| chr19_+_2631565 | 0.14 |
ENSDART00000171487
|
fam126a
|
family with sequence similarity 126, member A |
| chr8_-_38810233 | 0.14 |
ENSDART00000085304
|
pcsk5b
|
proprotein convertase subtilisin/kexin type 5b |
| chr17_+_50657509 | 0.14 |
ENSDART00000179957
|
ddhd1a
|
DDHD domain containing 1a |
| chr5_-_27994679 | 0.14 |
ENSDART00000132740
|
ppp3cca
|
protein phosphatase 3, catalytic subunit, gamma isozyme, a |
| chr14_+_30413312 | 0.13 |
ENSDART00000186864
|
cnot7
|
CCR4-NOT transcription complex, subunit 7 |
Network of associatons between targets according to the STRING database.
First level regulatory network of nr2f1a
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.6 | GO:0050847 | progesterone receptor signaling pathway(GO:0050847) |
| 0.2 | 0.5 | GO:0005991 | trehalose metabolic process(GO:0005991) |
| 0.1 | 0.6 | GO:0009097 | isoleucine metabolic process(GO:0006549) isoleucine biosynthetic process(GO:0009097) |
| 0.1 | 0.3 | GO:0060283 | negative regulation of oocyte development(GO:0060283) |
| 0.1 | 0.3 | GO:0051182 | biotin transport(GO:0015878) pantothenate transmembrane transport(GO:0015887) coenzyme transport(GO:0051182) |
| 0.1 | 0.2 | GO:0051026 | meiotic DNA repair synthesis(GO:0000711) chiasma assembly(GO:0051026) |
| 0.1 | 0.3 | GO:0060220 | retinal cone cell fate determination(GO:0042671) eye photoreceptor cell fate commitment(GO:0042706) photoreceptor cell fate determination(GO:0043703) retinal cone cell fate commitment(GO:0046551) photoreceptor cell fate commitment(GO:0046552) camera-type eye photoreceptor cell fate commitment(GO:0060220) |
| 0.1 | 0.2 | GO:0060571 | invagination involved in gastrulation with mouth forming second(GO:0055109) morphogenesis of an epithelial fold(GO:0060571) |
| 0.1 | 1.1 | GO:2000344 | binding of sperm to zona pellucida(GO:0007339) egg coat formation(GO:0035803) regulation of acrosome reaction(GO:0060046) positive regulation of acrosome reaction(GO:2000344) |
| 0.1 | 0.2 | GO:1990869 | response to chemokine(GO:1990868) cellular response to chemokine(GO:1990869) negative regulation of lymphocyte migration(GO:2000402) negative regulation of T cell migration(GO:2000405) |
| 0.1 | 0.3 | GO:0035461 | vitamin transmembrane transport(GO:0035461) |
| 0.1 | 0.2 | GO:0010961 | cellular magnesium ion homeostasis(GO:0010961) |
| 0.1 | 0.5 | GO:0046146 | tetrahydrobiopterin biosynthetic process(GO:0006729) tetrahydrobiopterin metabolic process(GO:0046146) |
| 0.1 | 0.2 | GO:0016332 | establishment or maintenance of polarity of embryonic epithelium(GO:0016332) |
| 0.0 | 0.3 | GO:0007289 | spermatid nucleus differentiation(GO:0007289) sperm chromatin condensation(GO:0035092) spermatogenesis, exchange of chromosomal proteins(GO:0035093) |
| 0.0 | 0.1 | GO:1901503 | ether lipid biosynthetic process(GO:0008611) glycerol ether biosynthetic process(GO:0046504) cellular lipid biosynthetic process(GO:0097384) ether biosynthetic process(GO:1901503) |
| 0.0 | 0.3 | GO:0060149 | negative regulation of posttranscriptional gene silencing(GO:0060149) negative regulation of gene silencing by miRNA(GO:0060965) negative regulation of gene silencing by RNA(GO:0060967) |
| 0.0 | 0.1 | GO:2001014 | skeletal muscle fiber differentiation(GO:0098528) regulation of skeletal muscle cell differentiation(GO:2001014) |
| 0.0 | 0.3 | GO:0045329 | carnitine biosynthetic process(GO:0045329) |
| 0.0 | 0.1 | GO:0042364 | water-soluble vitamin biosynthetic process(GO:0042364) pyridoxal phosphate metabolic process(GO:0042822) pyridoxal phosphate biosynthetic process(GO:0042823) |
| 0.0 | 0.2 | GO:0044210 | 'de novo' CTP biosynthetic process(GO:0044210) |
| 0.0 | 0.2 | GO:0008356 | asymmetric cell division(GO:0008356) |
| 0.0 | 0.2 | GO:0006750 | glutathione biosynthetic process(GO:0006750) |
| 0.0 | 0.2 | GO:0010481 | epidermal cell division(GO:0010481) regulation of epidermal cell division(GO:0010482) |
| 0.0 | 0.2 | GO:0016572 | histone phosphorylation(GO:0016572) |
| 0.0 | 0.3 | GO:0055129 | proline biosynthetic process(GO:0006561) L-proline biosynthetic process(GO:0055129) |
| 0.0 | 0.1 | GO:2000193 | regulation of plasma membrane long-chain fatty acid transport(GO:0010746) positive regulation of plasma membrane long-chain fatty acid transport(GO:0010747) plasma membrane long-chain fatty acid transport(GO:0015911) fatty acid transmembrane transport(GO:1902001) positive regulation of anion transmembrane transport(GO:1903961) regulation of fatty acid transport(GO:2000191) positive regulation of fatty acid transport(GO:2000193) |
| 0.0 | 0.2 | GO:0003319 | cardioblast migration to the midline involved in heart rudiment formation(GO:0003319) |
| 0.0 | 0.4 | GO:0006363 | termination of RNA polymerase I transcription(GO:0006363) |
| 0.0 | 0.1 | GO:0009211 | pyrimidine deoxyribonucleoside triphosphate metabolic process(GO:0009211) |
| 0.0 | 0.2 | GO:0098773 | ventricular trabecula myocardium morphogenesis(GO:0003222) trabecula formation(GO:0060343) heart trabecula formation(GO:0060347) skin epidermis development(GO:0098773) |
| 0.0 | 0.1 | GO:0046900 | tetrahydrofolylpolyglutamate metabolic process(GO:0046900) |
| 0.0 | 0.1 | GO:1900364 | negative regulation of mRNA 3'-end processing(GO:0031441) negative regulation of mRNA polyadenylation(GO:1900364) |
| 0.0 | 0.2 | GO:0033206 | meiotic cytokinesis(GO:0033206) polar body extrusion after meiotic divisions(GO:0040038) |
| 0.0 | 0.3 | GO:0035307 | positive regulation of dephosphorylation(GO:0035306) positive regulation of protein dephosphorylation(GO:0035307) |
| 0.0 | 0.1 | GO:0032755 | positive regulation of interleukin-6 production(GO:0032755) protein import into mitochondrial intermembrane space(GO:0045041) |
| 0.0 | 0.1 | GO:0002495 | antigen processing and presentation of peptide antigen via MHC class II(GO:0002495) |
| 0.0 | 0.4 | GO:0006003 | fructose 2,6-bisphosphate metabolic process(GO:0006003) |
| 0.0 | 0.3 | GO:0043928 | exonucleolytic nuclear-transcribed mRNA catabolic process involved in deadenylation-dependent decay(GO:0043928) |
| 0.0 | 0.1 | GO:0071423 | thiosulfate transport(GO:0015709) oxaloacetate transport(GO:0015729) malate transport(GO:0015743) succinate transport(GO:0015744) succinate transmembrane transport(GO:0071422) malate transmembrane transport(GO:0071423) |
| 0.0 | 0.1 | GO:0016191 | synaptic vesicle uncoating(GO:0016191) |
| 0.0 | 0.2 | GO:0045947 | negative regulation of translational initiation(GO:0045947) |
| 0.0 | 0.3 | GO:0030719 | P granule organization(GO:0030719) |
| 0.0 | 0.1 | GO:0098529 | neuromuscular junction development, skeletal muscle fiber(GO:0098529) |
| 0.0 | 0.1 | GO:0016267 | O-glycan processing, core 1(GO:0016267) |
| 0.0 | 0.1 | GO:0035513 | oxidative RNA demethylation(GO:0035513) |
| 0.0 | 0.2 | GO:0034063 | stress granule assembly(GO:0034063) |
| 0.0 | 0.2 | GO:0009109 | coenzyme catabolic process(GO:0009109) |
| 0.0 | 0.1 | GO:0007405 | neuroblast proliferation(GO:0007405) establishment of centrosome localization(GO:0051660) |
| 0.0 | 0.1 | GO:0051972 | regulation of telomerase activity(GO:0051972) |
| 0.0 | 0.1 | GO:2000434 | regulation of protein neddylation(GO:2000434) |
| 0.0 | 0.1 | GO:0010735 | positive regulation of transcription via serum response element binding(GO:0010735) |
| 0.0 | 0.1 | GO:0033615 | mitochondrial proton-transporting ATP synthase complex assembly(GO:0033615) |
| 0.0 | 0.3 | GO:0016486 | peptide hormone processing(GO:0016486) |
| 0.0 | 0.1 | GO:0045948 | positive regulation of translational initiation(GO:0045948) |
| 0.0 | 0.2 | GO:0003365 | establishment of cell polarity involved in ameboidal cell migration(GO:0003365) |
| 0.0 | 0.1 | GO:0018344 | protein geranylgeranylation(GO:0018344) |
| 0.0 | 0.1 | GO:0048210 | Golgi vesicle fusion to target membrane(GO:0048210) |
| 0.0 | 0.1 | GO:0060394 | negative regulation of pathway-restricted SMAD protein phosphorylation(GO:0060394) |
| 0.0 | 0.1 | GO:0043570 | maintenance of DNA repeat elements(GO:0043570) |
| 0.0 | 0.1 | GO:1900026 | positive regulation of substrate adhesion-dependent cell spreading(GO:1900026) |
| 0.0 | 0.1 | GO:0021785 | branchiomotor neuron axon guidance(GO:0021785) |
| 0.0 | 0.6 | GO:0048599 | oocyte development(GO:0048599) |
| 0.0 | 0.1 | GO:0031086 | nuclear-transcribed mRNA catabolic process, deadenylation-independent decay(GO:0031086) deadenylation-independent decapping of nuclear-transcribed mRNA(GO:0031087) |
| 0.0 | 0.2 | GO:0071539 | protein localization to centrosome(GO:0071539) |
| 0.0 | 0.2 | GO:0006622 | protein targeting to lysosome(GO:0006622) |
| 0.0 | 0.2 | GO:0045116 | protein neddylation(GO:0045116) |
| 0.0 | 0.3 | GO:0046501 | protoporphyrinogen IX biosynthetic process(GO:0006782) protoporphyrinogen IX metabolic process(GO:0046501) |
| 0.0 | 0.1 | GO:0032655 | interleukin-12 production(GO:0032615) regulation of interleukin-12 production(GO:0032655) |
| 0.0 | 0.2 | GO:0006048 | UDP-N-acetylglucosamine biosynthetic process(GO:0006048) |
| 0.0 | 0.2 | GO:0035283 | central nervous system segmentation(GO:0035283) brain segmentation(GO:0035284) |
| 0.0 | 0.1 | GO:0035066 | positive regulation of histone acetylation(GO:0035066) |
| 0.0 | 0.1 | GO:0036149 | phosphatidylinositol acyl-chain remodeling(GO:0036149) |
| 0.0 | 0.3 | GO:0097324 | melanocyte migration(GO:0097324) |
| 0.0 | 0.0 | GO:0032510 | endosome to lysosome transport via multivesicular body sorting pathway(GO:0032510) |
| 0.0 | 0.1 | GO:0006446 | regulation of translational initiation(GO:0006446) |
| 0.0 | 0.1 | GO:2000096 | positive regulation of Wnt signaling pathway, planar cell polarity pathway(GO:2000096) |
| 0.0 | 0.0 | GO:0007008 | outer mitochondrial membrane organization(GO:0007008) protein import into mitochondrial outer membrane(GO:0045040) |
| 0.0 | 0.1 | GO:2001295 | malonyl-CoA metabolic process(GO:2001293) malonyl-CoA biosynthetic process(GO:2001295) |
| 0.0 | 0.3 | GO:0009408 | response to heat(GO:0009408) |
| 0.0 | 0.6 | GO:0071391 | cellular response to estrogen stimulus(GO:0071391) |
| 0.0 | 0.2 | GO:0035435 | phosphate ion transmembrane transport(GO:0035435) |
| 0.0 | 0.4 | GO:0006998 | nuclear envelope organization(GO:0006998) |
| 0.0 | 0.2 | GO:0006689 | ganglioside catabolic process(GO:0006689) |
| 0.0 | 0.0 | GO:1904478 | regulation of intestinal absorption(GO:1904478) regulation of intestinal lipid absorption(GO:1904729) |
| 0.0 | 0.1 | GO:0000395 | mRNA 5'-splice site recognition(GO:0000395) |
| 0.0 | 0.1 | GO:0006999 | nuclear pore organization(GO:0006999) |
| 0.0 | 0.1 | GO:0003428 | growth plate cartilage morphogenesis(GO:0003422) chondrocyte intercalation involved in growth plate cartilage morphogenesis(GO:0003428) |
| 0.0 | 0.1 | GO:0021553 | olfactory nerve development(GO:0021553) olfactory nerve morphogenesis(GO:0021627) olfactory nerve formation(GO:0021628) |
| 0.0 | 0.1 | GO:1901841 | regulation of high voltage-gated calcium channel activity(GO:1901841) negative regulation of high voltage-gated calcium channel activity(GO:1901842) |
| 0.0 | 0.1 | GO:0045444 | fat cell differentiation(GO:0045444) |
| 0.0 | 0.0 | GO:0072003 | kidney rudiment formation(GO:0072003) pronephros formation(GO:0072116) |
| 0.0 | 0.0 | GO:0071926 | endocannabinoid signaling pathway(GO:0071926) retrograde trans-synaptic signaling by lipid(GO:0098920) retrograde trans-synaptic signaling by endocannabinoid(GO:0098921) |
| 0.0 | 0.1 | GO:1904825 | protein localization to microtubule(GO:0035372) protein localization to microtubule plus-end(GO:1904825) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.6 | GO:0005948 | acetolactate synthase complex(GO:0005948) |
| 0.1 | 0.5 | GO:0043073 | germ cell nucleus(GO:0043073) |
| 0.1 | 0.3 | GO:0033391 | chromatoid body(GO:0033391) |
| 0.1 | 0.3 | GO:0070545 | PeBoW complex(GO:0070545) |
| 0.1 | 0.3 | GO:1990111 | spermatoproteasome complex(GO:1990111) |
| 0.1 | 0.2 | GO:0031310 | integral component of vacuolar membrane(GO:0031166) intrinsic component of vacuolar membrane(GO:0031310) |
| 0.0 | 0.2 | GO:0060171 | stereocilium membrane(GO:0060171) |
| 0.0 | 0.2 | GO:0097268 | cytoophidium(GO:0097268) |
| 0.0 | 0.1 | GO:0005968 | Rab-protein geranylgeranyltransferase complex(GO:0005968) |
| 0.0 | 0.1 | GO:1990415 | Pex17p-Pex14p docking complex(GO:1990415) peroxisomal importomer complex(GO:1990429) |
| 0.0 | 0.1 | GO:0032301 | MutSalpha complex(GO:0032301) |
| 0.0 | 0.7 | GO:0030667 | secretory granule membrane(GO:0030667) |
| 0.0 | 0.3 | GO:0030015 | CCR4-NOT core complex(GO:0030015) |
| 0.0 | 0.1 | GO:0005955 | calcineurin complex(GO:0005955) |
| 0.0 | 0.2 | GO:0005826 | actomyosin contractile ring(GO:0005826) |
| 0.0 | 0.4 | GO:0034993 | microtubule organizing center attachment site(GO:0034992) LINC complex(GO:0034993) |
| 0.0 | 0.1 | GO:0032300 | mismatch repair complex(GO:0032300) |
| 0.0 | 0.1 | GO:0043034 | costamere(GO:0043034) |
| 0.0 | 0.1 | GO:0017177 | glucosidase II complex(GO:0017177) |
| 0.0 | 0.1 | GO:0016442 | RISC complex(GO:0016442) RNAi effector complex(GO:0031332) |
| 0.0 | 0.1 | GO:0044613 | nuclear pore central transport channel(GO:0044613) |
| 0.0 | 0.1 | GO:0035517 | PR-DUB complex(GO:0035517) |
| 0.0 | 0.2 | GO:1990124 | messenger ribonucleoprotein complex(GO:1990124) |
| 0.0 | 0.1 | GO:0033588 | Elongator holoenzyme complex(GO:0033588) |
| 0.0 | 0.1 | GO:0032021 | NELF complex(GO:0032021) |
| 0.0 | 0.1 | GO:0030670 | phagocytic vesicle membrane(GO:0030670) |
| 0.0 | 0.1 | GO:0045252 | oxoglutarate dehydrogenase complex(GO:0045252) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.6 | GO:0003984 | acetolactate synthase activity(GO:0003984) |
| 0.2 | 0.6 | GO:0070404 | 6,7-dihydropteridine reductase activity(GO:0004155) NADH binding(GO:0070404) |
| 0.2 | 0.5 | GO:0015927 | alpha,alpha-trehalase activity(GO:0004555) trehalase activity(GO:0015927) |
| 0.1 | 1.1 | GO:0035804 | structural constituent of egg coat(GO:0035804) |
| 0.1 | 0.3 | GO:0008523 | sodium-dependent multivitamin transmembrane transporter activity(GO:0008523) |
| 0.1 | 0.2 | GO:0008336 | gamma-butyrobetaine dioxygenase activity(GO:0008336) |
| 0.1 | 0.3 | GO:0030504 | inorganic diphosphate transmembrane transporter activity(GO:0030504) |
| 0.1 | 0.3 | GO:0004735 | pyrroline-5-carboxylate reductase activity(GO:0004735) |
| 0.1 | 0.4 | GO:0045735 | nutrient reservoir activity(GO:0045735) |
| 0.0 | 0.3 | GO:0070698 | type I activin receptor binding(GO:0070698) |
| 0.0 | 0.1 | GO:0071077 | coenzyme A transmembrane transporter activity(GO:0015228) adenosine 3',5'-bisphosphate transmembrane transporter activity(GO:0071077) AMP transmembrane transporter activity(GO:0080122) |
| 0.0 | 0.2 | GO:0071889 | 14-3-3 protein binding(GO:0071889) |
| 0.0 | 0.1 | GO:1990715 | mRNA CDS binding(GO:1990715) sequence-specific mRNA binding(GO:1990825) |
| 0.0 | 0.2 | GO:0033829 | O-fucosylpeptide 3-beta-N-acetylglucosaminyltransferase activity(GO:0033829) |
| 0.0 | 0.2 | GO:0003883 | CTP synthase activity(GO:0003883) |
| 0.0 | 0.5 | GO:0036312 | phosphatidylinositol 3-kinase regulatory subunit binding(GO:0036312) |
| 0.0 | 0.5 | GO:0003873 | 6-phosphofructo-2-kinase activity(GO:0003873) |
| 0.0 | 0.1 | GO:0000033 | alpha-1,3-mannosyltransferase activity(GO:0000033) |
| 0.0 | 0.1 | GO:0045145 | single-stranded DNA 5'-3' exodeoxyribonuclease activity(GO:0045145) |
| 0.0 | 0.2 | GO:0008190 | eukaryotic initiation factor 4E binding(GO:0008190) |
| 0.0 | 0.1 | GO:0004663 | Rab geranylgeranyltransferase activity(GO:0004663) |
| 0.0 | 0.1 | GO:0043812 | phosphatidylinositol-4-phosphate phosphatase activity(GO:0043812) |
| 0.0 | 0.4 | GO:0043495 | protein anchor(GO:0043495) |
| 0.0 | 0.1 | GO:0032357 | guanine/thymine mispair binding(GO:0032137) single base insertion or deletion binding(GO:0032138) single thymine insertion binding(GO:0032143) oxidized DNA binding(GO:0032356) oxidized purine DNA binding(GO:0032357) mismatch repair complex binding(GO:0032404) MutLalpha complex binding(GO:0032405) |
| 0.0 | 0.4 | GO:0004687 | myosin light chain kinase activity(GO:0004687) |
| 0.0 | 0.1 | GO:0009374 | biotin binding(GO:0009374) |
| 0.0 | 0.3 | GO:0016918 | retinal binding(GO:0016918) |
| 0.0 | 0.1 | GO:0035516 | oxidative DNA demethylase activity(GO:0035516) |
| 0.0 | 0.1 | GO:0043998 | H2A histone acetyltransferase activity(GO:0043998) |
| 0.0 | 0.1 | GO:0043878 | glyceraldehyde-3-phosphate dehydrogenase (NAD+) (non-phosphorylating) activity(GO:0043878) |
| 0.0 | 0.3 | GO:0016504 | peptidase activator activity(GO:0016504) |
| 0.0 | 0.1 | GO:0010521 | telomerase inhibitor activity(GO:0010521) |
| 0.0 | 0.1 | GO:0015140 | thiosulfate transmembrane transporter activity(GO:0015117) oxaloacetate transmembrane transporter activity(GO:0015131) malate transmembrane transporter activity(GO:0015140) |
| 0.0 | 0.1 | GO:0004064 | arylesterase activity(GO:0004064) |
| 0.0 | 0.1 | GO:0070412 | R-SMAD binding(GO:0070412) |
| 0.0 | 0.2 | GO:0004591 | oxoglutarate dehydrogenase (succinyl-transferring) activity(GO:0004591) |
| 0.0 | 0.1 | GO:0016314 | phosphatidylinositol-3,4,5-trisphosphate 3-phosphatase activity(GO:0016314) |
| 0.0 | 0.6 | GO:0004697 | protein kinase C activity(GO:0004697) calcium-dependent protein kinase C activity(GO:0004698) |
| 0.0 | 0.1 | GO:0016263 | glycoprotein-N-acetylgalactosamine 3-beta-galactosyltransferase activity(GO:0016263) beta-1,3-galactosyltransferase activity(GO:0048531) |
| 0.0 | 0.1 | GO:0004723 | calcium-dependent protein serine/threonine phosphatase activity(GO:0004723) calmodulin-dependent protein phosphatase activity(GO:0033192) |
| 0.0 | 0.2 | GO:0003854 | 3-beta-hydroxy-delta5-steroid dehydrogenase activity(GO:0003854) |
| 0.0 | 0.1 | GO:0042809 | vitamin D receptor binding(GO:0042809) |
| 0.0 | 0.3 | GO:0004711 | ribosomal protein S6 kinase activity(GO:0004711) |
| 0.0 | 0.1 | GO:0015189 | L-lysine transmembrane transporter activity(GO:0015189) |
| 0.0 | 0.1 | GO:0043914 | NADPH:sulfur oxidoreductase activity(GO:0043914) |
| 0.0 | 0.3 | GO:0004535 | poly(A)-specific ribonuclease activity(GO:0004535) |
| 0.0 | 0.1 | GO:0016863 | intramolecular oxidoreductase activity, transposing C=C bonds(GO:0016863) |
| 0.0 | 0.1 | GO:0042975 | peroxisome proliferator activated receptor binding(GO:0042975) |
| 0.0 | 0.1 | GO:0048156 | tau protein binding(GO:0048156) |
| 0.0 | 0.1 | GO:0004719 | protein-L-isoaspartate (D-aspartate) O-methyltransferase activity(GO:0004719) |
| 0.0 | 0.1 | GO:0003977 | UDP-N-acetylglucosamine diphosphorylase activity(GO:0003977) |
| 0.0 | 0.1 | GO:0043531 | ADP binding(GO:0043531) |
| 0.0 | 0.2 | GO:0031729 | CCR1 chemokine receptor binding(GO:0031726) CCR4 chemokine receptor binding(GO:0031729) |
| 0.0 | 0.2 | GO:0004028 | 3-chloroallyl aldehyde dehydrogenase activity(GO:0004028) |
| 0.0 | 0.2 | GO:0016840 | carbon-nitrogen lyase activity(GO:0016840) |
| 0.0 | 0.1 | GO:0004329 | formate-tetrahydrofolate ligase activity(GO:0004329) |
| 0.0 | 0.1 | GO:0010314 | phosphatidylinositol-5-phosphate binding(GO:0010314) |
| 0.0 | 0.1 | GO:0046592 | polyamine oxidase activity(GO:0046592) |
| 0.0 | 0.1 | GO:0008494 | translation activator activity(GO:0008494) |
| 0.0 | 0.2 | GO:0050291 | sphingosine N-acyltransferase activity(GO:0050291) |
| 0.0 | 0.2 | GO:0047676 | arachidonate-CoA ligase activity(GO:0047676) |
| 0.0 | 0.2 | GO:0005332 | gamma-aminobutyric acid:sodium symporter activity(GO:0005332) |
| 0.0 | 0.1 | GO:0035198 | miRNA binding(GO:0035198) |
| 0.0 | 0.1 | GO:0008432 | JUN kinase binding(GO:0008432) |
| 0.0 | 0.3 | GO:0015248 | sterol transporter activity(GO:0015248) |
| 0.0 | 0.1 | GO:0015166 | polyol transmembrane transporter activity(GO:0015166) |
| 0.0 | 0.2 | GO:0005247 | voltage-gated chloride channel activity(GO:0005247) |
| 0.0 | 0.1 | GO:0003989 | acetyl-CoA carboxylase activity(GO:0003989) |
| 0.0 | 0.3 | GO:0097602 | cullin family protein binding(GO:0097602) |
| 0.0 | 0.0 | GO:0004485 | methylcrotonoyl-CoA carboxylase activity(GO:0004485) |
| 0.0 | 0.1 | GO:0016494 | C-X-C chemokine receptor activity(GO:0016494) |
| 0.0 | 0.2 | GO:0047631 | ADP-ribose diphosphatase activity(GO:0047631) |
| 0.0 | 0.1 | GO:0004418 | hydroxymethylbilane synthase activity(GO:0004418) |
| 0.0 | 0.5 | GO:0051959 | dynein light intermediate chain binding(GO:0051959) |
| 0.0 | 0.1 | GO:0030274 | LIM domain binding(GO:0030274) |
| 0.0 | 0.4 | GO:0008195 | phosphatidate phosphatase activity(GO:0008195) |
| 0.0 | 0.1 | GO:0016922 | ligand-dependent nuclear receptor binding(GO:0016922) |
| 0.0 | 0.0 | GO:0031434 | mitogen-activated protein kinase kinase binding(GO:0031434) |
| 0.0 | 0.1 | GO:0031419 | cobalamin binding(GO:0031419) |
| 0.0 | 0.0 | GO:0071568 | UFM1 transferase activity(GO:0071568) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.6 | ST STAT3 PATHWAY | STAT3 Pathway |
| 0.0 | 0.2 | ST IL 13 PATHWAY | Interleukin 13 (IL-13) Pathway |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.6 | REACTOME SIGNAL REGULATORY PROTEIN SIRP FAMILY INTERACTIONS | Genes involved in Signal regulatory protein (SIRP) family interactions |
| 0.1 | 0.5 | REACTOME DIGESTION OF DIETARY CARBOHYDRATE | Genes involved in Digestion of dietary carbohydrate |
| 0.0 | 0.1 | REACTOME TRANSLOCATION OF ZAP 70 TO IMMUNOLOGICAL SYNAPSE | Genes involved in Translocation of ZAP-70 to Immunological synapse |
| 0.0 | 0.4 | REACTOME ACYL CHAIN REMODELLING OF PE | Genes involved in Acyl chain remodelling of PE |
| 0.0 | 0.4 | REACTOME DEADENYLATION OF MRNA | Genes involved in Deadenylation of mRNA |
| 0.0 | 0.1 | REACTOME CLASS C 3 METABOTROPIC GLUTAMATE PHEROMONE RECEPTORS | Genes involved in Class C/3 (Metabotropic glutamate/pheromone receptors) |
| 0.0 | 0.2 | REACTOME METABOLISM OF PORPHYRINS | Genes involved in Metabolism of porphyrins |
| 0.0 | 0.2 | REACTOME PRE NOTCH PROCESSING IN GOLGI | Genes involved in Pre-NOTCH Processing in Golgi |
| 0.0 | 0.1 | REACTOME ACYL CHAIN REMODELLING OF PG | Genes involved in Acyl chain remodelling of PG |
| 0.0 | 0.3 | REACTOME SULFUR AMINO ACID METABOLISM | Genes involved in Sulfur amino acid metabolism |
| 0.0 | 0.2 | REACTOME SIGNALING BY HIPPO | Genes involved in Signaling by Hippo |
| 0.0 | 0.3 | REACTOME MEIOTIC SYNAPSIS | Genes involved in Meiotic Synapsis |
| 0.0 | 0.4 | REACTOME ANTIGEN ACTIVATES B CELL RECEPTOR LEADING TO GENERATION OF SECOND MESSENGERS | Genes involved in Antigen Activates B Cell Receptor Leading to Generation of Second Messengers |