Project
PRJNA195909:zebrafish embryo and larva development
Navigation
Downloads
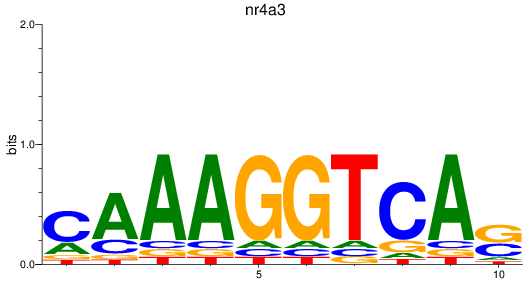
Results for nr4a3
Z-value: 1.15
Transcription factors associated with nr4a3
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
nr4a3
|
ENSDARG00000055854 | nuclear receptor subfamily 4, group A, member 3 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| nr4a3 | dr11_v1_chr16_+_27345383_27345383 | -0.78 | 1.3e-02 | Click! |
Activity profile of nr4a3 motif
Sorted Z-values of nr4a3 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr18_-_40708537 | 2.16 |
ENSDART00000077577
|
si:ch211-132b12.8
|
si:ch211-132b12.8 |
| chr4_-_77116266 | 1.82 |
ENSDART00000174249
|
CU467646.2
|
|
| chr4_-_77135076 | 1.75 |
ENSDART00000174184
|
zgc:173770
|
zgc:173770 |
| chr4_-_77130289 | 1.74 |
ENSDART00000174380
|
CU467646.7
|
|
| chr4_-_77135340 | 1.73 |
ENSDART00000180581
ENSDART00000179901 |
CU467646.7
|
|
| chr8_-_52909850 | 1.66 |
ENSDART00000161943
|
nr6a1a
|
nuclear receptor subfamily 6, group A, member 1a |
| chr3_-_3448095 | 1.55 |
ENSDART00000078886
|
A2ML1 (1 of many)
|
si:dkey-46g23.5 |
| chr5_-_14509137 | 1.53 |
ENSDART00000180742
|
si:ch211-244o22.2
|
si:ch211-244o22.2 |
| chr4_-_77120928 | 1.47 |
ENSDART00000174154
|
CU467646.1
|
|
| chr10_-_26202766 | 1.47 |
ENSDART00000136393
|
fhdc3
|
FH2 domain containing 3 |
| chr3_+_3454610 | 1.42 |
ENSDART00000024900
|
zgc:165453
|
zgc:165453 |
| chr23_+_25172682 | 1.37 |
ENSDART00000191197
ENSDART00000183497 |
si:dkey-151g10.3
|
si:dkey-151g10.3 |
| chr7_-_51775688 | 1.33 |
ENSDART00000149793
|
bmp15
|
bone morphogenetic protein 15 |
| chr1_+_24469313 | 1.31 |
ENSDART00000176581
|
fam160a1a
|
family with sequence similarity 160, member A1a |
| chr19_-_35439237 | 1.28 |
ENSDART00000145883
|
anln
|
anillin, actin binding protein |
| chr10_+_45031398 | 1.27 |
ENSDART00000160536
|
gnsb
|
glucosamine (N-acetyl)-6-sulfatase (Sanfilippo disease IIID), b |
| chr5_-_3839285 | 1.21 |
ENSDART00000122292
|
mlxipl
|
MLX interacting protein like |
| chr5_+_37406358 | 1.17 |
ENSDART00000162811
|
klhl13
|
kelch-like family member 13 |
| chr12_+_46634736 | 1.15 |
ENSDART00000008009
|
trim16
|
tripartite motif containing 16 |
| chr18_+_27515640 | 1.14 |
ENSDART00000181593
|
tp53i11b
|
tumor protein p53 inducible protein 11b |
| chr2_-_17115256 | 1.11 |
ENSDART00000190488
|
pif1
|
PIF1 5'-to-3' DNA helicase homolog (S. cerevisiae) |
| chr5_-_17876709 | 1.10 |
ENSDART00000141978
|
si:dkey-112e17.1
|
si:dkey-112e17.1 |
| chr20_+_18260358 | 1.10 |
ENSDART00000187734
ENSDART00000191947 ENSDART00000057039 |
taf4b
|
TAF4B RNA polymerase II, TATA box binding protein (TBP)-associated factor |
| chr21_+_18405585 | 1.07 |
ENSDART00000139318
|
si:dkey-1d7.3
|
si:dkey-1d7.3 |
| chr1_+_24557414 | 1.03 |
ENSDART00000076519
|
dctpp1
|
dCTP pyrophosphatase 1 |
| chr3_-_5829501 | 1.01 |
ENSDART00000091017
|
pkn1b
|
protein kinase N1b |
| chr5_+_36896933 | 0.98 |
ENSDART00000151984
|
srsf7a
|
serine/arginine-rich splicing factor 7a |
| chr12_-_33357655 | 0.98 |
ENSDART00000066233
ENSDART00000148165 |
slc16a3
|
solute carrier family 16 (monocarboxylate transporter), member 3 |
| chr22_-_38274188 | 0.95 |
ENSDART00000139420
ENSDART00000015117 |
elavl2
|
ELAV like neuron-specific RNA binding protein 2 |
| chr2_+_37897079 | 0.93 |
ENSDART00000141784
|
tep1
|
telomerase-associated protein 1 |
| chr11_-_44999858 | 0.93 |
ENSDART00000167759
ENSDART00000126845 |
ldb1b
|
LIM-domain binding 1b |
| chr9_+_500052 | 0.92 |
ENSDART00000166707
|
CU984600.1
|
|
| chr19_-_42045372 | 0.91 |
ENSDART00000144275
|
trioa
|
trio Rho guanine nucleotide exchange factor a |
| chr4_-_16451375 | 0.90 |
ENSDART00000192700
ENSDART00000128835 |
wu:fc23c09
|
wu:fc23c09 |
| chr19_+_2631565 | 0.87 |
ENSDART00000171487
|
fam126a
|
family with sequence similarity 126, member A |
| chr14_+_30413758 | 0.86 |
ENSDART00000092953
|
cnot7
|
CCR4-NOT transcription complex, subunit 7 |
| chr10_+_36441124 | 0.84 |
ENSDART00000185626
|
uspl1
|
ubiquitin specific peptidase like 1 |
| chr17_-_27200025 | 0.83 |
ENSDART00000192699
|
asap3
|
ArfGAP with SH3 domain, ankyrin repeat and PH domain 3 |
| chr2_-_58075414 | 0.82 |
ENSDART00000161920
|
nectin4
|
nectin cell adhesion molecule 4 |
| chr23_+_39346774 | 0.80 |
ENSDART00000190985
|
src
|
v-src avian sarcoma (Schmidt-Ruppin A-2) viral oncogene homolog |
| chr4_-_9191220 | 0.80 |
ENSDART00000156919
|
hcfc2
|
host cell factor C2 |
| chr18_-_43866526 | 0.79 |
ENSDART00000111309
|
treh
|
trehalase (brush-border membrane glycoprotein) |
| chr23_-_27050083 | 0.78 |
ENSDART00000142324
ENSDART00000133249 ENSDART00000138751 ENSDART00000128718 |
zgc:66440
|
zgc:66440 |
| chr3_+_7771420 | 0.78 |
ENSDART00000156809
ENSDART00000156309 |
hook2
|
hook microtubule-tethering protein 2 |
| chr7_+_34297271 | 0.77 |
ENSDART00000180342
|
bbox1
|
butyrobetaine (gamma), 2-oxoglutarate dioxygenase (gamma-butyrobetaine hydroxylase) 1 |
| chr24_-_26820698 | 0.76 |
ENSDART00000147788
|
fndc3bb
|
fibronectin type III domain containing 3Bb |
| chr2_+_44977889 | 0.76 |
ENSDART00000144024
|
alg3
|
asparagine-linked glycosylation 3 (alpha-1,3-mannosyltransferase) |
| chr21_+_1119046 | 0.75 |
ENSDART00000184678
|
CABZ01088049.1
|
|
| chr16_+_52343905 | 0.75 |
ENSDART00000131051
|
ifnlr1
|
interferon lambda receptor 1 |
| chr1_+_39859782 | 0.74 |
ENSDART00000149984
|
irf2a
|
interferon regulatory factor 2a |
| chr20_+_6535438 | 0.74 |
ENSDART00000145763
|
si:ch211-191a24.4
|
si:ch211-191a24.4 |
| chr3_-_58455289 | 0.74 |
ENSDART00000052179
|
cdr2a
|
cerebellar degeneration-related protein 2a |
| chr23_+_24272421 | 0.73 |
ENSDART00000029974
|
clcnk
|
chloride channel K |
| chr17_+_654759 | 0.73 |
ENSDART00000193703
|
CABZ01079748.1
|
|
| chr17_-_8592824 | 0.73 |
ENSDART00000127022
|
CU462878.1
|
|
| chr8_-_22542467 | 0.73 |
ENSDART00000182588
ENSDART00000134542 |
csde1
|
cold shock domain containing E1, RNA-binding |
| chr1_-_39859626 | 0.73 |
ENSDART00000053763
|
dctd
|
dCMP deaminase |
| chr20_+_6535176 | 0.73 |
ENSDART00000054652
|
si:ch211-191a24.4
|
si:ch211-191a24.4 |
| chr21_-_1640547 | 0.72 |
ENSDART00000151041
|
zgc:152948
|
zgc:152948 |
| chr23_+_39346930 | 0.72 |
ENSDART00000102843
|
src
|
v-src avian sarcoma (Schmidt-Ruppin A-2) viral oncogene homolog |
| chr2_-_31700396 | 0.71 |
ENSDART00000142366
|
lrrcc1
|
leucine rich repeat and coiled-coil centrosomal protein 1 |
| chr6_+_30533504 | 0.71 |
ENSDART00000155842
|
wwc3
|
WWC family member 3 |
| chr1_-_58868306 | 0.71 |
ENSDART00000166615
|
dnm2b
|
dynamin 2b |
| chr5_-_20491198 | 0.70 |
ENSDART00000183051
ENSDART00000144232 |
ficd
|
FIC domain containing |
| chr22_-_7050 | 0.70 |
ENSDART00000127829
|
atad3
|
ATPase family, AAA domain containing 3 |
| chr3_-_21166597 | 0.69 |
ENSDART00000175941
|
taok2a
|
TAO kinase 2a |
| chr3_-_34561624 | 0.67 |
ENSDART00000129313
|
sept9a
|
septin 9a |
| chr24_-_12770357 | 0.67 |
ENSDART00000060826
|
ipo4
|
importin 4 |
| chr7_+_41887429 | 0.67 |
ENSDART00000115090
|
gpt2
|
glutamic pyruvate transaminase (alanine aminotransferase) 2 |
| chr24_+_39034090 | 0.67 |
ENSDART00000185763
|
capn15
|
calpain 15 |
| chr3_-_18189283 | 0.67 |
ENSDART00000049240
|
tob1a
|
transducer of ERBB2, 1a |
| chr14_+_30413312 | 0.67 |
ENSDART00000186864
|
cnot7
|
CCR4-NOT transcription complex, subunit 7 |
| chr3_+_55100045 | 0.66 |
ENSDART00000113098
|
hbaa1
|
hemoglobin, alpha adult 1 |
| chr7_+_34296789 | 0.66 |
ENSDART00000052471
ENSDART00000173798 ENSDART00000173778 |
bbox1
|
butyrobetaine (gamma), 2-oxoglutarate dioxygenase (gamma-butyrobetaine hydroxylase) 1 |
| chr3_+_23047241 | 0.65 |
ENSDART00000103858
|
b4galnt2.2
|
beta-1,4-N-acetyl-galactosaminyl transferase 2, tandem duplicate 2 |
| chr23_-_20154422 | 0.64 |
ENSDART00000132025
|
usp19
|
ubiquitin specific peptidase 19 |
| chr18_+_44769027 | 0.64 |
ENSDART00000145190
|
ilvbl
|
ilvB (bacterial acetolactate synthase)-like |
| chr16_-_30655980 | 0.64 |
ENSDART00000146508
|
ldlrad4b
|
low density lipoprotein receptor class A domain containing 4b |
| chr3_+_35611625 | 0.63 |
ENSDART00000190995
|
traf7
|
TNF receptor-associated factor 7 |
| chr4_-_858434 | 0.62 |
ENSDART00000006961
|
sobpb
|
sine oculis binding protein homolog (Drosophila) b |
| chr6_-_15065376 | 0.61 |
ENSDART00000087797
|
tgfbrap1
|
transforming growth factor, beta receptor associated protein 1 |
| chr7_+_24520518 | 0.61 |
ENSDART00000173604
|
btr09
|
bloodthirsty-related gene family, member 9 |
| chr4_-_47304459 | 0.60 |
ENSDART00000169561
|
si:ch211-261d9.1
|
si:ch211-261d9.1 |
| chr18_+_45571378 | 0.59 |
ENSDART00000077251
|
kifc3
|
kinesin family member C3 |
| chr7_+_61764040 | 0.59 |
ENSDART00000056745
|
acox3
|
acyl-CoA oxidase 3, pristanoyl |
| chr15_+_40188076 | 0.59 |
ENSDART00000063779
|
efhd1
|
EF-hand domain family, member D1 |
| chr19_+_20787179 | 0.58 |
ENSDART00000193204
|
adnp2b
|
ADNP homeobox 2b |
| chr20_-_44575103 | 0.58 |
ENSDART00000192573
|
ubxn2a
|
UBX domain protein 2A |
| chr7_+_65398161 | 0.57 |
ENSDART00000166109
ENSDART00000157399 |
usp47
|
ubiquitin specific peptidase 47 |
| chr9_+_23770666 | 0.57 |
ENSDART00000182493
|
si:ch211-219a4.3
|
si:ch211-219a4.3 |
| chr22_-_24818066 | 0.56 |
ENSDART00000143443
|
vtg6
|
vitellogenin 6 |
| chr22_-_6361178 | 0.55 |
ENSDART00000150147
|
zgc:113298
|
zgc:113298 |
| chr13_-_25719628 | 0.55 |
ENSDART00000135383
|
si:dkey-192p21.6
|
si:dkey-192p21.6 |
| chr25_+_28693451 | 0.54 |
ENSDART00000148366
|
cnot2
|
CCR4-NOT transcription complex, subunit 2 |
| chr6_+_40952031 | 0.54 |
ENSDART00000189219
|
patz1
|
POZ (BTB) and AT hook containing zinc finger 1 |
| chr15_-_16946124 | 0.54 |
ENSDART00000154923
|
hip1
|
huntingtin interacting protein 1 |
| chr7_-_32021853 | 0.54 |
ENSDART00000134521
|
kif18a
|
kinesin family member 18A |
| chr7_+_24496894 | 0.53 |
ENSDART00000149994
|
nelfa
|
negative elongation factor complex member A |
| chr6_-_1587291 | 0.53 |
ENSDART00000067592
ENSDART00000178877 |
zgc:123305
|
zgc:123305 |
| chr9_-_7212973 | 0.53 |
ENSDART00000133638
|
mgat4a
|
mannosyl (alpha-1,3-)-glycoprotein beta-1,4-N-acetylglucosaminyltransferase, isozyme A |
| chr18_+_21273749 | 0.53 |
ENSDART00000143265
|
hydin
|
HYDIN, axonemal central pair apparatus protein |
| chr5_-_13076779 | 0.52 |
ENSDART00000192826
|
ypel1
|
yippee-like 1 |
| chr24_+_35183595 | 0.51 |
ENSDART00000075142
|
pcmtd1
|
protein-L-isoaspartate (D-aspartate) O-methyltransferase domain containing 1 |
| chr15_-_25365570 | 0.51 |
ENSDART00000152754
|
cluha
|
clustered mitochondria (cluA/CLU1) homolog a |
| chr5_+_29831235 | 0.50 |
ENSDART00000109660
|
f11r.1
|
F11 receptor, tandem duplicate 1 |
| chr2_-_4787566 | 0.50 |
ENSDART00000160663
ENSDART00000157808 |
tnk2b
|
tyrosine kinase, non-receptor, 2b |
| chr21_-_43398457 | 0.49 |
ENSDART00000166530
|
ccni2
|
cyclin I family, member 2 |
| chr19_-_1947403 | 0.49 |
ENSDART00000113951
ENSDART00000151293 ENSDART00000134074 |
znrf2a
|
zinc and ring finger 2a |
| chr25_-_10622449 | 0.49 |
ENSDART00000155375
|
ppp6r3
|
protein phosphatase 6, regulatory subunit 3 |
| chr7_+_13988075 | 0.47 |
ENSDART00000186812
|
furina
|
furin (paired basic amino acid cleaving enzyme) a |
| chr2_+_37110504 | 0.47 |
ENSDART00000132794
ENSDART00000042974 |
slc1a8b
|
solute carrier family 1 (glutamate transporter), member 8b |
| chr19_-_47997424 | 0.46 |
ENSDART00000081675
|
ctnnb2
|
catenin, beta 2 |
| chr9_+_23772516 | 0.46 |
ENSDART00000183126
|
si:ch211-219a4.3
|
si:ch211-219a4.3 |
| chr19_+_3056450 | 0.46 |
ENSDART00000141324
ENSDART00000082353 |
hsf1
|
heat shock transcription factor 1 |
| chr1_-_57129179 | 0.45 |
ENSDART00000157226
ENSDART00000152469 |
si:ch73-94k4.2
|
si:ch73-94k4.2 |
| chr7_+_14005111 | 0.45 |
ENSDART00000187365
|
furina
|
furin (paired basic amino acid cleaving enzyme) a |
| chr6_-_35046735 | 0.43 |
ENSDART00000143649
|
uap1
|
UDP-N-acetylglucosamine pyrophosphorylase 1 |
| chr2_+_50722439 | 0.43 |
ENSDART00000188927
|
fyco1b
|
FYVE and coiled-coil domain containing 1b |
| chr23_-_24247672 | 0.42 |
ENSDART00000141552
|
zbtb17
|
zinc finger and BTB domain containing 17 |
| chr2_-_41562868 | 0.42 |
ENSDART00000084597
|
d2hgdh
|
D-2-hydroxyglutarate dehydrogenase |
| chr1_-_53625142 | 0.42 |
ENSDART00000166852
|
USP34
|
ubiquitin specific peptidase 34 |
| chr19_-_79202 | 0.42 |
ENSDART00000166009
|
hnrnpr
|
heterogeneous nuclear ribonucleoprotein R |
| chr15_+_47618221 | 0.41 |
ENSDART00000168722
|
paf1
|
PAF1 homolog, Paf1/RNA polymerase II complex component |
| chr14_+_20941534 | 0.41 |
ENSDART00000185616
|
zgc:66433
|
zgc:66433 |
| chr21_-_25250594 | 0.40 |
ENSDART00000163862
|
nfrkb
|
nuclear factor related to kappaB binding protein |
| chr25_-_9805269 | 0.40 |
ENSDART00000192048
|
lrrc4c
|
leucine rich repeat containing 4C |
| chr4_-_2975461 | 0.40 |
ENSDART00000150794
|
plekha5
|
pleckstrin homology domain containing, family A member 5 |
| chr1_+_45839927 | 0.39 |
ENSDART00000148086
ENSDART00000180413 ENSDART00000048191 ENSDART00000179047 |
map2k7
|
mitogen-activated protein kinase kinase 7 |
| chr15_-_6615555 | 0.39 |
ENSDART00000152725
|
atm
|
ATM serine/threonine kinase |
| chr21_+_10076203 | 0.39 |
ENSDART00000190383
|
CABZ01043506.1
|
|
| chr19_+_2877079 | 0.39 |
ENSDART00000160847
|
hgh1
|
HGH1 homolog (S. cerevisiae) |
| chr20_+_36806398 | 0.39 |
ENSDART00000153317
|
abracl
|
ABRA C-terminal like |
| chr13_-_30143127 | 0.38 |
ENSDART00000146097
|
tysnd1
|
trypsin domain containing 1 |
| chr19_+_5146460 | 0.38 |
ENSDART00000150740
|
si:dkey-89b17.4
|
si:dkey-89b17.4 |
| chr6_-_40044111 | 0.38 |
ENSDART00000154347
|
si:dkey-197j19.5
|
si:dkey-197j19.5 |
| chr13_-_44842884 | 0.38 |
ENSDART00000184195
|
si:dkeyp-2e4.3
|
si:dkeyp-2e4.3 |
| chr8_-_21110233 | 0.37 |
ENSDART00000127371
ENSDART00000100276 |
tmco1
|
transmembrane and coiled-coil domains 1 |
| chr13_-_214122 | 0.37 |
ENSDART00000169273
|
ppp1r21
|
protein phosphatase 1, regulatory subunit 21 |
| chr22_-_31020690 | 0.37 |
ENSDART00000130604
|
ssuh2.4
|
ssu-2 homolog, tandem duplicate 4 |
| chr5_-_32489796 | 0.37 |
ENSDART00000168870
|
gpr107
|
G protein-coupled receptor 107 |
| chr5_-_39698224 | 0.37 |
ENSDART00000076929
|
prkg2
|
protein kinase, cGMP-dependent, type II |
| chr20_+_37794633 | 0.36 |
ENSDART00000022060
|
atf3
|
activating transcription factor 3 |
| chr2_-_56649883 | 0.36 |
ENSDART00000191786
|
gpx4b
|
glutathione peroxidase 4b |
| chr4_-_45337903 | 0.36 |
ENSDART00000159767
|
si:dkey-247i3.6
|
si:dkey-247i3.6 |
| chr21_-_15200556 | 0.36 |
ENSDART00000141809
|
sfswap
|
splicing factor SWAP |
| chr18_-_26510545 | 0.34 |
ENSDART00000135133
|
FRMD5
|
si:ch211-69m14.1 |
| chr25_+_35663933 | 0.34 |
ENSDART00000154979
|
slc2a13b
|
solute carrier family 2 (facilitated glucose transporter), member 13b |
| chr2_+_26060528 | 0.34 |
ENSDART00000058111
|
grin3ba
|
glutamate receptor, ionotropic, N-methyl-D-aspartate 3Ba |
| chr23_-_16737161 | 0.34 |
ENSDART00000132573
|
si:ch211-224l10.4
|
si:ch211-224l10.4 |
| chr22_+_29113796 | 0.34 |
ENSDART00000150264
|
pla2g6
|
phospholipase A2, group VI (cytosolic, calcium-independent) |
| chr8_+_8012570 | 0.34 |
ENSDART00000183429
|
si:ch211-169p10.1
|
si:ch211-169p10.1 |
| chr16_+_25342152 | 0.33 |
ENSDART00000156351
|
zfat
|
zinc finger and AT hook domain containing |
| chr13_-_49666615 | 0.33 |
ENSDART00000148083
|
tomm20a
|
translocase of outer mitochondrial membrane 20 |
| chr16_+_17252487 | 0.33 |
ENSDART00000063572
|
gtf3c6
|
general transcription factor IIIC, polypeptide 6, alpha |
| chr25_+_22017182 | 0.32 |
ENSDART00000156517
|
si:dkey-217l24.1
|
si:dkey-217l24.1 |
| chr25_+_418932 | 0.32 |
ENSDART00000059193
|
prtgb
|
protogenin homolog b (Gallus gallus) |
| chr11_+_45287541 | 0.32 |
ENSDART00000165321
ENSDART00000173116 |
pycr1b
|
pyrroline-5-carboxylate reductase 1b |
| chr12_+_23866368 | 0.32 |
ENSDART00000188652
ENSDART00000192478 |
svila
|
supervillin a |
| chr15_+_32405959 | 0.31 |
ENSDART00000177269
|
si:ch211-162k9.6
|
si:ch211-162k9.6 |
| chr17_-_25303486 | 0.31 |
ENSDART00000162235
|
ppie
|
peptidylprolyl isomerase E (cyclophilin E) |
| chr19_+_11214007 | 0.31 |
ENSDART00000127362
|
si:ch73-109i22.2
|
si:ch73-109i22.2 |
| chr9_-_33477588 | 0.31 |
ENSDART00000144150
|
caska
|
calcium/calmodulin-dependent serine protein kinase a |
| chr5_+_24156170 | 0.31 |
ENSDART00000136570
|
slc25a15b
|
solute carrier family 25 (mitochondrial carrier; ornithine transporter) member 15b |
| chr7_+_35068036 | 0.31 |
ENSDART00000022139
|
zgc:136461
|
zgc:136461 |
| chr9_-_44905867 | 0.31 |
ENSDART00000138316
ENSDART00000131252 ENSDART00000179383 ENSDART00000159337 |
zgc:66484
|
zgc:66484 |
| chr13_-_3879254 | 0.30 |
ENSDART00000166427
ENSDART00000171033 |
si:ch211-128m15.3
|
si:ch211-128m15.3 |
| chr1_+_38153944 | 0.30 |
ENSDART00000135666
|
galnt7
|
UDP-N-acetyl-alpha-D-galactosamine: polypeptide N-acetylgalactosaminyltransferase 7 |
| chr15_+_28268135 | 0.30 |
ENSDART00000152536
ENSDART00000188550 |
myo1cb
|
myosin Ic, paralog b |
| chr7_+_32021982 | 0.30 |
ENSDART00000173848
|
mettl15
|
methyltransferase like 15 |
| chr4_+_77957611 | 0.29 |
ENSDART00000156692
|
arfgap3
|
ADP-ribosylation factor GTPase activating protein 3 |
| chr20_+_37825804 | 0.29 |
ENSDART00000152865
|
tatdn3
|
TatD DNase domain containing 3 |
| chr23_-_27607039 | 0.29 |
ENSDART00000183639
|
phf8
|
PHD finger protein 8 |
| chr12_-_6551681 | 0.28 |
ENSDART00000145413
|
si:ch211-253p2.2
|
si:ch211-253p2.2 |
| chr6_+_52924103 | 0.28 |
ENSDART00000065693
|
or137-2
|
odorant receptor, family H, subfamily 137, member 2 |
| chr4_-_4535189 | 0.28 |
ENSDART00000057519
|
zgc:194209
|
zgc:194209 |
| chr10_+_20364009 | 0.28 |
ENSDART00000186139
ENSDART00000080395 |
golga7
|
golgin A7 |
| chr5_-_8907819 | 0.28 |
ENSDART00000188523
|
adamts12
|
ADAM metallopeptidase with thrombospondin type 1 motif, 12 |
| chr24_-_1021318 | 0.28 |
ENSDART00000181403
|
ralaa
|
v-ral simian leukemia viral oncogene homolog Aa (ras related) |
| chr5_-_38197080 | 0.28 |
ENSDART00000140708
|
si:ch211-284e13.9
|
si:ch211-284e13.9 |
| chr12_+_46791907 | 0.27 |
ENSDART00000110304
|
vcla
|
vinculin a |
| chr17_-_46850299 | 0.27 |
ENSDART00000154430
|
pimr23
|
Pim proto-oncogene, serine/threonine kinase, related 23 |
| chr3_-_18373425 | 0.27 |
ENSDART00000178522
|
spag9a
|
sperm associated antigen 9a |
| chr3_+_31662126 | 0.27 |
ENSDART00000113441
|
mylk5
|
myosin, light chain kinase 5 |
| chr2_-_7246848 | 0.27 |
ENSDART00000146434
|
zgc:153115
|
zgc:153115 |
| chr1_-_55196103 | 0.27 |
ENSDART00000140153
|
mri1
|
methylthioribose-1-phosphate isomerase 1 |
| chr16_-_11859309 | 0.27 |
ENSDART00000145754
|
cxcr3.1
|
chemokine (C-X-C motif) receptor 3, tandem duplicate 1 |
| chr22_-_6923347 | 0.26 |
ENSDART00000082048
|
CABZ01065328.1
|
|
| chr11_-_40647190 | 0.25 |
ENSDART00000173217
ENSDART00000173276 ENSDART00000147264 |
fam213b
|
family with sequence similarity 213, member B |
| chr7_+_29012033 | 0.25 |
ENSDART00000173909
ENSDART00000145762 |
dnaaf1
|
dynein, axonemal, assembly factor 1 |
| chr22_+_15720381 | 0.25 |
ENSDART00000128149
|
fam32a
|
family with sequence similarity 32, member A |
| chr17_-_45164291 | 0.25 |
ENSDART00000062109
|
noxred1
|
NADP-dependent oxidoreductase domain containing 1 |
| chr19_-_17972734 | 0.24 |
ENSDART00000126298
|
ints8
|
integrator complex subunit 8 |
| chr13_-_11578304 | 0.24 |
ENSDART00000171145
|
si:ch211-132e22.4
|
si:ch211-132e22.4 |
| chr18_-_46295132 | 0.24 |
ENSDART00000190940
|
pld3
|
phospholipase D family, member 3 |
| chr25_-_35095129 | 0.24 |
ENSDART00000099866
ENSDART00000099868 |
kif15
|
kinesin family member 15 |
| chr25_-_31898552 | 0.24 |
ENSDART00000156128
|
si:ch73-330k17.3
|
si:ch73-330k17.3 |
| chr22_-_5252005 | 0.23 |
ENSDART00000132942
ENSDART00000081801 |
ncln
|
nicalin |
| chr22_-_14262115 | 0.23 |
ENSDART00000168264
|
si:ch211-246m6.5
|
si:ch211-246m6.5 |
| chr10_-_36223005 | 0.23 |
ENSDART00000159962
|
or109-6
|
odorant receptor, family D, subfamily 109, member 6 |
| chr9_-_37613792 | 0.23 |
ENSDART00000138345
|
sema5ba
|
sema domain, seven thrombospondin repeats (type 1 and type 1-like), transmembrane domain (TM) and short cytoplasmic domain, (semaphorin) 5Ba |
Network of associatons between targets according to the STRING database.
First level regulatory network of nr4a3
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.5 | 1.5 | GO:0050847 | progesterone receptor signaling pathway(GO:0050847) |
| 0.4 | 1.8 | GO:0009219 | pyrimidine deoxyribonucleotide metabolic process(GO:0009219) |
| 0.3 | 1.3 | GO:0060283 | negative regulation of oocyte development(GO:0060283) |
| 0.3 | 1.3 | GO:0008356 | asymmetric cell division(GO:0008356) |
| 0.3 | 0.8 | GO:0005991 | trehalose metabolic process(GO:0005991) |
| 0.2 | 1.4 | GO:0045329 | carnitine biosynthetic process(GO:0045329) |
| 0.2 | 0.8 | GO:1903405 | nuclear body organization(GO:0030575) Cajal body organization(GO:0030576) protein localization to nuclear body(GO:1903405) protein localization to Cajal body(GO:1904867) protein localization to nucleoplasm(GO:1990173) |
| 0.2 | 0.7 | GO:0019276 | UDP-N-acetylgalactosamine metabolic process(GO:0019276) |
| 0.2 | 0.6 | GO:0009097 | isoleucine metabolic process(GO:0006549) isoleucine biosynthetic process(GO:0009097) |
| 0.1 | 0.6 | GO:0033540 | fatty acid beta-oxidation using acyl-CoA oxidase(GO:0033540) |
| 0.1 | 1.5 | GO:0043928 | exonucleolytic nuclear-transcribed mRNA catabolic process involved in deadenylation-dependent decay(GO:0043928) |
| 0.1 | 1.5 | GO:0035306 | positive regulation of dephosphorylation(GO:0035306) positive regulation of protein dephosphorylation(GO:0035307) |
| 0.1 | 0.3 | GO:1990575 | mitochondrial ornithine transport(GO:0000066) mitochondrial L-ornithine transmembrane transport(GO:1990575) |
| 0.1 | 0.3 | GO:0035964 | COPI-coated vesicle budding(GO:0035964) Golgi transport vesicle coating(GO:0048200) COPI coating of Golgi vesicle(GO:0048205) |
| 0.1 | 0.3 | GO:0061187 | regulation of chromatin silencing at rDNA(GO:0061187) negative regulation of chromatin silencing at rDNA(GO:0061188) |
| 0.1 | 1.1 | GO:0000002 | mitochondrial genome maintenance(GO:0000002) |
| 0.1 | 0.6 | GO:0060394 | negative regulation of pathway-restricted SMAD protein phosphorylation(GO:0060394) |
| 0.1 | 0.3 | GO:0019284 | L-methionine biosynthetic process from S-adenosylmethionine(GO:0019284) |
| 0.1 | 1.4 | GO:1902307 | positive regulation of sodium ion transport(GO:0010765) positive regulation of sodium ion transmembrane transport(GO:1902307) regulation of potassium ion import(GO:1903286) positive regulation of potassium ion import(GO:1903288) positive regulation of sodium ion transmembrane transporter activity(GO:2000651) |
| 0.1 | 0.4 | GO:1901098 | positive regulation of autophagosome maturation(GO:1901098) |
| 0.1 | 0.6 | GO:0034058 | endosomal vesicle fusion(GO:0034058) |
| 0.1 | 0.2 | GO:0033615 | mitochondrial proton-transporting ATP synthase complex assembly(GO:0033615) |
| 0.1 | 0.9 | GO:0060971 | embryonic heart tube left/right pattern formation(GO:0060971) |
| 0.1 | 0.4 | GO:0016572 | histone phosphorylation(GO:0016572) |
| 0.1 | 0.6 | GO:0006561 | proline biosynthetic process(GO:0006561) L-proline biosynthetic process(GO:0055129) |
| 0.1 | 0.4 | GO:0000395 | mRNA 5'-splice site recognition(GO:0000395) |
| 0.1 | 0.5 | GO:2000036 | regulation of stem cell population maintenance(GO:2000036) |
| 0.1 | 0.4 | GO:0032469 | endoplasmic reticulum calcium ion homeostasis(GO:0032469) |
| 0.0 | 0.6 | GO:0031468 | nuclear envelope reassembly(GO:0031468) |
| 0.0 | 0.2 | GO:1900145 | nodal signaling pathway involved in determination of left/right asymmetry(GO:0038107) regulation of nodal signaling pathway involved in determination of left/right asymmetry(GO:1900145) |
| 0.0 | 0.3 | GO:0086013 | membrane repolarization during cardiac muscle cell action potential(GO:0086013) |
| 0.0 | 0.5 | GO:0048312 | intracellular distribution of mitochondria(GO:0048312) |
| 0.0 | 0.5 | GO:0034244 | negative regulation of DNA-templated transcription, elongation(GO:0032785) negative regulation of transcription elongation from RNA polymerase II promoter(GO:0034244) |
| 0.0 | 0.1 | GO:0071470 | cellular response to osmotic stress(GO:0071470) |
| 0.0 | 0.2 | GO:0007060 | male meiosis chromosome segregation(GO:0007060) |
| 0.0 | 0.4 | GO:0031998 | regulation of fatty acid beta-oxidation(GO:0031998) |
| 0.0 | 0.5 | GO:0050892 | intestinal absorption(GO:0050892) |
| 0.0 | 0.2 | GO:0050911 | detection of chemical stimulus involved in sensory perception of smell(GO:0050911) |
| 0.0 | 0.7 | GO:0015671 | oxygen transport(GO:0015671) |
| 0.0 | 0.7 | GO:0043507 | activation of JUN kinase activity(GO:0007257) positive regulation of JUN kinase activity(GO:0043507) |
| 0.0 | 0.3 | GO:0070475 | rRNA base methylation(GO:0070475) |
| 0.0 | 0.1 | GO:1903817 | negative regulation of potassium ion transmembrane transporter activity(GO:1901017) negative regulation of potassium ion transmembrane transport(GO:1901380) negative regulation of delayed rectifier potassium channel activity(GO:1902260) negative regulation of voltage-gated potassium channel activity(GO:1903817) |
| 0.0 | 0.6 | GO:0021551 | central nervous system morphogenesis(GO:0021551) |
| 0.0 | 0.3 | GO:1900407 | regulation of cellular response to oxidative stress(GO:1900407) |
| 0.0 | 0.4 | GO:0032968 | positive regulation of transcription elongation from RNA polymerase II promoter(GO:0032968) |
| 0.0 | 0.7 | GO:0055064 | chloride ion homeostasis(GO:0055064) |
| 0.0 | 1.8 | GO:0030917 | midbrain-hindbrain boundary development(GO:0030917) |
| 0.0 | 0.2 | GO:0070816 | phosphorylation of RNA polymerase II C-terminal domain(GO:0070816) |
| 0.0 | 0.7 | GO:0072310 | glomerular visceral epithelial cell development(GO:0072015) glomerular epithelial cell development(GO:0072310) |
| 0.0 | 0.2 | GO:0050975 | sensory perception of touch(GO:0050975) |
| 0.0 | 0.2 | GO:0034720 | histone H3-K4 demethylation(GO:0034720) |
| 0.0 | 0.3 | GO:0044458 | motile cilium assembly(GO:0044458) |
| 0.0 | 1.1 | GO:0006367 | transcription initiation from RNA polymerase II promoter(GO:0006367) |
| 0.0 | 0.4 | GO:2000404 | regulation of T cell migration(GO:2000404) |
| 0.0 | 0.5 | GO:0043666 | regulation of phosphoprotein phosphatase activity(GO:0043666) |
| 0.0 | 0.6 | GO:0043249 | erythrocyte maturation(GO:0043249) |
| 0.0 | 0.5 | GO:0009408 | response to heat(GO:0009408) |
| 0.0 | 0.3 | GO:0035235 | ionotropic glutamate receptor signaling pathway(GO:0035235) |
| 0.0 | 0.5 | GO:0048263 | determination of dorsal identity(GO:0048263) |
| 0.0 | 0.1 | GO:0009450 | gamma-aminobutyric acid catabolic process(GO:0009450) |
| 0.0 | 0.6 | GO:0032355 | response to estradiol(GO:0032355) |
| 0.0 | 0.5 | GO:0048268 | clathrin coat assembly(GO:0048268) |
| 0.0 | 0.3 | GO:0043001 | Golgi to plasma membrane protein transport(GO:0043001) |
| 0.0 | 0.1 | GO:0016191 | synaptic vesicle uncoating(GO:0016191) |
| 0.0 | 0.5 | GO:0007019 | microtubule depolymerization(GO:0007019) |
| 0.0 | 1.3 | GO:0030203 | glycosaminoglycan metabolic process(GO:0030203) |
| 0.0 | 0.1 | GO:0060049 | regulation of protein glycosylation(GO:0060049) |
| 0.0 | 0.1 | GO:0045920 | negative regulation of exocytosis(GO:0045920) |
| 0.0 | 0.2 | GO:0006048 | UDP-N-acetylglucosamine biosynthetic process(GO:0006048) |
| 0.0 | 1.2 | GO:0006487 | protein N-linked glycosylation(GO:0006487) |
| 0.0 | 1.0 | GO:0015718 | monocarboxylic acid transport(GO:0015718) |
| 0.0 | 0.6 | GO:0031647 | regulation of protein stability(GO:0031647) |
| 0.0 | 0.8 | GO:0031122 | cytoplasmic microtubule organization(GO:0031122) |
| 0.0 | 0.0 | GO:0002637 | regulation of immunoglobulin production(GO:0002637) positive regulation of immunoglobulin production(GO:0002639) |
| 0.0 | 0.6 | GO:0009062 | fatty acid catabolic process(GO:0009062) |
| 0.0 | 0.2 | GO:0034472 | snRNA 3'-end processing(GO:0034472) |
| 0.0 | 0.2 | GO:0060999 | positive regulation of dendritic spine development(GO:0060999) |
| 0.0 | 0.3 | GO:0006383 | transcription from RNA polymerase III promoter(GO:0006383) |
| 0.0 | 0.3 | GO:0000413 | protein peptidyl-prolyl isomerization(GO:0000413) |
| 0.0 | 0.1 | GO:2001287 | caveolin-mediated endocytosis(GO:0072584) negative regulation of clathrin-mediated endocytosis(GO:1900186) regulation of caveolin-mediated endocytosis(GO:2001286) negative regulation of caveolin-mediated endocytosis(GO:2001287) |
| 0.0 | 0.3 | GO:0051014 | actin filament severing(GO:0051014) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.6 | GO:0005948 | acetolactate synthase complex(GO:0005948) |
| 0.2 | 1.3 | GO:0005826 | actomyosin contractile ring(GO:0005826) |
| 0.2 | 2.1 | GO:0030015 | CCR4-NOT core complex(GO:0030015) |
| 0.1 | 0.5 | GO:0098890 | extrinsic component of postsynaptic membrane(GO:0098890) |
| 0.1 | 0.5 | GO:0061673 | mitotic spindle astral microtubule(GO:0061673) |
| 0.1 | 0.4 | GO:0035327 | transcriptionally active chromatin(GO:0035327) |
| 0.1 | 0.5 | GO:0032021 | NELF complex(GO:0032021) |
| 0.1 | 0.7 | GO:0031838 | haptoglobin-hemoglobin complex(GO:0031838) |
| 0.1 | 1.1 | GO:0005669 | transcription factor TFIID complex(GO:0005669) |
| 0.1 | 0.3 | GO:0000127 | transcription factor TFIIIC complex(GO:0000127) |
| 0.0 | 1.0 | GO:0071339 | MLL1/2 complex(GO:0044665) MLL1 complex(GO:0071339) |
| 0.0 | 0.8 | GO:0015030 | Cajal body(GO:0015030) |
| 0.0 | 0.4 | GO:0033202 | Ino80 complex(GO:0031011) DNA helicase complex(GO:0033202) |
| 0.0 | 0.2 | GO:0000439 | core TFIIH complex(GO:0000439) |
| 0.0 | 0.3 | GO:0005742 | mitochondrial outer membrane translocase complex(GO:0005742) |
| 0.0 | 0.2 | GO:0000974 | Prp19 complex(GO:0000974) |
| 0.0 | 1.6 | GO:0031228 | integral component of Golgi membrane(GO:0030173) intrinsic component of Golgi membrane(GO:0031228) |
| 0.0 | 1.1 | GO:0005657 | replication fork(GO:0005657) |
| 0.0 | 0.1 | GO:0097519 | DNA recombinase complex(GO:0097519) |
| 0.0 | 0.3 | GO:0002178 | palmitoyltransferase complex(GO:0002178) |
| 0.0 | 0.7 | GO:0032156 | septin ring(GO:0005940) septin complex(GO:0031105) septin cytoskeleton(GO:0032156) |
| 0.0 | 2.1 | GO:0031234 | extrinsic component of cytoplasmic side of plasma membrane(GO:0031234) |
| 0.0 | 0.2 | GO:0032039 | integrator complex(GO:0032039) |
| 0.0 | 0.5 | GO:0000307 | cyclin-dependent protein kinase holoenzyme complex(GO:0000307) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.5 | 1.4 | GO:0008336 | gamma-butyrobetaine dioxygenase activity(GO:0008336) |
| 0.3 | 0.8 | GO:0015927 | alpha,alpha-trehalase activity(GO:0004555) trehalase activity(GO:0015927) |
| 0.2 | 1.1 | GO:0043139 | 5'-3' DNA helicase activity(GO:0043139) |
| 0.2 | 0.6 | GO:0003984 | acetolactate synthase activity(GO:0003984) |
| 0.2 | 1.3 | GO:0070698 | type I activin receptor binding(GO:0070698) |
| 0.2 | 0.8 | GO:0000033 | alpha-1,3-mannosyltransferase activity(GO:0000033) |
| 0.2 | 1.0 | GO:0047429 | nucleoside-triphosphate diphosphatase activity(GO:0047429) |
| 0.2 | 0.8 | GO:0032183 | SUMO binding(GO:0032183) |
| 0.1 | 0.9 | GO:0030274 | LIM domain binding(GO:0030274) |
| 0.1 | 0.6 | GO:0070412 | R-SMAD binding(GO:0070412) |
| 0.1 | 0.6 | GO:0003997 | acyl-CoA oxidase activity(GO:0003997) |
| 0.1 | 0.6 | GO:0004735 | pyrroline-5-carboxylate reductase activity(GO:0004735) |
| 0.1 | 1.5 | GO:0004535 | poly(A)-specific ribonuclease activity(GO:0004535) |
| 0.1 | 0.3 | GO:0016434 | rRNA (cytosine) methyltransferase activity(GO:0016434) |
| 0.1 | 0.4 | GO:0003977 | UDP-N-acetylglucosamine diphosphorylase activity(GO:0003977) |
| 0.1 | 1.4 | GO:0019870 | chloride channel inhibitor activity(GO:0019869) potassium channel inhibitor activity(GO:0019870) |
| 0.1 | 0.5 | GO:0004719 | protein-L-isoaspartate (D-aspartate) O-methyltransferase activity(GO:0004719) |
| 0.1 | 1.0 | GO:0015129 | lactate transmembrane transporter activity(GO:0015129) |
| 0.1 | 0.6 | GO:0045735 | nutrient reservoir activity(GO:0045735) |
| 0.1 | 0.4 | GO:0008545 | JUN kinase kinase activity(GO:0008545) |
| 0.1 | 0.3 | GO:0035575 | histone demethylase activity (H4-K20 specific)(GO:0035575) |
| 0.1 | 0.3 | GO:0016494 | C-X-C chemokine receptor activity(GO:0016494) |
| 0.1 | 0.6 | GO:0008569 | ATP-dependent microtubule motor activity, minus-end-directed(GO:0008569) |
| 0.1 | 0.3 | GO:0046592 | polyamine oxidase activity(GO:0046592) |
| 0.1 | 0.4 | GO:0004692 | cGMP-dependent protein kinase activity(GO:0004692) |
| 0.1 | 0.7 | GO:0031720 | haptoglobin binding(GO:0031720) |
| 0.1 | 1.1 | GO:0016251 | obsolete general RNA polymerase II transcription factor activity(GO:0016251) |
| 0.1 | 0.3 | GO:0008432 | JUN kinase binding(GO:0008432) |
| 0.1 | 0.5 | GO:0043325 | phosphatidylinositol-3,4-bisphosphate binding(GO:0043325) |
| 0.1 | 0.5 | GO:0045294 | alpha-catenin binding(GO:0045294) |
| 0.0 | 0.4 | GO:0010314 | phosphatidylinositol-5-phosphate binding(GO:0010314) |
| 0.0 | 0.3 | GO:0055056 | D-glucose transmembrane transporter activity(GO:0055056) |
| 0.0 | 0.2 | GO:0002151 | G-quadruplex RNA binding(GO:0002151) |
| 0.0 | 0.1 | GO:0050135 | NAD(P)+ nucleosidase activity(GO:0050135) |
| 0.0 | 0.7 | GO:0008449 | N-acetylglucosamine-6-sulfatase activity(GO:0008449) |
| 0.0 | 0.7 | GO:0005247 | voltage-gated chloride channel activity(GO:0005247) |
| 0.0 | 0.7 | GO:0061608 | nuclear import signal receptor activity(GO:0061608) |
| 0.0 | 0.7 | GO:0070566 | adenylyltransferase activity(GO:0070566) |
| 0.0 | 1.0 | GO:0004698 | protein kinase C activity(GO:0004697) calcium-dependent protein kinase C activity(GO:0004698) |
| 0.0 | 0.3 | GO:0000064 | L-ornithine transmembrane transporter activity(GO:0000064) |
| 0.0 | 0.2 | GO:0005549 | olfactory receptor activity(GO:0004984) odorant binding(GO:0005549) |
| 0.0 | 0.4 | GO:0002020 | protease binding(GO:0002020) |
| 0.0 | 0.1 | GO:1990238 | double-stranded DNA endodeoxyribonuclease activity(GO:1990238) |
| 0.0 | 0.2 | GO:0016314 | phosphatidylinositol-3,4,5-trisphosphate 3-phosphatase activity(GO:0016314) |
| 0.0 | 0.3 | GO:0016840 | carbon-nitrogen lyase activity(GO:0016840) |
| 0.0 | 1.9 | GO:0004715 | non-membrane spanning protein tyrosine kinase activity(GO:0004715) |
| 0.0 | 0.2 | GO:0072345 | NAADP-sensitive calcium-release channel activity(GO:0072345) |
| 0.0 | 0.2 | GO:0034648 | histone demethylase activity (H3-dimethyl-K4 specific)(GO:0034648) |
| 0.0 | 0.7 | GO:0004198 | calcium-dependent cysteine-type endopeptidase activity(GO:0004198) |
| 0.0 | 0.3 | GO:0004687 | myosin light chain kinase activity(GO:0004687) |
| 0.0 | 0.3 | GO:0016018 | cyclosporin A binding(GO:0016018) |
| 0.0 | 0.3 | GO:0016861 | intramolecular oxidoreductase activity, interconverting aldoses and ketoses(GO:0016861) |
| 0.0 | 0.4 | GO:0004972 | NMDA glutamate receptor activity(GO:0004972) |
| 0.0 | 0.3 | GO:0070840 | dynein complex binding(GO:0070840) |
| 0.0 | 0.7 | GO:0019239 | deaminase activity(GO:0019239) |
| 0.0 | 0.7 | GO:0051959 | dynein light intermediate chain binding(GO:0051959) |
| 0.0 | 0.4 | GO:0000993 | RNA polymerase II core binding(GO:0000993) |
| 0.0 | 0.5 | GO:1990939 | ATP-dependent microtubule motor activity, plus-end-directed(GO:0008574) ATP-dependent microtubule motor activity(GO:1990939) |
| 0.0 | 0.7 | GO:0008376 | acetylgalactosaminyltransferase activity(GO:0008376) |
| 0.0 | 1.8 | GO:0042803 | protein homodimerization activity(GO:0042803) |
| 0.0 | 0.2 | GO:0005220 | inositol 1,4,5-trisphosphate-sensitive calcium-release channel activity(GO:0005220) |
| 0.0 | 0.8 | GO:0030170 | pyridoxal phosphate binding(GO:0030170) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 1.5 | ST STAT3 PATHWAY | STAT3 Pathway |
| 0.0 | 0.4 | PID ANTHRAX PATHWAY | Cellular roles of Anthrax toxin |
| 0.0 | 0.6 | PID SMAD2 3PATHWAY | Regulation of cytoplasmic and nuclear SMAD2/3 signaling |
| 0.0 | 1.2 | PID AURORA B PATHWAY | Aurora B signaling |
| 0.0 | 1.2 | PID MYC REPRESS PATHWAY | Validated targets of C-MYC transcriptional repression |
| 0.0 | 1.3 | PID TELOMERASE PATHWAY | Regulation of Telomerase |
| 0.0 | 1.2 | PID AR PATHWAY | Coregulation of Androgen receptor activity |
| 0.0 | 1.1 | PID MYC ACTIV PATHWAY | Validated targets of C-MYC transcriptional activation |
| 0.0 | 0.6 | PID ECADHERIN STABILIZATION PATHWAY | Stabilization and expansion of the E-cadherin adherens junction |
| 0.0 | 0.2 | ST GA12 PATHWAY | G alpha 12 Pathway |
| 0.0 | 1.8 | NABA SECRETED FACTORS | Genes encoding secreted soluble factors |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 1.5 | REACTOME SIGNAL REGULATORY PROTEIN SIRP FAMILY INTERACTIONS | Genes involved in Signal regulatory protein (SIRP) family interactions |
| 0.1 | 0.8 | REACTOME DIGESTION OF DIETARY CARBOHYDRATE | Genes involved in Digestion of dietary carbohydrate |
| 0.1 | 2.1 | REACTOME DEADENYLATION OF MRNA | Genes involved in Deadenylation of mRNA |
| 0.1 | 0.7 | REACTOME AMINO ACID SYNTHESIS AND INTERCONVERSION TRANSAMINATION | Genes involved in Amino acid synthesis and interconversion (transamination) |
| 0.1 | 0.8 | REACTOME BILE SALT AND ORGANIC ANION SLC TRANSPORTERS | Genes involved in Bile salt and organic anion SLC transporters |
| 0.0 | 0.4 | REACTOME REGULATION OF THE FANCONI ANEMIA PATHWAY | Genes involved in Regulation of the Fanconi anemia pathway |
| 0.0 | 0.8 | REACTOME KINESINS | Genes involved in Kinesins |
| 0.0 | 0.7 | REACTOME PYRIMIDINE METABOLISM | Genes involved in Pyrimidine metabolism |
| 0.0 | 0.3 | REACTOME ACYL CHAIN REMODELLING OF PE | Genes involved in Acyl chain remodelling of PE |
| 0.0 | 1.3 | REACTOME RNA POL II TRANSCRIPTION PRE INITIATION AND PROMOTER OPENING | Genes involved in RNA Polymerase II Transcription Pre-Initiation And Promoter Opening |
| 0.0 | 0.6 | REACTOME PEROXISOMAL LIPID METABOLISM | Genes involved in Peroxisomal lipid metabolism |
| 0.0 | 0.4 | REACTOME CGMP EFFECTS | Genes involved in cGMP effects |
| 0.0 | 0.5 | REACTOME N GLYCAN ANTENNAE ELONGATION | Genes involved in N-Glycan antennae elongation |
| 0.0 | 0.3 | REACTOME METABOLISM OF POLYAMINES | Genes involved in Metabolism of polyamines |
| 0.0 | 0.4 | REACTOME ACTIVATION OF GENES BY ATF4 | Genes involved in Activation of Genes by ATF4 |
| 0.0 | 0.4 | REACTOME JNK C JUN KINASES PHOSPHORYLATION AND ACTIVATION MEDIATED BY ACTIVATED HUMAN TAK1 | Genes involved in JNK (c-Jun kinases) phosphorylation and activation mediated by activated human TAK1 |
| 0.0 | 0.5 | REACTOME BIOSYNTHESIS OF THE N GLYCAN PRECURSOR DOLICHOL LIPID LINKED OLIGOSACCHARIDE LLO AND TRANSFER TO A NASCENT PROTEIN | Genes involved in Biosynthesis of the N-glycan precursor (dolichol lipid-linked oligosaccharide, LLO) and transfer to a nascent protein |
| 0.0 | 0.6 | REACTOME ASSOCIATION OF TRIC CCT WITH TARGET PROTEINS DURING BIOSYNTHESIS | Genes involved in Association of TriC/CCT with target proteins during biosynthesis |
| 0.0 | 0.4 | REACTOME PYRUVATE METABOLISM AND CITRIC ACID TCA CYCLE | Genes involved in Pyruvate metabolism and Citric Acid (TCA) cycle |
| 0.0 | 1.2 | REACTOME INTEGRATION OF ENERGY METABOLISM | Genes involved in Integration of energy metabolism |