Project
PRJNA195909:zebrafish embryo and larva development
Navigation
Downloads
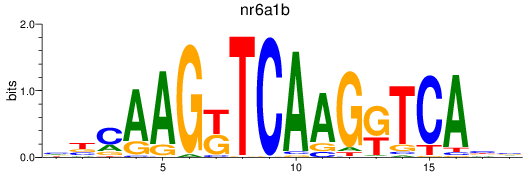
Results for nr6a1b
Z-value: 1.07
Transcription factors associated with nr6a1b
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
nr6a1b
|
ENSDARG00000014480 | nuclear receptor subfamily 6, group A, member 1b |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| nr6a1b | dr11_v1_chr21_+_8239544_8239544 | -0.16 | 6.7e-01 | Click! |
Activity profile of nr6a1b motif
Sorted Z-values of nr6a1b motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr2_-_37210397 | 1.53 |
ENSDART00000084938
|
apoda.1
|
apolipoprotein Da, duplicate 1 |
| chr3_-_7656059 | 1.34 |
ENSDART00000170917
|
junbb
|
JunB proto-oncogene, AP-1 transcription factor subunit b |
| chr16_-_45235947 | 1.22 |
ENSDART00000164436
|
si:dkey-33i11.4
|
si:dkey-33i11.4 |
| chr4_-_17409533 | 1.21 |
ENSDART00000011943
|
pah
|
phenylalanine hydroxylase |
| chr3_+_13469704 | 1.17 |
ENSDART00000166806
|
zgc:77748
|
zgc:77748 |
| chr19_-_5135345 | 1.17 |
ENSDART00000151787
|
gnb3b
|
guanine nucleotide binding protein (G protein), beta polypeptide 3b |
| chr1_-_55785722 | 1.16 |
ENSDART00000142069
ENSDART00000043933 |
ndufb7
|
NADH dehydrogenase (ubiquinone) 1 beta subcomplex, 7 |
| chr20_+_10539293 | 1.16 |
ENSDART00000182265
|
serpina1l
|
serine (or cysteine) proteinase inhibitor, clade A (alpha-1 antiproteinase, antitrypsin), member 1, like |
| chr12_-_30583668 | 1.07 |
ENSDART00000153406
|
casp7
|
caspase 7, apoptosis-related cysteine peptidase |
| chr5_-_28606916 | 1.01 |
ENSDART00000026107
ENSDART00000137717 |
tnc
|
tenascin C |
| chr6_+_39836474 | 0.93 |
ENSDART00000112637
|
smarcc2
|
SWI/SNF related, matrix associated, actin dependent regulator of chromatin, subfamily c, member 2 |
| chr14_-_5678457 | 0.92 |
ENSDART00000012116
|
tlx2
|
T cell leukemia homeobox 2 |
| chr9_-_1970071 | 0.90 |
ENSDART00000080608
|
hoxd10a
|
homeobox D10a |
| chr16_+_23811554 | 0.87 |
ENSDART00000114336
|
si:dkey-7f3.9
|
si:dkey-7f3.9 |
| chr20_+_4392687 | 0.87 |
ENSDART00000187271
|
im:7142702
|
im:7142702 |
| chr10_-_17284055 | 0.86 |
ENSDART00000167464
|
GNAZ
|
G protein subunit alpha z |
| chr15_+_19682013 | 0.86 |
ENSDART00000127368
|
si:dkey-4p15.5
|
si:dkey-4p15.5 |
| chr13_-_21688176 | 0.85 |
ENSDART00000063825
|
sprn
|
shadow of prion protein |
| chr25_-_225964 | 0.80 |
ENSDART00000193424
|
CABZ01113818.1
|
|
| chr7_-_15370042 | 0.74 |
ENSDART00000128183
|
ANPEP
|
si:ch211-276a23.5 |
| chr23_-_19953089 | 0.73 |
ENSDART00000153828
|
atp2b3b
|
ATPase plasma membrane Ca2+ transporting 3b |
| chr17_+_18117358 | 0.72 |
ENSDART00000144894
|
bcl11ba
|
B cell CLL/lymphoma 11Ba |
| chr25_-_210730 | 0.72 |
ENSDART00000187580
|
FP236318.1
|
|
| chr17_+_18117029 | 0.71 |
ENSDART00000154646
ENSDART00000179739 |
bcl11ba
|
B cell CLL/lymphoma 11Ba |
| chr20_-_53435483 | 0.69 |
ENSDART00000135091
|
mettl24
|
methyltransferase like 24 |
| chr23_+_23119008 | 0.69 |
ENSDART00000132418
|
samd11
|
sterile alpha motif domain containing 11 |
| chr6_+_41099787 | 0.68 |
ENSDART00000186884
|
fkbp5
|
FK506 binding protein 5 |
| chr21_+_43669943 | 0.65 |
ENSDART00000136025
|
tmlhe
|
trimethyllysine hydroxylase, epsilon |
| chr10_+_37927100 | 0.63 |
ENSDART00000172548
|
bhlha9
|
basic helix-loop-helix family, member a9 |
| chr19_+_30662529 | 0.62 |
ENSDART00000175662
|
fam49al
|
family with sequence similarity 49, member A-like |
| chr13_+_23157053 | 0.61 |
ENSDART00000162359
|
sorbs1
|
sorbin and SH3 domain containing 1 |
| chr21_-_43040780 | 0.61 |
ENSDART00000187037
|
jakmip2
|
janus kinase and microtubule interacting protein 2 |
| chr25_+_33939728 | 0.59 |
ENSDART00000148537
|
roraa
|
RAR-related orphan receptor A, paralog a |
| chr6_+_40671336 | 0.59 |
ENSDART00000111639
ENSDART00000186617 |
rereb
|
arginine-glutamic acid dipeptide (RE) repeats b |
| chr3_+_39540014 | 0.55 |
ENSDART00000074848
|
zgc:165423
|
zgc:165423 |
| chr7_-_31618166 | 0.55 |
ENSDART00000111388
|
igdcc3
|
immunoglobulin superfamily, DCC subclass, member 3 |
| chr7_-_20453661 | 0.55 |
ENSDART00000174001
|
ntn5
|
netrin 5 |
| chr6_+_48618512 | 0.55 |
ENSDART00000111190
|
FAM19A3
|
si:dkey-238f9.1 |
| chr25_+_14092871 | 0.55 |
ENSDART00000067239
|
guca1g
|
guanylate cyclase activator 1g |
| chr4_+_7827261 | 0.52 |
ENSDART00000129568
|
phyh
|
phytanoyl-CoA 2-hydroxylase |
| chr15_+_19681718 | 0.51 |
ENSDART00000164803
|
si:dkey-4p15.5
|
si:dkey-4p15.5 |
| chr18_+_8340886 | 0.50 |
ENSDART00000081132
|
cpt1b
|
carnitine palmitoyltransferase 1B (muscle) |
| chr17_-_37054959 | 0.47 |
ENSDART00000151921
|
dnmt3ab
|
DNA (cytosine-5-)-methyltransferase 3 alpha b |
| chr10_-_22057001 | 0.47 |
ENSDART00000016575
|
tlx3b
|
T cell leukemia homeobox 3b |
| chr6_+_39506043 | 0.45 |
ENSDART00000086260
|
CR388364.1
|
|
| chr8_-_14091886 | 0.43 |
ENSDART00000137857
|
si:ch211-229n2.7
|
si:ch211-229n2.7 |
| chr3_+_19299309 | 0.41 |
ENSDART00000046297
ENSDART00000146955 |
ldlra
|
low density lipoprotein receptor a |
| chr13_-_22843562 | 0.41 |
ENSDART00000142738
|
pbld
|
phenazine biosynthesis like protein domain containing |
| chr8_-_23578660 | 0.40 |
ENSDART00000039080
|
wasb
|
Wiskott-Aldrich syndrome (eczema-thrombocytopenia) b |
| chr10_+_35439952 | 0.39 |
ENSDART00000006284
|
hhla2a.2
|
HERV-H LTR-associating 2a, tandem duplicate 2 |
| chr23_-_10175898 | 0.39 |
ENSDART00000146185
|
krt5
|
keratin 5 |
| chr5_-_50992690 | 0.36 |
ENSDART00000149553
ENSDART00000097460 ENSDART00000192021 |
hmgcra
|
3-hydroxy-3-methylglutaryl-CoA reductase a |
| chr6_-_30932078 | 0.33 |
ENSDART00000028612
|
pde4ba
|
phosphodiesterase 4B, cAMP-specific a |
| chr5_+_20693724 | 0.31 |
ENSDART00000141368
|
si:ch211-240b21.2
|
si:ch211-240b21.2 |
| chr5_+_1079423 | 0.31 |
ENSDART00000172231
|
si:zfos-128g4.2
|
si:zfos-128g4.2 |
| chr25_+_10830269 | 0.30 |
ENSDART00000175736
|
si:ch211-147g22.5
|
si:ch211-147g22.5 |
| chr1_-_46632948 | 0.30 |
ENSDART00000148893
ENSDART00000053232 |
cdadc1
|
cytidine and dCMP deaminase domain containing 1 |
| chr19_+_20724347 | 0.29 |
ENSDART00000090757
|
kat2b
|
K(lysine) acetyltransferase 2B |
| chr21_-_5879897 | 0.27 |
ENSDART00000184034
|
rpl35
|
ribosomal protein L35 |
| chr6_+_2030703 | 0.27 |
ENSDART00000109679
ENSDART00000187502 ENSDART00000191165 ENSDART00000187544 |
quo
|
quattro |
| chr25_+_7492663 | 0.26 |
ENSDART00000166496
|
cat
|
catalase |
| chr15_+_46386261 | 0.26 |
ENSDART00000191793
|
igsf11
|
immunoglobulin superfamily member 11 |
| chr4_+_57192681 | 0.26 |
ENSDART00000190438
|
si:dkey-16p6.4
|
si:dkey-16p6.4 |
| chr12_+_387850 | 0.25 |
ENSDART00000182326
|
gsg1l2b
|
gsg1-like 2b |
| chr11_+_45287541 | 0.23 |
ENSDART00000165321
ENSDART00000173116 |
pycr1b
|
pyrroline-5-carboxylate reductase 1b |
| chr10_+_29260096 | 0.23 |
ENSDART00000088973
|
sytl2a
|
synaptotagmin-like 2a |
| chr23_-_19230627 | 0.23 |
ENSDART00000007122
|
guca1b
|
guanylate cyclase activator 1B |
| chr4_-_43616331 | 0.23 |
ENSDART00000183211
ENSDART00000150240 |
si:dkey-16p6.4
|
si:dkey-16p6.4 |
| chr2_+_38924975 | 0.19 |
ENSDART00000109219
|
rem2
|
RAS (RAD and GEM)-like GTP binding 2 |
| chr15_+_16886196 | 0.19 |
ENSDART00000139296
ENSDART00000049196 |
gdpd1
|
glycerophosphodiester phosphodiesterase domain containing 1 |
| chr11_-_17964525 | 0.19 |
ENSDART00000018948
|
cishb
|
cytokine inducible SH2-containing protein b |
| chr8_+_22472584 | 0.17 |
ENSDART00000138303
|
si:dkey-23c22.9
|
si:dkey-23c22.9 |
| chr7_+_35268054 | 0.16 |
ENSDART00000113842
|
dpep2
|
dipeptidase 2 |
| chr20_+_13175379 | 0.16 |
ENSDART00000025644
|
ppp2r5a
|
protein phosphatase 2, regulatory subunit B', alpha isoform |
| chr16_+_52751445 | 0.14 |
ENSDART00000191807
|
atp6v1c1a
|
ATPase H+ transporting V1 subunit C1a |
| chr4_+_69712223 | 0.14 |
ENSDART00000163035
ENSDART00000164475 |
si:dkey-238o14.1
zgc:174944
|
si:dkey-238o14.1 zgc:174944 |
| chr12_-_17024079 | 0.13 |
ENSDART00000129027
|
ifit11
|
interferon-induced protein with tetratricopeptide repeats 11 |
| chr8_+_18044257 | 0.13 |
ENSDART00000029255
|
glis1b
|
GLIS family zinc finger 1b |
| chr9_+_32301456 | 0.13 |
ENSDART00000078608
ENSDART00000185153 ENSDART00000144947 |
hspe1
|
heat shock 10 protein 1 |
| chr7_-_1918971 | 0.12 |
ENSDART00000181759
|
CABZ01040626.1
|
|
| chr9_+_24095677 | 0.11 |
ENSDART00000150443
|
lrrfip1a
|
leucine rich repeat (in FLII) interacting protein 1a |
| chr25_-_21609645 | 0.11 |
ENSDART00000087817
|
DOCK4 (1 of many)
|
si:dkey-81e3.1 |
| chr8_+_22405477 | 0.10 |
ENSDART00000148267
|
si:dkey-23c22.7
|
si:dkey-23c22.7 |
| chr4_-_63166758 | 0.10 |
ENSDART00000192765
|
CR450780.1
|
|
| chr3_-_40976288 | 0.08 |
ENSDART00000193553
|
cyp3c1
|
cytochrome P450, family 3, subfamily c, polypeptide 1 |
| chr15_+_44133972 | 0.08 |
ENSDART00000157846
ENSDART00000191082 |
CU655961.2
|
|
| chr7_-_30639385 | 0.08 |
ENSDART00000173618
|
myo1ea
|
myosin IE, a |
| chr17_+_33311784 | 0.07 |
ENSDART00000156229
|
si:ch211-132f19.7
|
si:ch211-132f19.7 |
| chr22_-_17781213 | 0.07 |
ENSDART00000137984
|
si:ch73-63e15.2
|
si:ch73-63e15.2 |
| chr21_+_11401247 | 0.06 |
ENSDART00000143952
|
cel.1
|
carboxyl ester lipase, tandem duplicate 1 |
| chr9_+_42270043 | 0.06 |
ENSDART00000137435
|
si:dkey-10c21.1
|
si:dkey-10c21.1 |
| chr4_-_70263609 | 0.05 |
ENSDART00000163868
|
si:dkey-3h2.3
|
si:dkey-3h2.3 |
| chr4_-_47096170 | 0.05 |
ENSDART00000161079
|
si:dkey-26m3.3
|
si:dkey-26m3.3 |
| chr22_+_10215558 | 0.04 |
ENSDART00000063274
|
kctd6a
|
potassium channel tetramerization domain containing 6a |
| chr4_-_63559311 | 0.04 |
ENSDART00000164415
|
si:dkey-16p6.1
|
si:dkey-16p6.1 |
| chr11_-_5563498 | 0.03 |
ENSDART00000160835
|
pex11g
|
peroxisomal biogenesis factor 11 gamma |
| chr4_+_58668661 | 0.03 |
ENSDART00000188815
|
CR388148.2
|
|
| chr4_+_76973771 | 0.02 |
ENSDART00000142978
|
si:dkey-240n22.3
|
si:dkey-240n22.3 |
| chr2_+_52232630 | 0.02 |
ENSDART00000006216
|
plpp2a
|
phospholipid phosphatase 2a |
| chr4_-_71267461 | 0.02 |
ENSDART00000161219
ENSDART00000151838 |
si:dkeyp-51g9.4
|
si:dkeyp-51g9.4 |
| chr3_-_39627412 | 0.01 |
ENSDART00000123292
|
si:dkey-27o4.1
|
si:dkey-27o4.1 |
| chr4_-_34775762 | 0.01 |
ENSDART00000167262
|
si:dkey-146m20.14
|
si:dkey-146m20.14 |
| chr4_-_47095583 | 0.01 |
ENSDART00000183595
ENSDART00000183279 ENSDART00000191638 |
si:dkey-26m3.3
|
si:dkey-26m3.3 |
| chr1_-_55118745 | 0.01 |
ENSDART00000133915
|
sertad2a
|
SERTA domain containing 2a |
| chr23_+_17102960 | 0.00 |
ENSDART00000053414
|
commd7
|
COMM domain containing 7 |
| chr12_+_46605600 | 0.00 |
ENSDART00000123533
|
FADS6
|
fatty acid desaturase 6 |
| chr1_-_49947290 | 0.00 |
ENSDART00000141476
|
sgms2
|
sphingomyelin synthase 2 |
Network of associatons between targets according to the STRING database.
First level regulatory network of nr6a1b
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 1.0 | GO:0008582 | regulation of synaptic growth at neuromuscular junction(GO:0008582) axon regeneration at neuromuscular junction(GO:0014814) positive regulation of synaptic growth at neuromuscular junction(GO:0045887) |
| 0.2 | 0.2 | GO:0070291 | N-acylethanolamine metabolic process(GO:0070291) |
| 0.1 | 1.2 | GO:0006558 | L-phenylalanine metabolic process(GO:0006558) L-phenylalanine catabolic process(GO:0006559) erythrose 4-phosphate/phosphoenolpyruvate family amino acid metabolic process(GO:1902221) erythrose 4-phosphate/phosphoenolpyruvate family amino acid catabolic process(GO:1902222) |
| 0.1 | 0.3 | GO:0090069 | regulation of ribosome biogenesis(GO:0090069) |
| 0.1 | 0.7 | GO:0045329 | carnitine biosynthetic process(GO:0045329) |
| 0.1 | 0.4 | GO:0070292 | N-acylphosphatidylethanolamine metabolic process(GO:0070292) |
| 0.1 | 0.5 | GO:0001561 | fatty acid alpha-oxidation(GO:0001561) |
| 0.1 | 0.4 | GO:0032367 | intracellular cholesterol transport(GO:0032367) |
| 0.0 | 1.5 | GO:0007568 | aging(GO:0007568) |
| 0.0 | 0.5 | GO:0009437 | carnitine metabolic process(GO:0009437) |
| 0.0 | 1.1 | GO:0097194 | execution phase of apoptosis(GO:0097194) |
| 0.0 | 0.3 | GO:0031295 | lymphocyte costimulation(GO:0031294) T cell costimulation(GO:0031295) |
| 0.0 | 0.4 | GO:0051127 | positive regulation of actin nucleation(GO:0051127) positive regulation of Arp2/3 complex-mediated actin nucleation(GO:2000601) |
| 0.0 | 0.7 | GO:0000413 | protein peptidyl-prolyl isomerization(GO:0000413) |
| 0.0 | 0.3 | GO:0006198 | cAMP catabolic process(GO:0006198) |
| 0.0 | 0.3 | GO:0000463 | maturation of LSU-rRNA from tricistronic rRNA transcript (SSU-rRNA, 5.8S rRNA, LSU-rRNA)(GO:0000463) |
| 0.0 | 0.9 | GO:0043044 | ATP-dependent chromatin remodeling(GO:0043044) |
| 0.0 | 1.7 | GO:0031101 | fin regeneration(GO:0031101) |
| 0.0 | 1.2 | GO:0010951 | negative regulation of endopeptidase activity(GO:0010951) |
| 0.0 | 0.5 | GO:0006306 | DNA alkylation(GO:0006305) DNA methylation(GO:0006306) |
| 0.0 | 0.3 | GO:0042542 | response to hydrogen peroxide(GO:0042542) response to copper ion(GO:0046688) |
| 0.0 | 0.3 | GO:0097324 | melanocyte migration(GO:0097324) |
| 0.0 | 0.4 | GO:0021587 | cerebellum morphogenesis(GO:0021587) |
| 0.0 | 0.3 | GO:0043171 | peptide catabolic process(GO:0043171) |
| 0.0 | 0.4 | GO:0006695 | cholesterol biosynthetic process(GO:0006695) secondary alcohol biosynthetic process(GO:1902653) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.9 | GO:0071564 | npBAF complex(GO:0071564) |
| 0.0 | 0.4 | GO:0045095 | keratin filament(GO:0045095) |
| 0.0 | 1.2 | GO:0031970 | mitochondrial intermembrane space(GO:0005758) organelle envelope lumen(GO:0031970) |
| 0.0 | 0.1 | GO:0000221 | vacuolar proton-transporting V-type ATPase, V1 domain(GO:0000221) |
| 0.0 | 1.0 | GO:0031225 | anchored component of membrane(GO:0031225) |
| 0.0 | 0.8 | GO:0005834 | heterotrimeric G-protein complex(GO:0005834) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.7 | GO:0005527 | macrolide binding(GO:0005527) FK506 binding(GO:0005528) |
| 0.1 | 0.4 | GO:0008970 | phosphatidylcholine 1-acylhydrolase activity(GO:0008970) |
| 0.1 | 1.2 | GO:0016714 | oxidoreductase activity, acting on paired donors, with incorporation or reduction of molecular oxygen, reduced pteridine as one donor, and incorporation of one atom of oxygen(GO:0016714) |
| 0.1 | 0.4 | GO:0030228 | lipoprotein particle receptor activity(GO:0030228) |
| 0.1 | 0.8 | GO:0008048 | calcium sensitive guanylate cyclase activator activity(GO:0008048) |
| 0.1 | 1.2 | GO:0031682 | G-protein gamma-subunit binding(GO:0031682) |
| 0.1 | 0.5 | GO:0016416 | carnitine O-palmitoyltransferase activity(GO:0004095) O-palmitoyltransferase activity(GO:0016416) |
| 0.1 | 1.1 | GO:0097200 | cysteine-type endopeptidase activity involved in execution phase of apoptosis(GO:0097200) |
| 0.1 | 1.2 | GO:0008137 | NADH dehydrogenase (ubiquinone) activity(GO:0008137) NADH dehydrogenase (quinone) activity(GO:0050136) |
| 0.0 | 0.2 | GO:0004735 | pyrroline-5-carboxylate reductase activity(GO:0004735) |
| 0.0 | 0.3 | GO:0004126 | cytidine deaminase activity(GO:0004126) |
| 0.0 | 0.5 | GO:0003886 | DNA (cytosine-5-)-methyltransferase activity(GO:0003886) |
| 0.0 | 0.3 | GO:0004145 | diamine N-acetyltransferase activity(GO:0004145) |
| 0.0 | 0.2 | GO:0070573 | metallodipeptidase activity(GO:0070573) |
| 0.0 | 0.3 | GO:0004096 | catalase activity(GO:0004096) |
| 0.0 | 0.7 | GO:0005388 | calcium-transporting ATPase activity(GO:0005388) |
| 0.0 | 0.1 | GO:0004771 | sterol esterase activity(GO:0004771) |
| 0.0 | 1.2 | GO:0016706 | oxidoreductase activity, acting on paired donors, with incorporation or reduction of molecular oxygen, 2-oxoglutarate as one donor, and incorporation of one atom each of oxygen into both donors(GO:0016706) |
| 0.0 | 0.3 | GO:0004115 | 3',5'-cyclic-AMP phosphodiesterase activity(GO:0004115) |
| 0.0 | 0.2 | GO:0042043 | neurexin family protein binding(GO:0042043) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.9 | PID ALK2 PATHWAY | ALK2 signaling events |
| 0.1 | 1.0 | PID INTEGRIN A9B1 PATHWAY | Alpha9 beta1 integrin signaling events |
| 0.1 | 1.1 | SA CASPASE CASCADE | Apoptosis is mediated by caspases, cysteine proteases arranged in a proteolytic cascade. |
| 0.0 | 0.5 | PID S1P S1P4 PATHWAY | S1P4 pathway |
| 0.0 | 1.8 | PID REG GR PATHWAY | Glucocorticoid receptor regulatory network |
| 0.0 | 0.3 | PID RETINOIC ACID PATHWAY | Retinoic acid receptors-mediated signaling |
| 0.0 | 0.2 | PID CONE PATHWAY | Visual signal transduction: Cones |
| 0.0 | 0.6 | SIG INSULIN RECEPTOR PATHWAY IN CARDIAC MYOCYTES | Genes related to the insulin receptor pathway |
| 0.0 | 0.5 | PID HNF3B PATHWAY | FOXA2 and FOXA3 transcription factor networks |
| 0.0 | 0.7 | PID CMYB PATHWAY | C-MYB transcription factor network |
| 0.0 | 0.5 | NABA BASEMENT MEMBRANES | Genes encoding structural components of basement membranes |
| 0.0 | 0.3 | PID FOXO PATHWAY | FoxO family signaling |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 1.1 | REACTOME CASPASE MEDIATED CLEAVAGE OF CYTOSKELETAL PROTEINS | Genes involved in Caspase-mediated cleavage of cytoskeletal proteins |
| 0.1 | 0.3 | REACTOME PURINE CATABOLISM | Genes involved in Purine catabolism |
| 0.0 | 0.5 | REACTOME ACTIVATED AMPK STIMULATES FATTY ACID OXIDATION IN MUSCLE | Genes involved in Activated AMPK stimulates fatty-acid oxidation in muscle |
| 0.0 | 0.6 | REACTOME SMOOTH MUSCLE CONTRACTION | Genes involved in Smooth Muscle Contraction |
| 0.0 | 0.3 | REACTOME NOTCH HLH TRANSCRIPTION PATHWAY | Genes involved in Notch-HLH transcription pathway |
| 0.0 | 0.5 | REACTOME PEROXISOMAL LIPID METABOLISM | Genes involved in Peroxisomal lipid metabolism |
| 0.0 | 0.5 | REACTOME G PROTEIN ACTIVATION | Genes involved in G-protein activation |
| 0.0 | 1.2 | REACTOME RESPIRATORY ELECTRON TRANSPORT | Genes involved in Respiratory electron transport |
| 0.0 | 0.2 | REACTOME CTLA4 INHIBITORY SIGNALING | Genes involved in CTLA4 inhibitory signaling |
| 0.0 | 1.0 | REACTOME INTEGRIN CELL SURFACE INTERACTIONS | Genes involved in Integrin cell surface interactions |