Project
PRJNA195909:zebrafish embryo and larva development
Navigation
Downloads
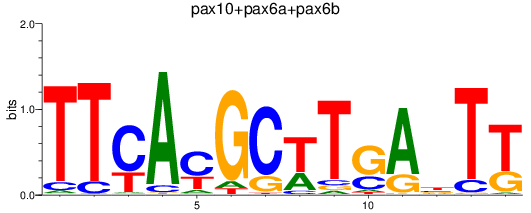
Results for pax10+pax6a+pax6b
Z-value: 0.60
Transcription factors associated with pax10+pax6a+pax6b
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
pax6b
|
ENSDARG00000045936 | paired box 6b |
|
pax10
|
ENSDARG00000053364 | paired box 10 |
|
pax6a
|
ENSDARG00000103379 | paired box 6a |
|
pax10
|
ENSDARG00000111379 | paired box 10 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| pax6b | dr11_v1_chr7_+_15871156_15871156 | -0.97 | 2.0e-05 | Click! |
| pax6a | dr11_v1_chr25_-_15049694_15049781 | -0.79 | 1.2e-02 | Click! |
| pax10 | dr11_v1_chr3_+_32553714_32553714 | -0.60 | 8.6e-02 | Click! |
Activity profile of pax10+pax6a+pax6b motif
Sorted Z-values of pax10+pax6a+pax6b motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr21_-_2322102 | 1.43 |
ENSDART00000162867
|
zgc:66483
|
zgc:66483 |
| chr8_+_52442622 | 1.40 |
ENSDART00000012758
|
zgc:77112
|
zgc:77112 |
| chr25_+_22320738 | 1.33 |
ENSDART00000073566
|
cyp11a1
|
cytochrome P450, family 11, subfamily A, polypeptide 1 |
| chr5_+_57924611 | 1.23 |
ENSDART00000050949
|
btg4
|
B-cell translocation gene 4 |
| chr18_+_14277003 | 1.16 |
ENSDART00000006628
|
zgc:173742
|
zgc:173742 |
| chr2_+_6255434 | 1.10 |
ENSDART00000139429
|
zp3b
|
zona pellucida glycoprotein 3b |
| chr12_-_35582683 | 0.91 |
ENSDART00000167933
|
sec24c
|
SEC24 homolog C, COPII coat complex component |
| chr8_+_52442785 | 0.90 |
ENSDART00000189958
|
zgc:77112
|
zgc:77112 |
| chr23_+_17926279 | 0.89 |
ENSDART00000012540
|
chia.4
|
chitinase, acidic.4 |
| chr21_-_22117085 | 0.81 |
ENSDART00000146673
|
slc35f2
|
solute carrier family 35, member F2 |
| chr8_+_41038141 | 0.80 |
ENSDART00000075620
|
gpat2
|
glycerol-3-phosphate acyltransferase 2, mitochondrial |
| chr11_-_40257225 | 0.80 |
ENSDART00000139009
|
TRIM62 (1 of many)
|
si:ch211-193i15.2 |
| chr24_+_26379441 | 0.78 |
ENSDART00000137786
|
si:ch211-230g15.5
|
si:ch211-230g15.5 |
| chr16_+_39159752 | 0.77 |
ENSDART00000122081
|
sybu
|
syntabulin (syntaxin-interacting) |
| chr9_-_2936017 | 0.74 |
ENSDART00000102823
|
zak
|
sterile alpha motif and leucine zipper containing kinase AZK |
| chr10_-_8046764 | 0.69 |
ENSDART00000099031
|
zgc:136254
|
zgc:136254 |
| chr8_+_41037541 | 0.69 |
ENSDART00000129344
|
gpat2
|
glycerol-3-phosphate acyltransferase 2, mitochondrial |
| chr10_-_8053385 | 0.68 |
ENSDART00000142714
|
si:ch211-251f6.7
|
si:ch211-251f6.7 |
| chr12_+_47794089 | 0.67 |
ENSDART00000160726
|
polr3a
|
polymerase (RNA) III (DNA directed) polypeptide A |
| chr13_-_32726178 | 0.66 |
ENSDART00000012232
|
pdss2
|
prenyl (decaprenyl) diphosphate synthase, subunit 2 |
| chr7_-_45852270 | 0.66 |
ENSDART00000170224
|
shcbp1
|
SHC SH2-domain binding protein 1 |
| chr5_+_40837539 | 0.65 |
ENSDART00000188279
|
si:dkey-3h3.3
|
si:dkey-3h3.3 |
| chr7_-_20582842 | 0.65 |
ENSDART00000169750
ENSDART00000111719 |
si:dkey-19b23.11
|
si:dkey-19b23.11 |
| chr24_+_40905100 | 0.64 |
ENSDART00000167854
|
scn12ab
|
sodium channel, voltage gated, type XII, alpha b |
| chr5_-_26247973 | 0.64 |
ENSDART00000098527
|
erap1b
|
endoplasmic reticulum aminopeptidase 1b |
| chr3_+_43086548 | 0.64 |
ENSDART00000163579
|
si:dkey-43p13.5
|
si:dkey-43p13.5 |
| chr21_-_3700334 | 0.63 |
ENSDART00000137844
|
atp8b1
|
ATPase phospholipid transporting 8B1 |
| chr23_-_40194732 | 0.63 |
ENSDART00000164931
|
tgm1l2
|
transglutaminase 1 like 2 |
| chr18_+_36782930 | 0.62 |
ENSDART00000004129
|
si:ch211-160d20.3
|
si:ch211-160d20.3 |
| chr5_+_1109098 | 0.58 |
ENSDART00000166268
|
LO017790.1
|
|
| chr19_-_4793263 | 0.57 |
ENSDART00000147510
ENSDART00000141336 ENSDART00000110551 ENSDART00000146684 |
st3gal1
|
ST3 beta-galactoside alpha-2,3-sialyltransferase 1 |
| chr14_-_14659023 | 0.57 |
ENSDART00000170355
ENSDART00000159888 ENSDART00000172241 |
nsdhl
|
NAD(P) dependent steroid dehydrogenase-like |
| chr10_-_8053753 | 0.56 |
ENSDART00000162289
|
si:ch211-251f6.7
|
si:ch211-251f6.7 |
| chr16_+_11779534 | 0.55 |
ENSDART00000133497
|
si:dkey-250k15.4
|
si:dkey-250k15.4 |
| chr12_-_35582521 | 0.53 |
ENSDART00000162175
ENSDART00000168958 |
sec24c
|
SEC24 homolog C, COPII coat complex component |
| chr1_-_29653472 | 0.52 |
ENSDART00000109224
|
si:dkey-1h24.2
|
si:dkey-1h24.2 |
| chr24_-_2381143 | 0.52 |
ENSDART00000144307
|
rreb1a
|
ras responsive element binding protein 1a |
| chr9_+_8365398 | 0.52 |
ENSDART00000138713
ENSDART00000136847 |
si:dkey-90l23.2
|
si:dkey-90l23.2 |
| chr2_+_25657958 | 0.52 |
ENSDART00000161407
|
tnika
|
TRAF2 and NCK interacting kinase a |
| chr25_+_19870603 | 0.52 |
ENSDART00000047251
|
gramd4b
|
GRAM domain containing 4b |
| chr13_-_4018888 | 0.51 |
ENSDART00000058238
|
tjap1
|
tight junction associated protein 1 (peripheral) |
| chr11_-_8126223 | 0.50 |
ENSDART00000091617
ENSDART00000192391 ENSDART00000101561 |
ttll7
|
tubulin tyrosine ligase-like family, member 7 |
| chr16_+_11779761 | 0.49 |
ENSDART00000140297
|
si:dkey-250k15.4
|
si:dkey-250k15.4 |
| chr3_-_34717882 | 0.49 |
ENSDART00000151127
|
thraa
|
thyroid hormone receptor alpha a |
| chr7_-_8738827 | 0.49 |
ENSDART00000172807
ENSDART00000173026 |
si:ch211-1o7.3
|
si:ch211-1o7.3 |
| chr3_+_17346502 | 0.48 |
ENSDART00000187786
ENSDART00000131584 |
si:ch211-210g13.5
|
si:ch211-210g13.5 |
| chr6_+_33076839 | 0.47 |
ENSDART00000073755
ENSDART00000122242 |
pomgnt1
|
protein O-linked mannose N-acetylglucosaminyltransferase 1 (beta 1,2-) |
| chr7_-_41512999 | 0.46 |
ENSDART00000173577
|
si:dkey-10f23.2
|
si:dkey-10f23.2 |
| chr23_-_24542156 | 0.46 |
ENSDART00000132265
|
atp13a2
|
ATPase 13A2 |
| chr5_-_69523816 | 0.46 |
ENSDART00000112692
|
si:ch211-157p22.10
|
si:ch211-157p22.10 |
| chr4_-_13613148 | 0.45 |
ENSDART00000067164
ENSDART00000111247 |
irf5
|
interferon regulatory factor 5 |
| chr3_-_54607166 | 0.44 |
ENSDART00000021977
|
dnmt1
|
DNA (cytosine-5-)-methyltransferase 1 |
| chr15_-_1001177 | 0.44 |
ENSDART00000160730
|
zgc:162936
|
zgc:162936 |
| chr7_-_30624435 | 0.44 |
ENSDART00000173828
|
rnf111
|
ring finger protein 111 |
| chr5_-_33039670 | 0.43 |
ENSDART00000141361
|
glipr2
|
GLI pathogenesis-related 2 |
| chr14_-_45967981 | 0.43 |
ENSDART00000188062
|
macrod1
|
MACRO domain containing 1 |
| chr7_-_1101071 | 0.42 |
ENSDART00000176053
|
dctn1a
|
dynactin 1a |
| chr10_-_14920989 | 0.42 |
ENSDART00000184617
|
smad2
|
SMAD family member 2 |
| chr5_+_41477954 | 0.41 |
ENSDART00000185871
|
pias2
|
protein inhibitor of activated STAT, 2 |
| chr20_-_47348116 | 0.41 |
ENSDART00000162087
ENSDART00000160769 ENSDART00000164484 |
dtnba
|
dystrobrevin, beta a |
| chr23_+_38957738 | 0.40 |
ENSDART00000193480
|
ATP9A
|
ATPase phospholipid transporting 9A (putative) |
| chr15_+_39977461 | 0.40 |
ENSDART00000063786
|
cab39
|
calcium binding protein 39 |
| chr10_+_20392083 | 0.40 |
ENSDART00000166867
|
r3hcc1
|
R3H domain and coiled-coil containing 1 |
| chr5_+_32141790 | 0.40 |
ENSDART00000041504
|
tescb
|
tescalcin b |
| chr2_+_46032678 | 0.40 |
ENSDART00000184382
ENSDART00000125971 |
gpc1b
|
glypican 1b |
| chr9_-_53537989 | 0.39 |
ENSDART00000114022
|
slitrk5b
|
SLIT and NTRK-like family, member 5b |
| chr5_-_47975440 | 0.39 |
ENSDART00000145665
ENSDART00000007057 |
ccnh
|
cyclin H |
| chr17_+_43926523 | 0.39 |
ENSDART00000121550
ENSDART00000041447 |
ktn1
|
kinectin 1 |
| chr10_-_19497914 | 0.39 |
ENSDART00000132084
|
si:ch211-127i16.2
|
si:ch211-127i16.2 |
| chr3_+_25914064 | 0.39 |
ENSDART00000162558
ENSDART00000166843 |
tom1
|
target of myb1 membrane trafficking protein |
| chr23_-_27505825 | 0.38 |
ENSDART00000137229
ENSDART00000013797 |
asb8
|
ankyrin repeat and SOCS box containing 8 |
| chr16_+_48460873 | 0.38 |
ENSDART00000159902
|
ext1a
|
exostosin glycosyltransferase 1a |
| chr5_-_28016805 | 0.38 |
ENSDART00000078642
|
vps37b
|
vacuolar protein sorting 37 homolog B (S. cerevisiae) |
| chr15_-_43238220 | 0.38 |
ENSDART00000027019
|
wdfy1
|
WD repeat and FYVE domain containing 1 |
| chr21_-_32781612 | 0.38 |
ENSDART00000031028
|
cnot6a
|
CCR4-NOT transcription complex, subunit 6a |
| chr20_+_51833030 | 0.38 |
ENSDART00000074330
|
taf1a
|
TATA box binding protein (TBP)-associated factor, RNA polymerase I, A |
| chr17_-_15029258 | 0.37 |
ENSDART00000168332
ENSDART00000012877 ENSDART00000109190 |
ero1a
|
endoplasmic reticulum oxidoreductase alpha |
| chr20_+_4222357 | 0.37 |
ENSDART00000188331
|
ipcef1
|
interaction protein for cytohesin exchange factors 1 |
| chr15_-_25319663 | 0.37 |
ENSDART00000042218
|
pafah1b1a
|
platelet-activating factor acetylhydrolase 1b, regulatory subunit 1a |
| chr20_-_3238110 | 0.37 |
ENSDART00000008077
|
spint1b
|
serine peptidase inhibitor, Kunitz type 1 b |
| chr7_+_26545502 | 0.37 |
ENSDART00000140528
|
tnk1
|
tyrosine kinase, non-receptor, 1 |
| chr3_-_8542577 | 0.37 |
ENSDART00000183527
|
rbfox3b
|
RNA binding fox-1 homolog 3b |
| chr18_-_20466061 | 0.37 |
ENSDART00000060311
|
paqr5a
|
progestin and adipoQ receptor family member Va |
| chr14_-_30918662 | 0.37 |
ENSDART00000176631
|
si:ch211-126c2.4
|
si:ch211-126c2.4 |
| chr11_-_44931962 | 0.36 |
ENSDART00000170345
|
pfklb
|
phosphofructokinase, liver b |
| chr23_-_15916316 | 0.36 |
ENSDART00000134096
ENSDART00000042469 ENSDART00000146605 |
mrgbp
|
MRG/MORF4L binding protein |
| chr8_-_8489886 | 0.36 |
ENSDART00000183334
|
abt1
|
activator of basal transcription 1 |
| chr14_+_31529958 | 0.36 |
ENSDART00000053026
|
fam122b
|
family with sequence similarity 122B |
| chr4_-_5370468 | 0.36 |
ENSDART00000133423
ENSDART00000184963 |
si:dkey-14d8.1
|
si:dkey-14d8.1 |
| chr8_-_43456025 | 0.36 |
ENSDART00000001092
ENSDART00000140618 |
ncor2
|
nuclear receptor corepressor 2 |
| chr17_-_41798856 | 0.36 |
ENSDART00000156031
ENSDART00000192801 ENSDART00000180172 ENSDART00000084745 ENSDART00000175577 |
ralgapa2
|
Ral GTPase activating protein, alpha subunit 2 (catalytic) |
| chr16_+_21918503 | 0.36 |
ENSDART00000167919
|
znf687a
|
zinc finger protein 687a |
| chr7_+_26545911 | 0.36 |
ENSDART00000135313
|
tnk1
|
tyrosine kinase, non-receptor, 1 |
| chr23_+_38957472 | 0.35 |
ENSDART00000193836
|
ATP9A
|
ATPase phospholipid transporting 9A (putative) |
| chr4_+_76466751 | 0.35 |
ENSDART00000164709
|
zgc:153116
|
zgc:153116 |
| chr3_-_26184018 | 0.34 |
ENSDART00000191604
|
si:ch211-11k18.4
|
si:ch211-11k18.4 |
| chr9_-_746317 | 0.34 |
ENSDART00000129632
ENSDART00000130720 |
usp37
|
ubiquitin specific peptidase 37 |
| chr25_+_7532811 | 0.34 |
ENSDART00000161593
|
ptdss2
|
phosphatidylserine synthase 2 |
| chr12_-_3077395 | 0.34 |
ENSDART00000002867
ENSDART00000126315 |
rfng
|
RFNG O-fucosylpeptide 3-beta-N-acetylglucosaminyltransferase |
| chr7_-_58244220 | 0.34 |
ENSDART00000180450
|
unm_hu7910
|
un-named hu7910 |
| chr19_-_15192840 | 0.33 |
ENSDART00000151337
|
phactr4a
|
phosphatase and actin regulator 4a |
| chr6_+_27923054 | 0.33 |
ENSDART00000136833
ENSDART00000145533 |
cep63
|
centrosomal protein 63 |
| chr17_+_17804752 | 0.33 |
ENSDART00000123350
|
sptlc2a
|
serine palmitoyltransferase, long chain base subunit 2a |
| chr7_+_24729558 | 0.33 |
ENSDART00000111542
ENSDART00000170100 |
shroom4
|
shroom family member 4 |
| chr20_+_27712714 | 0.33 |
ENSDART00000008306
|
zbtb1
|
zinc finger and BTB domain containing 1 |
| chr5_-_55914268 | 0.33 |
ENSDART00000014049
|
wdr36
|
WD repeat domain 36 |
| chr19_-_25114701 | 0.33 |
ENSDART00000149035
|
ptp4a3
|
protein tyrosine phosphatase type IVA, member 3 |
| chr8_-_8489685 | 0.33 |
ENSDART00000131849
ENSDART00000064113 |
abt1
|
activator of basal transcription 1 |
| chr3_-_36152974 | 0.32 |
ENSDART00000002568
|
engase
|
endo-beta-N-acetylglucosaminidase |
| chr19_-_42503143 | 0.32 |
ENSDART00000007642
|
zgc:110239
|
zgc:110239 |
| chr18_-_45761868 | 0.32 |
ENSDART00000025423
|
cstf3
|
cleavage stimulation factor, 3' pre-RNA, subunit 3 |
| chr9_-_31915423 | 0.32 |
ENSDART00000060051
|
fgf14
|
fibroblast growth factor 14 |
| chr19_+_34311374 | 0.32 |
ENSDART00000086617
|
gabbr2
|
gamma-aminobutyric acid (GABA) B receptor, 2 |
| chr11_-_11336986 | 0.32 |
ENSDART00000016677
|
zgc:77929
|
zgc:77929 |
| chr16_+_13993285 | 0.32 |
ENSDART00000139130
ENSDART00000130353 |
si:dkey-85k15.7
fdps
|
si:dkey-85k15.7 farnesyl diphosphate synthase (farnesyl pyrophosphate synthetase, dimethylallyltranstransferase, geranyltranstransferase) |
| chr12_+_23812530 | 0.32 |
ENSDART00000066331
|
svila
|
supervillin a |
| chr15_+_19990068 | 0.31 |
ENSDART00000154033
ENSDART00000054428 |
zgc:112083
|
zgc:112083 |
| chr5_-_69316142 | 0.31 |
ENSDART00000157238
ENSDART00000144570 |
smtnb
|
smoothelin b |
| chr8_-_28357177 | 0.31 |
ENSDART00000182319
|
KLHL12 (1 of many)
|
kelch like family member 12 |
| chr24_-_1010467 | 0.31 |
ENSDART00000114544
|
cdk13
|
cyclin-dependent kinase 13 |
| chr11_-_4023940 | 0.30 |
ENSDART00000058728
ENSDART00000171245 |
nek4
|
NIMA-related kinase 4 |
| chr9_-_50000144 | 0.30 |
ENSDART00000123416
|
scn1a
|
sodium channel, voltage-gated, type I, alpha |
| chr11_+_2902592 | 0.30 |
ENSDART00000093108
|
kif21b
|
kinesin family member 21B |
| chr13_-_31164749 | 0.30 |
ENSDART00000049180
|
mapk8a
|
mitogen-activated protein kinase 8a |
| chr25_-_37284370 | 0.30 |
ENSDART00000103222
|
nudt7
|
nudix (nucleoside diphosphate linked moiety X)-type motif 7 |
| chr4_+_5341592 | 0.29 |
ENSDART00000123375
ENSDART00000067371 |
zgc:113263
|
zgc:113263 |
| chr18_-_16315822 | 0.29 |
ENSDART00000136479
|
rassf9
|
Ras association (RalGDS/AF-6) domain family (N-terminal) member 9 |
| chr1_+_2431956 | 0.29 |
ENSDART00000183832
|
farp1
|
FERM, RhoGEF (ARHGEF) and pleckstrin domain protein 1 (chondrocyte-derived) |
| chr22_+_18389271 | 0.29 |
ENSDART00000088270
|
yjefn3
|
YjeF N-terminal domain containing 3 |
| chr4_-_9196291 | 0.29 |
ENSDART00000153963
|
hcfc2
|
host cell factor C2 |
| chr22_-_35063526 | 0.29 |
ENSDART00000162211
|
zgc:63733
|
zgc:63733 |
| chr7_+_48555626 | 0.28 |
ENSDART00000125483
ENSDART00000083514 |
kcnq1
|
potassium voltage-gated channel, KQT-like subfamily, member 1 |
| chr2_+_11031360 | 0.28 |
ENSDART00000180020
ENSDART00000145093 |
acot11a
|
acyl-CoA thioesterase 11a |
| chr10_-_31015535 | 0.28 |
ENSDART00000146116
|
panx3
|
pannexin 3 |
| chr22_+_21398508 | 0.27 |
ENSDART00000089408
ENSDART00000186091 |
shdb
|
Src homology 2 domain containing transforming protein D, b |
| chr16_-_25680666 | 0.27 |
ENSDART00000132693
ENSDART00000140539 ENSDART00000015302 |
tomm40
|
translocase of outer mitochondrial membrane 40 homolog (yeast) |
| chr13_-_25196758 | 0.26 |
ENSDART00000184722
|
adka
|
adenosine kinase a |
| chr1_+_45839927 | 0.26 |
ENSDART00000148086
ENSDART00000180413 ENSDART00000048191 ENSDART00000179047 |
map2k7
|
mitogen-activated protein kinase kinase 7 |
| chr6_-_10034145 | 0.26 |
ENSDART00000185999
|
nudt15
|
nudix (nucleoside diphosphate linked moiety X)-type motif 15 |
| chr5_+_62374092 | 0.26 |
ENSDART00000082965
|
CU928117.1
|
|
| chr7_+_24496894 | 0.26 |
ENSDART00000149994
|
nelfa
|
negative elongation factor complex member A |
| chr9_+_7732714 | 0.25 |
ENSDART00000145853
|
si:ch1073-349o24.2
|
si:ch1073-349o24.2 |
| chr15_+_1534644 | 0.25 |
ENSDART00000130413
|
smc4
|
structural maintenance of chromosomes 4 |
| chr1_-_26045560 | 0.25 |
ENSDART00000172737
ENSDART00000076120 ENSDART00000193593 |
gpank1
|
G patch domain and ankyrin repeats 1 |
| chr10_+_20180163 | 0.24 |
ENSDART00000080016
|
ppp3ccb
|
protein phosphatase 3, catalytic subunit, gamma isozyme, b |
| chr25_+_5015019 | 0.24 |
ENSDART00000127600
|
hdac10
|
histone deacetylase 10 |
| chr22_+_8365905 | 0.24 |
ENSDART00000105945
|
si:dkey-19a16.11
|
si:dkey-19a16.11 |
| chr22_+_22438783 | 0.24 |
ENSDART00000147825
|
kif14
|
kinesin family member 14 |
| chr12_-_48374728 | 0.24 |
ENSDART00000153403
ENSDART00000188117 |
dnajb12b
|
DnaJ (Hsp40) homolog, subfamily B, member 12b |
| chr24_+_28383278 | 0.24 |
ENSDART00000018095
|
sh3glb1a
|
SH3-domain GRB2-like endophilin B1a |
| chr5_-_38438613 | 0.24 |
ENSDART00000131912
|
pld2
|
phospholipase D2 |
| chr13_-_31167461 | 0.23 |
ENSDART00000190640
ENSDART00000192002 |
mapk8a
|
mitogen-activated protein kinase 8a |
| chr1_+_26105141 | 0.23 |
ENSDART00000102379
ENSDART00000127154 |
toporsa
|
topoisomerase I binding, arginine/serine-rich a |
| chr19_+_32201991 | 0.23 |
ENSDART00000022667
|
fam8a1a
|
family with sequence similarity 8, member A1a |
| chr14_-_5407555 | 0.23 |
ENSDART00000001424
|
pcgf1
|
polycomb group ring finger 1 |
| chr18_-_42313798 | 0.23 |
ENSDART00000098639
|
cntn5
|
contactin 5 |
| chr25_+_25516016 | 0.23 |
ENSDART00000188980
|
phrf1
|
PHD and ring finger domains 1 |
| chr7_-_19526721 | 0.23 |
ENSDART00000114203
|
man2b2
|
mannosidase, alpha, class 2B, member 2 |
| chr25_-_8201983 | 0.23 |
ENSDART00000006579
|
saal1
|
serum amyloid A-like 1 |
| chr8_+_25892319 | 0.23 |
ENSDART00000187167
ENSDART00000078163 |
tmem115
|
transmembrane protein 115 |
| chr3_+_60828813 | 0.22 |
ENSDART00000128260
|
CABZ01087514.1
|
|
| chr23_-_18381361 | 0.22 |
ENSDART00000016891
|
hsd17b10
|
hydroxysteroid (17-beta) dehydrogenase 10 |
| chr23_-_33680265 | 0.22 |
ENSDART00000138416
|
tfcp2
|
transcription factor CP2 |
| chr5_+_26807142 | 0.22 |
ENSDART00000188981
|
rabl6b
|
RAB, member RAS oncogene family-like 6b |
| chr5_-_26330313 | 0.22 |
ENSDART00000148656
|
arvcfb
|
ARVCF, delta catenin family member b |
| chr5_+_41477526 | 0.22 |
ENSDART00000153567
|
pias2
|
protein inhibitor of activated STAT, 2 |
| chr13_+_23093743 | 0.22 |
ENSDART00000148034
|
pik3ap1
|
phosphoinositide-3-kinase adaptor protein 1 |
| chr13_-_48388726 | 0.22 |
ENSDART00000169473
|
fbxo11a
|
F-box protein 11a |
| chr6_-_51811095 | 0.22 |
ENSDART00000161048
|
psmf1
|
proteasome inhibitor subunit 1 |
| chr7_+_48555400 | 0.21 |
ENSDART00000174474
|
kcnq1
|
potassium voltage-gated channel, KQT-like subfamily, member 1 |
| chr11_-_45152702 | 0.21 |
ENSDART00000168066
|
afmid
|
arylformamidase |
| chr17_+_8799451 | 0.21 |
ENSDART00000189814
ENSDART00000191577 |
tonsl
|
tonsoku-like, DNA repair protein |
| chr24_+_25032340 | 0.20 |
ENSDART00000005845
|
mtmr6
|
myotubularin related protein 6 |
| chr5_+_26686279 | 0.20 |
ENSDART00000193543
|
tango2
|
transport and golgi organization 2 homolog (Drosophila) |
| chr12_+_33361948 | 0.20 |
ENSDART00000124982
|
fasn
|
fatty acid synthase |
| chr23_-_7125494 | 0.20 |
ENSDART00000111929
|
slco4a1
|
solute carrier organic anion transporter family, member 4A1 |
| chr18_+_50924556 | 0.20 |
ENSDART00000159287
|
pik3c2g
|
phosphatidylinositol-4-phosphate 3-kinase catalytic subunit type 2 gamma |
| chr13_-_11378127 | 0.20 |
ENSDART00000158632
|
akt3a
|
v-akt murine thymoma viral oncogene homolog 3a |
| chr7_-_17814118 | 0.20 |
ENSDART00000179688
|
ecsit
|
ECSIT signalling integrator |
| chr21_-_1644414 | 0.20 |
ENSDART00000105736
ENSDART00000124904 |
zgc:152948
|
zgc:152948 |
| chr5_+_62052750 | 0.19 |
ENSDART00000192103
ENSDART00000181866 |
si:dkey-35m8.1
|
si:dkey-35m8.1 |
| chr11_+_6115621 | 0.19 |
ENSDART00000165031
ENSDART00000027666 ENSDART00000161458 |
nr2f6b
|
nuclear receptor subfamily 2, group F, member 6b |
| chr10_+_37500234 | 0.19 |
ENSDART00000132096
ENSDART00000099473 |
msi2a
|
musashi RNA-binding protein 2a |
| chr1_+_53714734 | 0.19 |
ENSDART00000114689
|
pus10
|
pseudouridylate synthase 10 |
| chr18_-_17513426 | 0.19 |
ENSDART00000146725
|
b3gnt9
|
UDP-GlcNAc:betaGal beta-1,3-N-acetylglucosaminyltransferase 9 |
| chr24_-_36270855 | 0.19 |
ENSDART00000154858
|
si:ch211-40k21.5
|
si:ch211-40k21.5 |
| chr8_+_26083808 | 0.18 |
ENSDART00000099283
|
dalrd3
|
DALR anticodon binding domain containing 3 |
| chr15_-_704408 | 0.18 |
ENSDART00000156200
ENSDART00000166404 ENSDART00000131040 |
zgc:174574
|
zgc:174574 |
| chr18_+_3064237 | 0.18 |
ENSDART00000161848
|
ints4
|
integrator complex subunit 4 |
| chr1_+_12068800 | 0.18 |
ENSDART00000055566
|
xpa
|
xeroderma pigmentosum, complementation group A |
| chr14_-_7375049 | 0.18 |
ENSDART00000054809
|
trappc11
|
trafficking protein particle complex 11 |
| chr19_-_20163197 | 0.18 |
ENSDART00000148179
ENSDART00000151646 |
fam221a
|
family with sequence similarity 221, member A |
| chr5_-_19036164 | 0.18 |
ENSDART00000099378
|
fam214b
|
family with sequence similarity 214, member B |
| chr2_-_43739559 | 0.18 |
ENSDART00000138947
|
kif5ba
|
kinesin family member 5B, a |
| chr23_+_30707837 | 0.18 |
ENSDART00000016096
|
dnajc11a
|
DnaJ (Hsp40) homolog, subfamily C, member 11a |
| chr23_-_2901167 | 0.18 |
ENSDART00000165955
ENSDART00000190616 |
zhx3
|
zinc fingers and homeoboxes 3 |
| chr9_+_40546677 | 0.17 |
ENSDART00000132911
|
vwc2l
|
von Willebrand factor C domain containing protein 2-like |
| chr13_+_21919786 | 0.17 |
ENSDART00000182440
|
ndst2a
|
N-deacetylase/N-sulfotransferase (heparan glucosaminyl) 2a |
| chr11_-_45152912 | 0.17 |
ENSDART00000167540
ENSDART00000173050 ENSDART00000170795 |
afmid
|
arylformamidase |
| chr17_+_8799661 | 0.17 |
ENSDART00000105326
|
tonsl
|
tonsoku-like, DNA repair protein |
Network of associatons between targets according to the STRING database.
First level regulatory network of pax10+pax6a+pax6b
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 1.3 | GO:0006700 | C21-steroid hormone biosynthetic process(GO:0006700) |
| 0.2 | 0.7 | GO:0000480 | endonucleolytic cleavage to generate mature 5'-end of SSU-rRNA from (SSU-rRNA, 5.8S rRNA, LSU-rRNA)(GO:0000472) endonucleolytic cleavage in 5'-ETS of tricistronic rRNA transcript (SSU-rRNA, 5.8S rRNA, LSU-rRNA)(GO:0000480) rRNA 5'-end processing(GO:0000967) ncRNA 5'-end processing(GO:0034471) |
| 0.2 | 1.5 | GO:1990511 | piRNA biosynthetic process(GO:1990511) |
| 0.2 | 0.8 | GO:0060074 | synapse maturation(GO:0060074) |
| 0.1 | 0.5 | GO:0034164 | regulation of toll-like receptor 9 signaling pathway(GO:0034163) negative regulation of toll-like receptor 9 signaling pathway(GO:0034164) |
| 0.1 | 2.3 | GO:0005979 | regulation of glycogen biosynthetic process(GO:0005979) regulation of glucan biosynthetic process(GO:0010962) |
| 0.1 | 1.4 | GO:0090110 | cargo loading into COPII-coated vesicle(GO:0090110) |
| 0.1 | 0.4 | GO:0051012 | microtubule sliding(GO:0051012) |
| 0.1 | 0.4 | GO:0032776 | DNA methylation on cytosine(GO:0032776) |
| 0.1 | 0.9 | GO:0006032 | chitin catabolic process(GO:0006032) |
| 0.1 | 0.3 | GO:0010874 | regulation of cholesterol efflux(GO:0010874) |
| 0.1 | 0.4 | GO:0030579 | ubiquitin-dependent SMAD protein catabolic process(GO:0030579) |
| 0.1 | 0.4 | GO:0060282 | positive regulation of oocyte development(GO:0060282) |
| 0.1 | 1.2 | GO:0006265 | DNA topological change(GO:0006265) |
| 0.1 | 0.3 | GO:0018197 | peptidyl-aspartic acid modification(GO:0018197) peptidyl-aspartic acid hydroxylation(GO:0042264) |
| 0.1 | 0.5 | GO:0016266 | O-glycan processing(GO:0016266) |
| 0.1 | 0.3 | GO:0098535 | de novo centriole assembly(GO:0098535) |
| 0.1 | 1.1 | GO:0035803 | binding of sperm to zona pellucida(GO:0007339) egg coat formation(GO:0035803) regulation of acrosome reaction(GO:0060046) positive regulation of acrosome reaction(GO:2000344) |
| 0.1 | 0.4 | GO:0034975 | protein folding in endoplasmic reticulum(GO:0034975) |
| 0.1 | 0.2 | GO:0070914 | UV-damage excision repair(GO:0070914) nucleotide-excision repair involved in interstrand cross-link repair(GO:1901255) |
| 0.1 | 0.6 | GO:0019883 | antigen processing and presentation of endogenous peptide antigen(GO:0002483) antigen processing and presentation of endogenous antigen(GO:0019883) antigen processing and presentation of endogenous peptide antigen via MHC class I(GO:0019885) |
| 0.1 | 0.5 | GO:0021588 | cerebellum formation(GO:0021588) |
| 0.1 | 0.3 | GO:0045337 | farnesyl diphosphate biosynthetic process(GO:0045337) |
| 0.0 | 0.5 | GO:0002154 | thyroid hormone mediated signaling pathway(GO:0002154) |
| 0.0 | 0.5 | GO:0006659 | phosphatidylserine biosynthetic process(GO:0006659) |
| 0.0 | 0.3 | GO:2001045 | negative regulation of integrin-mediated signaling pathway(GO:2001045) |
| 0.0 | 0.4 | GO:0019441 | tryptophan catabolic process to kynurenine(GO:0019441) |
| 0.0 | 0.7 | GO:0070816 | phosphorylation of RNA polymerase II C-terminal domain(GO:0070816) |
| 0.0 | 0.1 | GO:0010847 | regulation of chromatin assembly(GO:0010847) |
| 0.0 | 0.1 | GO:0046125 | deoxyribonucleoside metabolic process(GO:0009120) thymidine metabolic process(GO:0046104) pyrimidine deoxyribonucleoside metabolic process(GO:0046125) |
| 0.0 | 0.3 | GO:0044209 | AMP salvage(GO:0044209) |
| 0.0 | 0.3 | GO:0006517 | protein deglycosylation(GO:0006517) |
| 0.0 | 0.3 | GO:0032732 | positive regulation of interleukin-1 production(GO:0032732) |
| 0.0 | 0.4 | GO:0008105 | asymmetric protein localization(GO:0008105) apical protein localization(GO:0045176) |
| 0.0 | 0.2 | GO:0006013 | mannose metabolic process(GO:0006013) |
| 0.0 | 0.3 | GO:0035871 | protein K11-linked deubiquitination(GO:0035871) |
| 0.0 | 0.1 | GO:0071548 | response to dexamethasone(GO:0071548) |
| 0.0 | 0.3 | GO:0009109 | coenzyme catabolic process(GO:0009109) |
| 0.0 | 0.7 | GO:0006743 | ubiquinone metabolic process(GO:0006743) ubiquinone biosynthetic process(GO:0006744) quinone biosynthetic process(GO:1901663) |
| 0.0 | 0.3 | GO:0006203 | dGTP catabolic process(GO:0006203) purine nucleoside triphosphate catabolic process(GO:0009146) purine deoxyribonucleoside triphosphate catabolic process(GO:0009217) dGTP metabolic process(GO:0046070) |
| 0.0 | 0.2 | GO:0031119 | tRNA pseudouridine synthesis(GO:0031119) |
| 0.0 | 0.3 | GO:0035283 | central nervous system segmentation(GO:0035283) brain segmentation(GO:0035284) |
| 0.0 | 0.3 | GO:0034312 | diol biosynthetic process(GO:0034312) sphingosine biosynthetic process(GO:0046512) |
| 0.0 | 0.6 | GO:0018149 | peptide cross-linking(GO:0018149) |
| 0.0 | 0.2 | GO:0061635 | regulation of protein complex stability(GO:0061635) |
| 0.0 | 0.7 | GO:0016233 | telomere capping(GO:0016233) |
| 0.0 | 0.5 | GO:0018095 | protein polyglutamylation(GO:0018095) |
| 0.0 | 0.2 | GO:0098971 | dendritic transport(GO:0098935) anterograde dendritic transport(GO:0098937) anterograde dendritic transport of neurotransmitter receptor complex(GO:0098971) |
| 0.0 | 0.1 | GO:0015862 | uridine transport(GO:0015862) pyrimidine nucleoside transport(GO:0015864) |
| 0.0 | 0.3 | GO:0061718 | glucose catabolic process(GO:0006007) NADH regeneration(GO:0006735) glycolytic process through fructose-6-phosphate(GO:0061615) glycolytic process through glucose-6-phosphate(GO:0061620) canonical glycolysis(GO:0061621) glucose catabolic process to pyruvate(GO:0061718) |
| 0.0 | 0.3 | GO:1905150 | regulation of voltage-gated sodium channel activity(GO:1905150) |
| 0.0 | 0.1 | GO:0006546 | glycine catabolic process(GO:0006546) glycine decarboxylation via glycine cleavage system(GO:0019464) |
| 0.0 | 0.6 | GO:0016925 | protein sumoylation(GO:0016925) |
| 0.0 | 0.1 | GO:0061469 | regulation of type B pancreatic cell proliferation(GO:0061469) |
| 0.0 | 0.1 | GO:0051754 | meiotic sister chromatid cohesion, centromeric(GO:0051754) |
| 0.0 | 0.1 | GO:0070131 | regulation of mitochondrial translation(GO:0070129) positive regulation of mitochondrial translation(GO:0070131) |
| 0.0 | 0.3 | GO:0032785 | negative regulation of DNA-templated transcription, elongation(GO:0032785) negative regulation of transcription elongation from RNA polymerase II promoter(GO:0034244) |
| 0.0 | 0.2 | GO:0007076 | mitotic chromosome condensation(GO:0007076) |
| 0.0 | 0.1 | GO:2000580 | positive regulation of microtubule motor activity(GO:2000576) regulation of ATP-dependent microtubule motor activity, plus-end-directed(GO:2000580) positive regulation of ATP-dependent microtubule motor activity, plus-end-directed(GO:2000582) |
| 0.0 | 0.2 | GO:0046473 | phosphatidic acid biosynthetic process(GO:0006654) phosphatidic acid metabolic process(GO:0046473) |
| 0.0 | 0.4 | GO:0031297 | replication fork processing(GO:0031297) |
| 0.0 | 0.2 | GO:0036353 | histone H2A monoubiquitination(GO:0035518) histone H2A-K119 monoubiquitination(GO:0036353) |
| 0.0 | 0.2 | GO:0050975 | sensory perception of touch(GO:0050975) |
| 0.0 | 0.2 | GO:0015014 | heparan sulfate proteoglycan biosynthetic process, polysaccharide chain biosynthetic process(GO:0015014) |
| 0.0 | 0.6 | GO:0001878 | response to yeast(GO:0001878) |
| 0.0 | 0.4 | GO:0015012 | heparan sulfate proteoglycan biosynthetic process(GO:0015012) |
| 0.0 | 1.4 | GO:0007030 | Golgi organization(GO:0007030) |
| 0.0 | 0.4 | GO:0030225 | macrophage differentiation(GO:0030225) |
| 0.0 | 0.3 | GO:0030150 | protein import into mitochondrial matrix(GO:0030150) |
| 0.0 | 0.1 | GO:0051315 | attachment of mitotic spindle microtubules to kinetochore(GO:0051315) |
| 0.0 | 0.2 | GO:0030309 | poly-N-acetyllactosamine metabolic process(GO:0030309) poly-N-acetyllactosamine biosynthetic process(GO:0030311) |
| 0.0 | 0.3 | GO:0007257 | activation of JUN kinase activity(GO:0007257) positive regulation of JUN kinase activity(GO:0043507) |
| 0.0 | 0.1 | GO:0071479 | cellular response to ionizing radiation(GO:0071479) |
| 0.0 | 0.2 | GO:0060324 | face development(GO:0060324) |
| 0.0 | 0.4 | GO:0006360 | transcription from RNA polymerase I promoter(GO:0006360) |
| 0.0 | 0.2 | GO:0042407 | cristae formation(GO:0042407) |
| 0.0 | 0.2 | GO:0006513 | protein monoubiquitination(GO:0006513) |
| 0.0 | 0.2 | GO:0051788 | response to misfolded protein(GO:0051788) cellular response to misfolded protein(GO:0071218) |
| 0.0 | 0.1 | GO:0097531 | mast cell chemotaxis(GO:0002551) regulation of mast cell chemotaxis(GO:0060753) positive regulation of mast cell chemotaxis(GO:0060754) mast cell migration(GO:0097531) |
| 0.0 | 1.1 | GO:0031098 | stress-activated protein kinase signaling cascade(GO:0031098) |
| 0.0 | 0.1 | GO:0043928 | exonucleolytic nuclear-transcribed mRNA catabolic process involved in deadenylation-dependent decay(GO:0043928) |
| 0.0 | 0.2 | GO:0036092 | phosphatidylinositol-3-phosphate biosynthetic process(GO:0036092) |
| 0.0 | 0.4 | GO:0000289 | nuclear-transcribed mRNA poly(A) tail shortening(GO:0000289) |
| 0.0 | 0.3 | GO:0051014 | actin filament severing(GO:0051014) |
| 0.0 | 0.3 | GO:0040037 | negative regulation of fibroblast growth factor receptor signaling pathway(GO:0040037) |
| 0.0 | 0.2 | GO:0043252 | sodium-independent organic anion transport(GO:0043252) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 2.3 | GO:0000164 | protein phosphatase type 1 complex(GO:0000164) |
| 0.1 | 0.4 | GO:0000120 | RNA polymerase I transcription factor complex(GO:0000120) |
| 0.1 | 0.4 | GO:0008247 | 1-alkyl-2-acetylglycerophosphocholine esterase complex(GO:0008247) |
| 0.1 | 1.4 | GO:0030127 | COPII vesicle coat(GO:0030127) |
| 0.1 | 0.4 | GO:0005662 | DNA replication factor A complex(GO:0005662) |
| 0.1 | 1.9 | GO:0000421 | autophagosome membrane(GO:0000421) |
| 0.0 | 0.3 | GO:0034388 | Pwp2p-containing subcomplex of 90S preribosome(GO:0034388) |
| 0.0 | 0.2 | GO:0000110 | nucleotide-excision repair factor 1 complex(GO:0000110) |
| 0.0 | 0.7 | GO:0070187 | telosome(GO:0070187) |
| 0.0 | 1.0 | GO:0035102 | PRC1 complex(GO:0035102) |
| 0.0 | 0.2 | GO:0005955 | calcineurin complex(GO:0005955) |
| 0.0 | 0.4 | GO:0035145 | exon-exon junction complex(GO:0035145) |
| 0.0 | 0.3 | GO:0097648 | G-protein coupled receptor dimeric complex(GO:0038037) G-protein coupled receptor heterodimeric complex(GO:0038039) G-protein coupled receptor complex(GO:0097648) |
| 0.0 | 0.3 | GO:0000347 | THO complex(GO:0000347) THO complex part of transcription export complex(GO:0000445) |
| 0.0 | 0.4 | GO:0005675 | holo TFIIH complex(GO:0005675) |
| 0.0 | 0.3 | GO:0019908 | cyclin/CDK positive transcription elongation factor complex(GO:0008024) nuclear cyclin-dependent protein kinase holoenzyme complex(GO:0019908) |
| 0.0 | 0.3 | GO:0031211 | serine C-palmitoyltransferase complex(GO:0017059) endoplasmic reticulum palmitoyltransferase complex(GO:0031211) |
| 0.0 | 0.1 | GO:0030896 | checkpoint clamp complex(GO:0030896) |
| 0.0 | 0.3 | GO:0032021 | NELF complex(GO:0032021) |
| 0.0 | 0.1 | GO:0005960 | glycine cleavage complex(GO:0005960) |
| 0.0 | 0.6 | GO:0005666 | DNA-directed RNA polymerase III complex(GO:0005666) |
| 0.0 | 0.3 | GO:0005742 | mitochondrial outer membrane translocase complex(GO:0005742) |
| 0.0 | 0.3 | GO:0005945 | 6-phosphofructokinase complex(GO:0005945) |
| 0.0 | 0.5 | GO:0030014 | CCR4-NOT complex(GO:0030014) |
| 0.0 | 0.4 | GO:0071144 | SMAD2-SMAD3 protein complex(GO:0071144) |
| 0.0 | 0.8 | GO:0005881 | cytoplasmic microtubule(GO:0005881) |
| 0.0 | 0.1 | GO:0055087 | Ski complex(GO:0055087) |
| 0.0 | 0.4 | GO:1902562 | NuA4 histone acetyltransferase complex(GO:0035267) H4/H2A histone acetyltransferase complex(GO:0043189) H4 histone acetyltransferase complex(GO:1902562) |
| 0.0 | 0.3 | GO:0035371 | microtubule plus-end(GO:0035371) |
| 0.0 | 0.1 | GO:0032783 | ELL-EAF complex(GO:0032783) |
| 0.0 | 0.4 | GO:0046658 | anchored component of plasma membrane(GO:0046658) |
| 0.0 | 0.2 | GO:0061617 | MICOS complex(GO:0061617) |
| 0.0 | 0.3 | GO:0044665 | MLL1/2 complex(GO:0044665) MLL1 complex(GO:0071339) |
| 0.0 | 0.1 | GO:0031080 | nuclear pore outer ring(GO:0031080) |
| 0.0 | 0.4 | GO:0016605 | PML body(GO:0016605) |
| 0.0 | 0.3 | GO:0005921 | gap junction(GO:0005921) |
| 0.0 | 0.8 | GO:0031228 | integral component of Golgi membrane(GO:0030173) intrinsic component of Golgi membrane(GO:0031228) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.4 | 1.3 | GO:0016713 | cholesterol monooxygenase (side-chain-cleaving) activity(GO:0008386) oxidoreductase activity, acting on paired donors, with incorporation or reduction of molecular oxygen, reduced iron-sulfur protein as one donor, and incorporation of one atom of oxygen(GO:0016713) |
| 0.2 | 1.5 | GO:0004366 | glycerol-3-phosphate O-acyltransferase activity(GO:0004366) |
| 0.2 | 2.3 | GO:2001069 | glycogen binding(GO:2001069) |
| 0.1 | 0.4 | GO:0032184 | SUMO polymer binding(GO:0032184) |
| 0.1 | 0.4 | GO:0004061 | arylformamidase activity(GO:0004061) |
| 0.1 | 0.3 | GO:0004161 | dimethylallyltranstransferase activity(GO:0004161) geranyltranstransferase activity(GO:0004337) |
| 0.1 | 1.1 | GO:0035804 | structural constituent of egg coat(GO:0035804) |
| 0.1 | 0.4 | GO:0050508 | glucuronosyl-N-acetylglucosaminyl-proteoglycan 4-alpha-N-acetylglucosaminyltransferase activity(GO:0050508) |
| 0.1 | 1.2 | GO:0003917 | DNA topoisomerase type I activity(GO:0003917) |
| 0.1 | 0.3 | GO:0033829 | O-fucosylpeptide 3-beta-N-acetylglucosaminyltransferase activity(GO:0033829) |
| 0.1 | 0.3 | GO:0004597 | peptide-aspartate beta-dioxygenase activity(GO:0004597) |
| 0.1 | 0.6 | GO:0003836 | beta-galactoside (CMP) alpha-2,3-sialyltransferase activity(GO:0003836) |
| 0.1 | 0.9 | GO:0004568 | chitinase activity(GO:0004568) |
| 0.1 | 0.6 | GO:0003854 | 3-beta-hydroxy-delta5-steroid dehydrogenase activity(GO:0003854) |
| 0.1 | 0.2 | GO:0047611 | acetylspermidine deacetylase activity(GO:0047611) |
| 0.1 | 1.9 | GO:0032266 | phosphatidylinositol-3-phosphate binding(GO:0032266) |
| 0.1 | 0.5 | GO:0004705 | JUN kinase activity(GO:0004705) SAP kinase activity(GO:0016909) |
| 0.1 | 1.2 | GO:0019894 | kinesin binding(GO:0019894) |
| 0.1 | 0.3 | GO:0004001 | adenosine kinase activity(GO:0004001) |
| 0.1 | 0.3 | GO:0008545 | JUN kinase kinase activity(GO:0008545) |
| 0.0 | 0.6 | GO:0061665 | SUMO ligase activity(GO:0061665) |
| 0.0 | 0.3 | GO:0004758 | serine C-palmitoyltransferase activity(GO:0004758) C-palmitoyltransferase activity(GO:0016454) |
| 0.0 | 0.3 | GO:0001784 | phosphotyrosine binding(GO:0001784) |
| 0.0 | 0.3 | GO:0047429 | nucleoside-triphosphate diphosphatase activity(GO:0047429) |
| 0.0 | 0.2 | GO:0004723 | calcium-dependent protein serine/threonine phosphatase activity(GO:0004723) calmodulin-dependent protein phosphatase activity(GO:0033192) |
| 0.0 | 0.3 | GO:0004965 | G-protein coupled GABA receptor activity(GO:0004965) |
| 0.0 | 0.3 | GO:0036042 | long-chain fatty acyl-CoA binding(GO:0036042) |
| 0.0 | 0.5 | GO:0070739 | protein-glutamic acid ligase activity(GO:0070739) tubulin-glutamic acid ligase activity(GO:0070740) |
| 0.0 | 0.4 | GO:0003886 | DNA (cytosine-5-)-methyltransferase activity(GO:0003886) |
| 0.0 | 0.2 | GO:0004630 | phospholipase D activity(GO:0004630) |
| 0.0 | 0.6 | GO:0003810 | protein-glutamine gamma-glutamyltransferase activity(GO:0003810) |
| 0.0 | 0.3 | GO:0030332 | cyclin binding(GO:0030332) |
| 0.0 | 0.7 | GO:0004659 | prenyltransferase activity(GO:0004659) |
| 0.0 | 0.4 | GO:0016864 | protein disulfide isomerase activity(GO:0003756) intramolecular oxidoreductase activity, transposing S-S bonds(GO:0016864) |
| 0.0 | 0.1 | GO:0001042 | RNA polymerase I core binding(GO:0001042) |
| 0.0 | 0.4 | GO:0070840 | dynein complex binding(GO:0070840) |
| 0.0 | 0.4 | GO:0004535 | poly(A)-specific ribonuclease activity(GO:0004535) |
| 0.0 | 0.7 | GO:0003899 | DNA-directed RNA polymerase activity(GO:0003899) RNA polymerase activity(GO:0034062) |
| 0.0 | 0.1 | GO:0030331 | estrogen receptor binding(GO:0030331) |
| 0.0 | 0.1 | GO:0042806 | fucose binding(GO:0042806) |
| 0.0 | 0.1 | GO:0070016 | armadillo repeat domain binding(GO:0070016) |
| 0.0 | 0.3 | GO:0003872 | 6-phosphofructokinase activity(GO:0003872) |
| 0.0 | 0.2 | GO:0015016 | [heparan sulfate]-glucosamine N-sulfotransferase activity(GO:0015016) |
| 0.0 | 0.4 | GO:0070411 | I-SMAD binding(GO:0070411) |
| 0.0 | 0.1 | GO:0004694 | eukaryotic translation initiation factor 2alpha kinase activity(GO:0004694) |
| 0.0 | 0.6 | GO:0070006 | metalloaminopeptidase activity(GO:0070006) |
| 0.0 | 0.1 | GO:0019136 | deoxynucleoside kinase activity(GO:0019136) |
| 0.0 | 0.1 | GO:0070004 | cysteine-type exopeptidase activity(GO:0070004) |
| 0.0 | 0.3 | GO:0008320 | protein transmembrane transporter activity(GO:0008320) |
| 0.0 | 0.3 | GO:0072542 | phosphatase activator activity(GO:0019211) protein phosphatase activator activity(GO:0072542) |
| 0.0 | 0.2 | GO:0036312 | phosphatidylinositol 3-kinase regulatory subunit binding(GO:0036312) |
| 0.0 | 0.1 | GO:0001222 | transcription corepressor binding(GO:0001222) |
| 0.0 | 0.2 | GO:0030544 | Hsp70 protein binding(GO:0030544) |
| 0.0 | 0.3 | GO:0016289 | CoA hydrolase activity(GO:0016289) |
| 0.0 | 0.6 | GO:0004435 | phosphatidylinositol phospholipase C activity(GO:0004435) phospholipase C activity(GO:0004629) |
| 0.0 | 0.1 | GO:0016286 | small conductance calcium-activated potassium channel activity(GO:0016286) |
| 0.0 | 0.2 | GO:0004559 | alpha-mannosidase activity(GO:0004559) |
| 0.0 | 0.2 | GO:0004526 | ribonuclease P activity(GO:0004526) |
| 0.0 | 0.4 | GO:0051010 | microtubule plus-end binding(GO:0051010) |
| 0.0 | 0.2 | GO:0008532 | N-acetyllactosaminide beta-1,3-N-acetylglucosaminyltransferase activity(GO:0008532) |
| 0.0 | 0.3 | GO:0016854 | racemase and epimerase activity(GO:0016854) |
| 0.0 | 0.4 | GO:0043539 | protein serine/threonine kinase activator activity(GO:0043539) |
| 0.0 | 0.3 | GO:0017080 | sodium channel regulator activity(GO:0017080) |
| 0.0 | 0.1 | GO:0005172 | vascular endothelial growth factor receptor binding(GO:0005172) |
| 0.0 | 0.2 | GO:0035005 | 1-phosphatidylinositol-4-phosphate 3-kinase activity(GO:0035005) |
| 0.0 | 0.3 | GO:0022829 | wide pore channel activity(GO:0022829) |
| 0.0 | 0.2 | GO:0015347 | sodium-independent organic anion transmembrane transporter activity(GO:0015347) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.7 | PID P38 GAMMA DELTA PATHWAY | Signaling mediated by p38-gamma and p38-delta |
| 0.0 | 0.6 | PID IL12 STAT4 PATHWAY | IL12 signaling mediated by STAT4 |
| 0.0 | 0.3 | PID ANTHRAX PATHWAY | Cellular roles of Anthrax toxin |
| 0.0 | 0.8 | PID RETINOIC ACID PATHWAY | Retinoic acid receptors-mediated signaling |
| 0.0 | 0.2 | PID ARF6 DOWNSTREAM PATHWAY | Arf6 downstream pathway |
| 0.0 | 0.4 | PID SMAD2 3PATHWAY | Regulation of cytoplasmic and nuclear SMAD2/3 signaling |
| 0.0 | 0.4 | PID RB 1PATHWAY | Regulation of retinoblastoma protein |
| 0.0 | 0.4 | SA PTEN PATHWAY | PTEN is a tumor suppressor that dephosphorylates the lipid messenger phosphatidylinositol triphosphate. |
| 0.0 | 0.4 | PID HDAC CLASSII PATHWAY | Signaling events mediated by HDAC Class II |
| 0.0 | 0.4 | ST GA13 PATHWAY | G alpha 13 Pathway |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 1.3 | REACTOME ENDOGENOUS STEROLS | Genes involved in Endogenous sterols |
| 0.1 | 1.7 | REACTOME SYNTHESIS OF PA | Genes involved in Synthesis of PA |
| 0.1 | 1.4 | REACTOME ANTIGEN PRESENTATION FOLDING ASSEMBLY AND PEPTIDE LOADING OF CLASS I MHC | Genes involved in Antigen Presentation: Folding, assembly and peptide loading of class I MHC |
| 0.1 | 0.6 | REACTOME TERMINATION OF O GLYCAN BIOSYNTHESIS | Genes involved in Termination of O-glycan biosynthesis |
| 0.0 | 0.4 | REACTOME SIGNALING BY NODAL | Genes involved in Signaling by NODAL |
| 0.0 | 0.8 | REACTOME RNA POL I TRANSCRIPTION TERMINATION | Genes involved in RNA Polymerase I Transcription Termination |
| 0.0 | 0.3 | REACTOME CLASS C 3 METABOTROPIC GLUTAMATE PHEROMONE RECEPTORS | Genes involved in Class C/3 (Metabotropic glutamate/pheromone receptors) |
| 0.0 | 0.5 | REACTOME ENERGY DEPENDENT REGULATION OF MTOR BY LKB1 AMPK | Genes involved in Energy dependent regulation of mTOR by LKB1-AMPK |
| 0.0 | 0.6 | REACTOME RNA POL III CHAIN ELONGATION | Genes involved in RNA Polymerase III Chain Elongation |
| 0.0 | 0.9 | REACTOME CHOLESTEROL BIOSYNTHESIS | Genes involved in Cholesterol biosynthesis |
| 0.0 | 0.4 | REACTOME MEMBRANE BINDING AND TARGETTING OF GAG PROTEINS | Genes involved in Membrane binding and targetting of GAG proteins |
| 0.0 | 0.4 | REACTOME TRYPTOPHAN CATABOLISM | Genes involved in Tryptophan catabolism |
| 0.0 | 0.3 | REACTOME PRE NOTCH PROCESSING IN GOLGI | Genes involved in Pre-NOTCH Processing in Golgi |
| 0.0 | 0.2 | REACTOME TRANSPORT OF ORGANIC ANIONS | Genes involved in Transport of organic anions |
| 0.0 | 0.4 | REACTOME DOWNREGULATION OF SMAD2 3 SMAD4 TRANSCRIPTIONAL ACTIVITY | Genes involved in Downregulation of SMAD2/3:SMAD4 transcriptional activity |
| 0.0 | 1.2 | REACTOME ION TRANSPORT BY P TYPE ATPASES | Genes involved in Ion transport by P-type ATPases |
| 0.0 | 0.3 | REACTOME ABCA TRANSPORTERS IN LIPID HOMEOSTASIS | Genes involved in ABCA transporters in lipid homeostasis |
| 0.0 | 0.4 | REACTOME INTERFERON GAMMA SIGNALING | Genes involved in Interferon gamma signaling |
| 0.0 | 0.3 | REACTOME PROCESSING OF INTRONLESS PRE MRNAS | Genes involved in Processing of Intronless Pre-mRNAs |
| 0.0 | 0.5 | REACTOME VOLTAGE GATED POTASSIUM CHANNELS | Genes involved in Voltage gated Potassium channels |
| 0.0 | 0.2 | REACTOME FATTY ACYL COA BIOSYNTHESIS | Genes involved in Fatty Acyl-CoA Biosynthesis |
| 0.0 | 0.4 | REACTOME SYNTHESIS OF PIPS AT THE PLASMA MEMBRANE | Genes involved in Synthesis of PIPs at the plasma membrane |
| 0.0 | 0.1 | REACTOME ACTIVATION OF CHAPERONES BY ATF6 ALPHA | Genes involved in Activation of Chaperones by ATF6-alpha |
| 0.0 | 0.2 | REACTOME FORMATION OF INCISION COMPLEX IN GG NER | Genes involved in Formation of incision complex in GG-NER |
| 0.0 | 0.2 | REACTOME BRANCHED CHAIN AMINO ACID CATABOLISM | Genes involved in Branched-chain amino acid catabolism |
| 0.0 | 0.3 | REACTOME JNK C JUN KINASES PHOSPHORYLATION AND ACTIVATION MEDIATED BY ACTIVATED HUMAN TAK1 | Genes involved in JNK (c-Jun kinases) phosphorylation and activation mediated by activated human TAK1 |