Project
PRJNA195909:zebrafish embryo and larva development
Navigation
Downloads
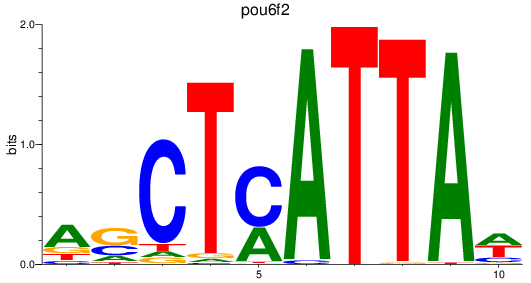
Results for pou6f2
Z-value: 1.21
Transcription factors associated with pou6f2
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
pou6f2
|
ENSDARG00000086362 | POU class 6 homeobox 2 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| pou6f2 | dr11_v1_chr24_+_10202718_10202718 | -0.66 | 5.4e-02 | Click! |
Activity profile of pou6f2 motif
Sorted Z-values of pou6f2 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr12_-_17698517 | 3.41 |
ENSDART00000105974
|
pvalb9
|
parvalbumin 9 |
| chr3_-_61162750 | 2.78 |
ENSDART00000055064
|
pvalb8
|
parvalbumin 8 |
| chr18_-_1185772 | 2.77 |
ENSDART00000143245
|
nptnb
|
neuroplastin b |
| chr12_-_35830625 | 2.75 |
ENSDART00000180028
|
CU459056.1
|
|
| chr12_-_17698669 | 2.66 |
ENSDART00000191384
|
pvalb9
|
parvalbumin 9 |
| chr12_-_26406323 | 2.49 |
ENSDART00000131896
|
myoz1b
|
myozenin 1b |
| chr8_+_33035709 | 2.06 |
ENSDART00000131660
|
angptl2b
|
angiopoietin-like 2b |
| chr3_-_43356082 | 2.04 |
ENSDART00000171213
|
uncx
|
UNC homeobox |
| chr2_-_35566938 | 1.94 |
ENSDART00000029006
ENSDART00000077178 ENSDART00000125298 |
tnn
|
tenascin N |
| chr4_+_12615836 | 1.91 |
ENSDART00000003583
|
lmo3
|
LIM domain only 3 |
| chr2_-_44255537 | 1.89 |
ENSDART00000011188
ENSDART00000093298 |
atp1a2a
|
ATPase Na+/K+ transporting subunit alpha 2 |
| chr13_+_25449681 | 1.84 |
ENSDART00000101328
|
atoh7
|
atonal bHLH transcription factor 7 |
| chr20_-_48485354 | 1.81 |
ENSDART00000124040
ENSDART00000148437 |
insm1a
|
insulinoma-associated 1a |
| chr3_-_46817499 | 1.73 |
ENSDART00000013717
|
elavl3
|
ELAV like neuron-specific RNA binding protein 3 |
| chr3_+_23737795 | 1.72 |
ENSDART00000182247
|
hoxb3a
|
homeobox B3a |
| chr16_-_2414063 | 1.69 |
ENSDART00000073621
|
zgc:152945
|
zgc:152945 |
| chr16_-_31717851 | 1.64 |
ENSDART00000169109
|
rbp5
|
retinol binding protein 1a, cellular |
| chr24_-_21923930 | 1.62 |
ENSDART00000131944
|
tagln3b
|
transgelin 3b |
| chr3_+_23738215 | 1.62 |
ENSDART00000143981
|
hoxb3a
|
homeobox B3a |
| chr22_-_13350240 | 1.61 |
ENSDART00000154095
ENSDART00000155118 |
si:ch211-227m13.1
|
si:ch211-227m13.1 |
| chr19_+_30662529 | 1.52 |
ENSDART00000175662
|
fam49al
|
family with sequence similarity 49, member A-like |
| chr16_-_31718013 | 1.39 |
ENSDART00000190716
|
rbp5
|
retinol binding protein 1a, cellular |
| chr13_-_31435137 | 1.38 |
ENSDART00000057441
|
rtn1a
|
reticulon 1a |
| chr24_+_9744012 | 1.37 |
ENSDART00000129656
|
tmem108
|
transmembrane protein 108 |
| chr4_-_16341801 | 1.36 |
ENSDART00000140190
|
kera
|
keratocan |
| chr13_+_24662238 | 1.36 |
ENSDART00000014176
|
msx3
|
muscle segment homeobox 3 |
| chr12_+_20352400 | 1.31 |
ENSDART00000066383
|
hbae5
|
hemoglobin, alpha embryonic 5 |
| chr4_-_17629444 | 1.25 |
ENSDART00000108814
|
nrip2
|
nuclear receptor interacting protein 2 |
| chr11_+_41981959 | 1.24 |
ENSDART00000055707
|
her15.1
|
hairy and enhancer of split-related 15, tandem duplicate 1 |
| chr13_+_35339182 | 1.23 |
ENSDART00000019323
|
jag1b
|
jagged 1b |
| chr16_+_46111849 | 1.20 |
ENSDART00000172232
|
sv2a
|
synaptic vesicle glycoprotein 2A |
| chr20_+_4060839 | 1.19 |
ENSDART00000178565
|
TRIM67
|
tripartite motif containing 67 |
| chr1_+_25696798 | 1.18 |
ENSDART00000054228
|
lrata
|
lecithin retinol acyltransferase a |
| chr9_-_14504834 | 1.15 |
ENSDART00000056103
|
nrp2b
|
neuropilin 2b |
| chr22_-_13042992 | 1.15 |
ENSDART00000028787
|
ahr1b
|
aryl hydrocarbon receptor 1b |
| chr19_-_32641725 | 1.08 |
ENSDART00000165006
ENSDART00000188185 |
hpca
|
hippocalcin |
| chr25_+_35553542 | 1.07 |
ENSDART00000113723
|
spi1a
|
Spi-1 proto-oncogene a |
| chr3_+_33341640 | 1.06 |
ENSDART00000186352
|
pyya
|
peptide YYa |
| chr1_+_40023640 | 1.05 |
ENSDART00000101623
|
lgi2b
|
leucine-rich repeat LGI family, member 2b |
| chr17_-_49977966 | 1.04 |
ENSDART00000183735
|
col12a1a
|
collagen, type XII, alpha 1a |
| chr19_-_31402429 | 1.03 |
ENSDART00000137292
|
tmem106bb
|
transmembrane protein 106Bb |
| chr25_+_6186823 | 0.98 |
ENSDART00000153526
|
oaz2a
|
ornithine decarboxylase antizyme 2a |
| chr3_-_28250722 | 0.97 |
ENSDART00000165936
|
rbfox1
|
RNA binding fox-1 homolog 1 |
| chr20_-_9436521 | 0.95 |
ENSDART00000133000
|
zgc:101840
|
zgc:101840 |
| chr4_+_12612145 | 0.94 |
ENSDART00000181201
|
lmo3
|
LIM domain only 3 |
| chr23_+_28731379 | 0.92 |
ENSDART00000047378
|
cort
|
cortistatin |
| chr2_-_50372467 | 0.92 |
ENSDART00000108900
|
cntnap2b
|
contactin associated protein like 2b |
| chr23_+_31107685 | 0.90 |
ENSDART00000103448
|
tbx18
|
T-box 18 |
| chr12_-_10275320 | 0.87 |
ENSDART00000170078
|
mpp2b
|
membrane protein, palmitoylated 2b (MAGUK p55 subfamily member 2) |
| chr15_-_17870090 | 0.83 |
ENSDART00000155066
|
atf5b
|
activating transcription factor 5b |
| chr16_-_29702447 | 0.83 |
ENSDART00000150617
|
tnfaip8l2b
|
tumor necrosis factor, alpha-induced protein 8-like 2b |
| chr6_+_4033832 | 0.82 |
ENSDART00000159952
|
sema5bb
|
sema domain, seven thrombospondin repeats (type 1 and type 1-like), transmembrane domain (TM) and short cytoplasmic domain, (semaphorin) 5Bb |
| chr4_+_22365061 | 0.80 |
ENSDART00000039277
|
lhfpl3
|
LHFPL tetraspan subfamily member 3 |
| chr1_+_50968908 | 0.78 |
ENSDART00000150353
ENSDART00000012842 |
mdh1aa
|
malate dehydrogenase 1Aa, NAD (soluble) |
| chr22_-_30881738 | 0.77 |
ENSDART00000059965
|
si:dkey-49n23.1
|
si:dkey-49n23.1 |
| chr8_-_23776399 | 0.77 |
ENSDART00000114800
|
INAVA
|
si:ch211-163l21.4 |
| chr11_+_20057182 | 0.77 |
ENSDART00000184979
|
fezf2
|
FEZ family zinc finger 2 |
| chr5_-_48268049 | 0.76 |
ENSDART00000187454
|
mef2cb
|
myocyte enhancer factor 2cb |
| chr9_-_32753535 | 0.76 |
ENSDART00000060006
|
olig2
|
oligodendrocyte lineage transcription factor 2 |
| chr13_+_22476742 | 0.76 |
ENSDART00000078759
ENSDART00000130101 ENSDART00000137220 ENSDART00000133065 ENSDART00000147348 |
ldb3a
|
LIM domain binding 3a |
| chr7_-_31759602 | 0.75 |
ENSDART00000113467
|
igdcc4
|
immunoglobulin superfamily, DCC subclass, member 4 |
| chr10_-_24391716 | 0.75 |
ENSDART00000141332
ENSDART00000100772 |
slc43a2b
|
solute carrier family 43 (amino acid system L transporter), member 2b |
| chr13_-_42560662 | 0.74 |
ENSDART00000124898
|
CR792417.1
|
|
| chr3_-_21288202 | 0.74 |
ENSDART00000191766
ENSDART00000187319 |
fam171a2a
|
family with sequence similarity 171, member A2a |
| chr7_+_58699900 | 0.74 |
ENSDART00000144009
|
sdr16c5b
|
short chain dehydrogenase/reductase family 16C, member 5b |
| chr7_-_27686021 | 0.73 |
ENSDART00000079112
ENSDART00000100989 |
calca
|
calcitonin/calcitonin-related polypeptide, alpha |
| chr21_+_25231160 | 0.73 |
ENSDART00000063089
ENSDART00000139127 |
gng8
|
guanine nucleotide binding protein (G protein), gamma 8 |
| chr17_-_37052622 | 0.72 |
ENSDART00000186408
|
dnmt3ab
|
DNA (cytosine-5-)-methyltransferase 3 alpha b |
| chr3_+_57038033 | 0.72 |
ENSDART00000162930
|
bahcc1a
|
BAH domain and coiled-coil containing 1a |
| chr6_+_27146671 | 0.72 |
ENSDART00000156792
|
kif1aa
|
kinesin family member 1Aa |
| chr13_+_30912117 | 0.70 |
ENSDART00000133138
|
drgx
|
dorsal root ganglia homeobox |
| chr15_+_7057050 | 0.70 |
ENSDART00000061828
|
foxl2a
|
forkhead box L2a |
| chr6_+_3004972 | 0.70 |
ENSDART00000186750
ENSDART00000183862 ENSDART00000191485 ENSDART00000171014 |
ptprfa
|
protein tyrosine phosphatase, receptor type, f, a |
| chr6_+_16406723 | 0.69 |
ENSDART00000040035
|
ccdc80l1
|
coiled-coil domain containing 80 like 1 |
| chr17_-_48705993 | 0.68 |
ENSDART00000030934
|
kcnk5a
|
potassium channel, subfamily K, member 5a |
| chr16_-_12173554 | 0.66 |
ENSDART00000110567
ENSDART00000155935 |
clstn3
|
calsyntenin 3 |
| chr17_-_24916889 | 0.66 |
ENSDART00000156157
|
si:ch211-195o20.7
|
si:ch211-195o20.7 |
| chr20_+_28245164 | 0.65 |
ENSDART00000103320
|
dll4
|
delta-like 4 (Drosophila) |
| chr1_+_25801648 | 0.65 |
ENSDART00000129471
|
gucy1b1
|
guanylate cyclase 1 soluble subunit beta 1 |
| chr11_+_38280454 | 0.64 |
ENSDART00000171496
|
CDK18
|
si:dkey-166c18.1 |
| chr11_-_45138857 | 0.64 |
ENSDART00000166501
|
cant1b
|
calcium activated nucleotidase 1b |
| chr16_+_43152727 | 0.64 |
ENSDART00000125590
ENSDART00000154493 |
adam22
|
ADAM metallopeptidase domain 22 |
| chr12_+_35654749 | 0.64 |
ENSDART00000169889
ENSDART00000167873 |
baiap2b
|
BAI1-associated protein 2b |
| chr4_+_72723304 | 0.64 |
ENSDART00000186791
ENSDART00000158902 ENSDART00000191925 |
rab3ip
|
RAB3A interacting protein (rabin3) |
| chr19_-_26869103 | 0.63 |
ENSDART00000089699
|
prrt1
|
proline-rich transmembrane protein 1 |
| chr20_-_22476255 | 0.63 |
ENSDART00000103510
|
pdgfra
|
platelet-derived growth factor receptor, alpha polypeptide |
| chr7_-_30174882 | 0.62 |
ENSDART00000110409
|
frmd5
|
FERM domain containing 5 |
| chr21_-_26918901 | 0.62 |
ENSDART00000100685
|
lrfn4a
|
leucine rich repeat and fibronectin type III domain containing 4a |
| chr17_+_11675362 | 0.61 |
ENSDART00000157911
|
kif26ba
|
kinesin family member 26Ba |
| chr9_-_38036984 | 0.61 |
ENSDART00000134574
|
hacd2
|
3-hydroxyacyl-CoA dehydratase 2 |
| chr11_+_20056732 | 0.60 |
ENSDART00000103997
|
fezf2
|
FEZ family zinc finger 2 |
| chr10_-_27199135 | 0.60 |
ENSDART00000189511
ENSDART00000180314 |
auts2a
|
autism susceptibility candidate 2a |
| chr5_-_23362602 | 0.58 |
ENSDART00000137120
|
gria3a
|
glutamate receptor, ionotropic, AMPA 3a |
| chr11_-_42918971 | 0.58 |
ENSDART00000052912
|
pcdh20
|
protocadherin 20 |
| chr7_-_27685365 | 0.57 |
ENSDART00000188342
|
calca
|
calcitonin/calcitonin-related polypeptide, alpha |
| chr3_-_56541723 | 0.57 |
ENSDART00000156398
ENSDART00000050576 ENSDART00000184874 |
si:ch211-189a21.1
cyth1a
|
si:ch211-189a21.1 cytohesin 1a |
| chr8_+_21588067 | 0.57 |
ENSDART00000172190
|
ajap1
|
adherens junctions associated protein 1 |
| chr23_+_41912151 | 0.55 |
ENSDART00000191115
|
podn
|
podocan |
| chr7_-_22941472 | 0.53 |
ENSDART00000190334
|
tnfsf10l
|
TNF superfamily member 10, like |
| chr4_+_12612723 | 0.53 |
ENSDART00000133767
|
lmo3
|
LIM domain only 3 |
| chr10_-_26738209 | 0.53 |
ENSDART00000188590
|
fgf13b
|
fibroblast growth factor 13b |
| chr20_+_28245587 | 0.52 |
ENSDART00000191703
|
dll4
|
delta-like 4 (Drosophila) |
| chr25_+_13498188 | 0.51 |
ENSDART00000015710
|
snrkb
|
SNF related kinase b |
| chr15_+_21276735 | 0.49 |
ENSDART00000111213
|
ubash3bb
|
ubiquitin associated and SH3 domain containing Bb |
| chr13_+_1089942 | 0.48 |
ENSDART00000054322
|
cnrip1b
|
cannabinoid receptor interacting protein 1b |
| chr22_+_9472814 | 0.47 |
ENSDART00000112125
ENSDART00000138850 |
cacna2d2b
|
calcium channel, voltage-dependent, alpha 2/delta subunit 2b |
| chr7_-_31759394 | 0.47 |
ENSDART00000193040
|
igdcc4
|
immunoglobulin superfamily, DCC subclass, member 4 |
| chr13_+_16279890 | 0.47 |
ENSDART00000101775
ENSDART00000057948 |
anxa11a
|
annexin A11a |
| chr25_+_32755485 | 0.47 |
ENSDART00000162188
|
etfa
|
electron-transfer-flavoprotein, alpha polypeptide |
| chr7_-_39203799 | 0.47 |
ENSDART00000173727
|
chrm4a
|
cholinergic receptor, muscarinic 4a |
| chr9_-_40765868 | 0.46 |
ENSDART00000138634
|
abca12
|
ATP-binding cassette, sub-family A (ABC1), member 12 |
| chr21_-_20733615 | 0.46 |
ENSDART00000145544
|
si:ch211-22d5.2
|
si:ch211-22d5.2 |
| chr15_-_14469704 | 0.44 |
ENSDART00000185077
|
numbl
|
numb homolog (Drosophila)-like |
| chr11_-_27702778 | 0.44 |
ENSDART00000045942
ENSDART00000125352 |
phf2
|
PHD finger protein 2 |
| chr17_-_29902187 | 0.43 |
ENSDART00000009104
|
esrrb
|
estrogen-related receptor beta |
| chr16_-_12173399 | 0.43 |
ENSDART00000142574
|
clstn3
|
calsyntenin 3 |
| chr15_+_5360407 | 0.42 |
ENSDART00000110420
|
or112-1
|
odorant receptor, family A, subfamily 112, member 1 |
| chr4_+_5642696 | 0.42 |
ENSDART00000028941
|
mrps18a
|
mitochondrial ribosomal protein S18A |
| chr8_+_37755099 | 0.42 |
ENSDART00000075708
ENSDART00000140966 |
got1l1
|
glutamic-oxaloacetic transaminase 1 like 1 |
| chr4_+_3980247 | 0.42 |
ENSDART00000049194
|
gpr37b
|
G protein-coupled receptor 37b |
| chr23_-_28141419 | 0.42 |
ENSDART00000133039
|
tac3a
|
tachykinin 3a |
| chr24_-_39186185 | 0.42 |
ENSDART00000123019
ENSDART00000191114 |
nubp2
|
nucleotide binding protein 2 (MinD homolog, E. coli) |
| chr3_-_58798377 | 0.42 |
ENSDART00000161248
|
si:ch73-281f12.4
|
si:ch73-281f12.4 |
| chr6_-_40722480 | 0.41 |
ENSDART00000188187
|
kbtbd12
|
kelch repeat and BTB (POZ) domain containing 12 |
| chr21_-_37973819 | 0.41 |
ENSDART00000133405
|
ripply1
|
ripply transcriptional repressor 1 |
| chr15_-_13208607 | 0.40 |
ENSDART00000184151
|
zgc:172282
|
zgc:172282 |
| chr3_-_58798815 | 0.40 |
ENSDART00000082920
|
si:ch73-281f12.4
|
si:ch73-281f12.4 |
| chr6_-_39344259 | 0.40 |
ENSDART00000104074
|
zgc:158846
|
zgc:158846 |
| chr6_-_40722200 | 0.39 |
ENSDART00000035101
|
kbtbd12
|
kelch repeat and BTB (POZ) domain containing 12 |
| chr3_+_17537352 | 0.38 |
ENSDART00000104549
|
hcrt
|
hypocretin (orexin) neuropeptide precursor |
| chr18_+_43365890 | 0.38 |
ENSDART00000173113
|
si:ch211-129p13.1
|
si:ch211-129p13.1 |
| chr17_+_51499789 | 0.38 |
ENSDART00000187701
|
CABZ01067581.1
|
|
| chr18_+_7283283 | 0.37 |
ENSDART00000141493
|
ANO2 (1 of many)
|
si:ch73-86n2.1 |
| chr21_-_37790727 | 0.36 |
ENSDART00000162907
|
gabrb4
|
gamma-aminobutyric acid (GABA) A receptor, beta 4 |
| chr4_+_2619132 | 0.36 |
ENSDART00000128807
|
gpr22a
|
G protein-coupled receptor 22a |
| chr20_+_37294112 | 0.35 |
ENSDART00000076293
ENSDART00000140450 |
cx23
|
connexin 23 |
| chr6_+_23712911 | 0.35 |
ENSDART00000167795
|
zgc:158654
|
zgc:158654 |
| chr13_-_45155792 | 0.34 |
ENSDART00000163556
|
runx3
|
runt-related transcription factor 3 |
| chr1_-_55270966 | 0.34 |
ENSDART00000152807
|
si:ch211-286b5.5
|
si:ch211-286b5.5 |
| chr21_+_40232910 | 0.33 |
ENSDART00000075915
|
or115-11
|
odorant receptor, family F, subfamily 115, member 11 |
| chr4_-_76370630 | 0.33 |
ENSDART00000168831
ENSDART00000174313 |
si:ch73-158p21.3
|
si:ch73-158p21.3 |
| chr3_-_49110710 | 0.33 |
ENSDART00000160404
|
trim35-12
|
tripartite motif containing 35-12 |
| chr19_-_657439 | 0.32 |
ENSDART00000167100
|
slc6a18
|
solute carrier family 6 (neutral amino acid transporter), member 18 |
| chr10_-_9192450 | 0.32 |
ENSDART00000139783
|
si:dkeyp-41f9.4
|
si:dkeyp-41f9.4 |
| chr8_+_51050554 | 0.32 |
ENSDART00000166249
|
DISP3
|
si:dkey-32e23.6 |
| chr23_+_21405201 | 0.31 |
ENSDART00000144409
|
iffo2a
|
intermediate filament family orphan 2a |
| chr1_-_9123465 | 0.31 |
ENSDART00000081337
|
ndufab1a
|
NADH dehydrogenase (ubiquinone) 1, alpha/beta subcomplex, 1a |
| chr13_+_18533005 | 0.31 |
ENSDART00000136024
|
ftr14l
|
finTRIM family, member 14-like |
| chr13_-_44630111 | 0.30 |
ENSDART00000110092
|
mdga1
|
MAM domain containing glycosylphosphatidylinositol anchor 1 |
| chr18_-_2433011 | 0.30 |
ENSDART00000181922
ENSDART00000193276 |
CR769778.1
|
|
| chr10_+_3507861 | 0.30 |
ENSDART00000092684
|
rph3aa
|
rabphilin 3A homolog (mouse), a |
| chr8_+_20776654 | 0.30 |
ENSDART00000135850
|
nfic
|
nuclear factor I/C |
| chr12_+_41697664 | 0.29 |
ENSDART00000162302
|
bnip3
|
BCL2 interacting protein 3 |
| chr20_-_48516765 | 0.27 |
ENSDART00000150200
|
si:zfos-223e1.2
|
si:zfos-223e1.2 |
| chr14_+_41345175 | 0.27 |
ENSDART00000086104
|
nox1
|
NADPH oxidase 1 |
| chr18_-_40773413 | 0.26 |
ENSDART00000133797
|
vaspb
|
vasodilator stimulated phosphoprotein b |
| chr7_+_36898850 | 0.26 |
ENSDART00000113342
|
tox3
|
TOX high mobility group box family member 3 |
| chr10_-_34870667 | 0.26 |
ENSDART00000161272
|
dclk1a
|
doublecortin-like kinase 1a |
| chr12_+_28117365 | 0.25 |
ENSDART00000066290
|
UTS2R
|
urotensin 2 receptor |
| chr24_+_40473032 | 0.25 |
ENSDART00000084238
ENSDART00000178508 |
CABZ01076968.1
|
|
| chr16_+_42481447 | 0.25 |
ENSDART00000037401
|
herpud2
|
HERPUD family member 2 |
| chr2_-_17044959 | 0.22 |
ENSDART00000090260
|
clcn2a
|
chloride channel, voltage-sensitive 2a |
| chr1_-_57501299 | 0.22 |
ENSDART00000080600
|
zgc:171470
|
zgc:171470 |
| chr7_+_41303572 | 0.21 |
ENSDART00000012692
|
rgs9bp
|
regulator of G protein signaling 9 binding protein |
| chr2_-_10906854 | 0.21 |
ENSDART00000150227
|
cdcp2
|
CUB domain containing protein 2 |
| chr14_+_45471642 | 0.20 |
ENSDART00000126979
ENSDART00000172952 ENSDART00000173284 |
ubxn1
|
UBX domain protein 1 |
| chr15_-_38154616 | 0.20 |
ENSDART00000099392
|
irgq2
|
immunity-related GTPase family, q2 |
| chr11_+_33284837 | 0.20 |
ENSDART00000166239
ENSDART00000111412 |
nkpd1
|
NTPase, KAP family P-loop domain containing 1 |
| chr5_-_30481263 | 0.19 |
ENSDART00000086734
|
phldb1a
|
pleckstrin homology-like domain, family B, member 1a |
| chr7_-_4315859 | 0.19 |
ENSDART00000172762
|
si:ch211-63p21.8
|
si:ch211-63p21.8 |
| chr17_-_45150763 | 0.19 |
ENSDART00000155043
ENSDART00000156786 ENSDART00000191147 |
tmed8
|
transmembrane p24 trafficking protein 8 |
| chr22_+_25814941 | 0.19 |
ENSDART00000161120
|
vasna
|
vasorin a |
| chr5_+_21211135 | 0.18 |
ENSDART00000088492
|
bmp10
|
bone morphogenetic protein 10 |
| chr20_-_40755614 | 0.18 |
ENSDART00000061247
|
cx32.3
|
connexin 32.3 |
| chr24_-_28648949 | 0.18 |
ENSDART00000180227
|
tecrl2a
|
trans-2,3-enoyl-CoA reductase-like 2a |
| chr18_-_8398972 | 0.18 |
ENSDART00000136512
|
BEND7
|
si:ch211-220f12.4 |
| chr9_-_44953664 | 0.17 |
ENSDART00000188558
ENSDART00000185210 |
vil1
|
villin 1 |
| chr8_+_8927870 | 0.17 |
ENSDART00000081985
|
pim2
|
Pim-2 proto-oncogene, serine/threonine kinase |
| chr9_-_19699728 | 0.17 |
ENSDART00000166780
|
si:ch211-141e20.2
|
si:ch211-141e20.2 |
| chr9_-_6624663 | 0.16 |
ENSDART00000092537
|
GPR45
|
si:dkeyp-118h3.5 |
| chr11_+_24620742 | 0.16 |
ENSDART00000182471
ENSDART00000048365 |
syt6b
|
synaptotagmin VIb |
| chr24_-_21404367 | 0.16 |
ENSDART00000152093
|
atp8a2
|
ATPase phospholipid transporting 8A2 |
| chr18_+_34362608 | 0.15 |
ENSDART00000131478
|
kcnab1a
|
potassium voltage-gated channel, shaker-related subfamily, beta member 1 a |
| chr5_-_50641129 | 0.15 |
ENSDART00000190090
|
mctp1a
|
multiple C2 domains, transmembrane 1a |
| chr5_-_26566435 | 0.15 |
ENSDART00000146070
|
arvcfb
|
ARVCF, delta catenin family member b |
| chr2_+_39021282 | 0.15 |
ENSDART00000056577
|
RBP1 (1 of many)
|
si:ch211-119o8.7 |
| chr11_-_37425407 | 0.15 |
ENSDART00000160911
|
erc2
|
ELKS/RAB6-interacting/CAST family member 2 |
| chr2_+_29249204 | 0.14 |
ENSDART00000168957
|
cdh18a
|
cadherin 18, type 2a |
| chr13_-_3370638 | 0.14 |
ENSDART00000029649
|
prkn
|
parkin RBR E3 ubiquitin protein ligase |
| chr16_+_16849220 | 0.13 |
ENSDART00000047409
ENSDART00000142155 |
myh14
|
myosin, heavy chain 14, non-muscle |
| chr14_+_23717165 | 0.12 |
ENSDART00000006373
|
ndfip1
|
Nedd4 family interacting protein 1 |
| chr23_+_6077503 | 0.12 |
ENSDART00000081714
ENSDART00000139834 |
mybpha
|
myosin binding protein Ha |
| chr5_-_35953472 | 0.12 |
ENSDART00000143448
|
rxfp2l
|
relaxin/insulin-like family peptide receptor 2, like |
| chr10_+_19528321 | 0.12 |
ENSDART00000184816
|
vsig8a
|
V-set and immunoglobulin domain containing 8a |
| chr18_-_43993614 | 0.11 |
ENSDART00000148535
ENSDART00000077357 |
bcl9l
|
B cell CLL/lymphoma 9-like |
| chr16_+_26012569 | 0.11 |
ENSDART00000148846
|
prss59.1
|
protease, serine, 59, tandem duplicate 1 |
| chr8_+_6576940 | 0.11 |
ENSDART00000138135
|
vsig8b
|
V-set and immunoglobulin domain containing 8b |
| chr18_+_14529005 | 0.10 |
ENSDART00000186379
|
kcng4a
|
potassium voltage-gated channel, subfamily G, member 4a |
| chr22_+_2830703 | 0.10 |
ENSDART00000145463
ENSDART00000144785 |
si:dkey-20i20.8
|
si:dkey-20i20.8 |
| chr12_-_28983584 | 0.09 |
ENSDART00000112374
|
zgc:171713
|
zgc:171713 |
Network of associatons between targets according to the STRING database.
First level regulatory network of pou6f2
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.6 | 1.9 | GO:0065001 | specification of axis polarity(GO:0065001) |
| 0.4 | 1.2 | GO:0010633 | negative regulation of epithelial cell migration(GO:0010633) |
| 0.3 | 1.8 | GO:0060221 | retinal rod cell differentiation(GO:0060221) |
| 0.2 | 0.7 | GO:0048496 | maintenance of organ identity(GO:0048496) |
| 0.2 | 1.4 | GO:0021767 | mammillary body development(GO:0021767) |
| 0.2 | 0.8 | GO:0021742 | abducens nucleus development(GO:0021742) |
| 0.2 | 0.9 | GO:0099612 | protein localization to juxtaparanode region of axon(GO:0071205) protein localization to axon(GO:0099612) |
| 0.2 | 1.4 | GO:0097106 | postsynaptic density organization(GO:0097106) |
| 0.2 | 0.6 | GO:0099543 | trans-synaptic signaling by soluble gas(GO:0099543) trans-synaptic signaling by nitric oxide(GO:0099548) |
| 0.2 | 0.8 | GO:0006107 | oxaloacetate metabolic process(GO:0006107) |
| 0.1 | 0.4 | GO:0061187 | regulation of chromatin silencing at rDNA(GO:0061187) negative regulation of chromatin silencing at rDNA(GO:0061188) |
| 0.1 | 0.6 | GO:0003261 | cardiac muscle progenitor cell migration to the midline involved in heart field formation(GO:0003261) |
| 0.1 | 1.1 | GO:0051965 | positive regulation of synapse assembly(GO:0051965) |
| 0.1 | 2.1 | GO:0045747 | positive regulation of Notch signaling pathway(GO:0045747) |
| 0.1 | 0.4 | GO:0006531 | aspartate metabolic process(GO:0006531) |
| 0.1 | 1.2 | GO:0071678 | olfactory bulb axon guidance(GO:0071678) |
| 0.1 | 0.4 | GO:0042755 | eating behavior(GO:0042755) |
| 0.1 | 2.8 | GO:0070593 | dendrite self-avoidance(GO:0070593) |
| 0.1 | 1.2 | GO:0042572 | retinol metabolic process(GO:0042572) |
| 0.1 | 1.2 | GO:0060325 | face morphogenesis(GO:0060325) |
| 0.1 | 0.4 | GO:0007217 | tachykinin receptor signaling pathway(GO:0007217) |
| 0.1 | 0.7 | GO:0097374 | sensory neuron axon guidance(GO:0097374) |
| 0.1 | 0.6 | GO:2000251 | positive regulation of actin cytoskeleton reorganization(GO:2000251) |
| 0.1 | 0.5 | GO:0070584 | mitochondrion morphogenesis(GO:0070584) |
| 0.1 | 1.3 | GO:0015671 | oxygen transport(GO:0015671) |
| 0.1 | 0.2 | GO:0072673 | lamellipodium morphogenesis(GO:0072673) regulation of lamellipodium morphogenesis(GO:2000392) |
| 0.1 | 1.0 | GO:0032418 | lysosome localization(GO:0032418) |
| 0.1 | 1.0 | GO:0070570 | regulation of axon regeneration(GO:0048679) regulation of neuron projection regeneration(GO:0070570) |
| 0.1 | 0.5 | GO:0048730 | epidermis morphogenesis(GO:0048730) |
| 0.1 | 0.5 | GO:2000272 | negative regulation of receptor activity(GO:2000272) |
| 0.1 | 0.5 | GO:2001238 | positive regulation of extrinsic apoptotic signaling pathway(GO:2001238) |
| 0.0 | 0.5 | GO:0060217 | hemangioblast cell differentiation(GO:0060217) |
| 0.0 | 1.1 | GO:0002761 | regulation of myeloid leukocyte differentiation(GO:0002761) |
| 0.0 | 1.1 | GO:0007631 | feeding behavior(GO:0007631) |
| 0.0 | 0.5 | GO:0007197 | adenylate cyclase-inhibiting G-protein coupled acetylcholine receptor signaling pathway(GO:0007197) |
| 0.0 | 1.3 | GO:0055064 | chloride ion homeostasis(GO:0055064) |
| 0.0 | 1.0 | GO:0006595 | polyamine metabolic process(GO:0006595) |
| 0.0 | 1.4 | GO:0043049 | otic placode formation(GO:0043049) |
| 0.0 | 1.8 | GO:0036269 | swimming behavior(GO:0036269) |
| 0.0 | 0.5 | GO:1905150 | regulation of voltage-gated sodium channel activity(GO:1905150) |
| 0.0 | 0.1 | GO:1901017 | negative regulation of potassium ion transmembrane transporter activity(GO:1901017) negative regulation of potassium ion transmembrane transport(GO:1901380) negative regulation of delayed rectifier potassium channel activity(GO:1902260) negative regulation of voltage-gated potassium channel activity(GO:1903817) |
| 0.0 | 0.3 | GO:0042554 | superoxide anion generation(GO:0042554) |
| 0.0 | 0.3 | GO:0097345 | mitochondrial outer membrane permeabilization(GO:0097345) |
| 0.0 | 0.4 | GO:0032525 | somite rostral/caudal axis specification(GO:0032525) |
| 0.0 | 1.0 | GO:0021761 | limbic system development(GO:0021761) hypothalamus development(GO:0021854) |
| 0.0 | 0.6 | GO:0021884 | forebrain neuron development(GO:0021884) |
| 0.0 | 0.7 | GO:0030322 | stabilization of membrane potential(GO:0030322) |
| 0.0 | 1.6 | GO:1902668 | negative regulation of axon extension involved in axon guidance(GO:0048843) negative regulation of axon guidance(GO:1902668) |
| 0.0 | 0.6 | GO:0030497 | fatty acid elongation(GO:0030497) |
| 0.0 | 0.4 | GO:0031163 | iron-sulfur cluster assembly(GO:0016226) metallo-sulfur cluster assembly(GO:0031163) |
| 0.0 | 0.7 | GO:0015804 | neutral amino acid transport(GO:0015804) |
| 0.0 | 0.1 | GO:0010387 | COP9 signalosome assembly(GO:0010387) |
| 0.0 | 0.8 | GO:0060914 | heart formation(GO:0060914) |
| 0.0 | 1.1 | GO:0006805 | xenobiotic metabolic process(GO:0006805) |
| 0.0 | 0.0 | GO:0003214 | cardiac left ventricle morphogenesis(GO:0003214) cardiac left ventricle formation(GO:0003218) |
| 0.0 | 0.5 | GO:0032506 | cytokinetic process(GO:0032506) |
| 0.0 | 1.9 | GO:0007160 | cell-matrix adhesion(GO:0007160) |
| 0.0 | 0.4 | GO:0060319 | primitive erythrocyte differentiation(GO:0060319) |
| 0.0 | 1.8 | GO:0001756 | somitogenesis(GO:0001756) |
| 0.0 | 0.1 | GO:1900407 | regulation of cellular response to oxidative stress(GO:1900407) |
| 0.0 | 0.1 | GO:0048790 | maintenance of presynaptic active zone structure(GO:0048790) maintenance of synapse structure(GO:0099558) |
| 0.0 | 0.2 | GO:0050919 | negative chemotaxis(GO:0050919) |
| 0.0 | 0.6 | GO:0032012 | ARF protein signal transduction(GO:0032011) regulation of ARF protein signal transduction(GO:0032012) |
| 0.0 | 0.4 | GO:0071910 | determination of liver left/right asymmetry(GO:0071910) |
| 0.0 | 0.5 | GO:0042149 | cellular response to glucose starvation(GO:0042149) |
| 0.0 | 0.3 | GO:0001843 | neural tube closure(GO:0001843) |
| 0.0 | 0.1 | GO:0007190 | activation of adenylate cyclase activity(GO:0007190) positive regulation of cAMP metabolic process(GO:0030816) positive regulation of cAMP biosynthetic process(GO:0030819) positive regulation of adenylate cyclase activity(GO:0045762) |
| 0.0 | 0.2 | GO:0031397 | negative regulation of protein ubiquitination(GO:0031397) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.6 | GO:0070319 | Golgi to plasma membrane transport vesicle(GO:0070319) |
| 0.1 | 0.6 | GO:0089717 | spanning component of plasma membrane(GO:0044214) spanning component of membrane(GO:0089717) |
| 0.1 | 0.9 | GO:0033010 | paranodal junction(GO:0033010) |
| 0.1 | 1.3 | GO:0031838 | haptoglobin-hemoglobin complex(GO:0031838) |
| 0.1 | 1.1 | GO:0034751 | aryl hydrocarbon receptor complex(GO:0034751) |
| 0.1 | 1.1 | GO:0031680 | G-protein beta/gamma-subunit complex(GO:0031680) |
| 0.0 | 0.2 | GO:0032433 | filopodium tip(GO:0032433) |
| 0.0 | 0.4 | GO:0045095 | keratin filament(GO:0045095) |
| 0.0 | 0.3 | GO:0043020 | NADPH oxidase complex(GO:0043020) |
| 0.0 | 1.2 | GO:0017053 | transcriptional repressor complex(GO:0017053) |
| 0.0 | 3.3 | GO:0030018 | Z disc(GO:0030018) |
| 0.0 | 0.7 | GO:0031941 | filamentous actin(GO:0031941) |
| 0.0 | 0.4 | GO:0005763 | organellar small ribosomal subunit(GO:0000314) mitochondrial small ribosomal subunit(GO:0005763) |
| 0.0 | 1.3 | GO:0030672 | synaptic vesicle membrane(GO:0030672) exocytic vesicle membrane(GO:0099501) |
| 0.0 | 4.8 | GO:0030424 | axon(GO:0030424) |
| 0.0 | 0.5 | GO:0030426 | growth cone(GO:0030426) |
| 0.0 | 0.6 | GO:0032281 | AMPA glutamate receptor complex(GO:0032281) |
| 0.0 | 0.8 | GO:0048786 | presynaptic active zone(GO:0048786) |
| 0.0 | 0.9 | GO:0031902 | late endosome membrane(GO:0031902) |
| 0.0 | 1.0 | GO:0005581 | collagen trimer(GO:0005581) |
| 0.0 | 0.4 | GO:1902711 | GABA receptor complex(GO:1902710) GABA-A receptor complex(GO:1902711) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.4 | 1.3 | GO:0031716 | calcitonin receptor binding(GO:0031716) |
| 0.4 | 1.2 | GO:0047173 | phosphatidylcholine-retinol O-acyltransferase activity(GO:0047173) |
| 0.3 | 3.0 | GO:0016918 | retinal binding(GO:0016918) |
| 0.2 | 2.5 | GO:0051373 | telethonin binding(GO:0031433) FATZ binding(GO:0051373) |
| 0.2 | 1.0 | GO:0008073 | ornithine decarboxylase inhibitor activity(GO:0008073) |
| 0.2 | 1.1 | GO:0031841 | neuropeptide Y receptor binding(GO:0031841) type 2 neuropeptide Y receptor binding(GO:0031843) |
| 0.2 | 1.9 | GO:0005391 | sodium:potassium-exchanging ATPase activity(GO:0005391) |
| 0.2 | 0.6 | GO:0005017 | platelet-derived growth factor-activated receptor activity(GO:0005017) platelet-derived growth factor binding(GO:0048407) |
| 0.2 | 0.8 | GO:0030060 | L-malate dehydrogenase activity(GO:0030060) |
| 0.2 | 0.6 | GO:0102345 | 3-hydroxyacyl-CoA dehydratase activity(GO:0018812) 3-hydroxy-behenoyl-CoA dehydratase activity(GO:0102344) 3-hydroxy-lignoceroyl-CoA dehydratase activity(GO:0102345) |
| 0.1 | 1.3 | GO:0031720 | haptoglobin binding(GO:0031720) |
| 0.1 | 0.4 | GO:0004069 | L-aspartate:2-oxoglutarate aminotransferase activity(GO:0004069) |
| 0.1 | 0.3 | GO:0000035 | acyl binding(GO:0000035) |
| 0.1 | 2.4 | GO:0005112 | Notch binding(GO:0005112) |
| 0.1 | 1.2 | GO:0005021 | vascular endothelial growth factor-activated receptor activity(GO:0005021) |
| 0.1 | 2.8 | GO:0098632 | protein binding involved in cell-cell adhesion(GO:0098632) |
| 0.1 | 1.2 | GO:0070001 | aspartic-type endopeptidase activity(GO:0004190) aspartic-type peptidase activity(GO:0070001) |
| 0.1 | 0.4 | GO:0070181 | small ribosomal subunit rRNA binding(GO:0070181) |
| 0.1 | 0.7 | GO:0003886 | DNA (cytosine-5-)-methyltransferase activity(GO:0003886) |
| 0.1 | 1.1 | GO:0031681 | G-protein beta-subunit binding(GO:0031681) |
| 0.0 | 0.5 | GO:0016907 | G-protein coupled acetylcholine receptor activity(GO:0016907) |
| 0.0 | 0.6 | GO:0004971 | AMPA glutamate receptor activity(GO:0004971) |
| 0.0 | 0.8 | GO:0051393 | muscle alpha-actinin binding(GO:0051371) alpha-actinin binding(GO:0051393) |
| 0.0 | 0.4 | GO:0032454 | histone demethylase activity (H3-K9 specific)(GO:0032454) |
| 0.0 | 0.6 | GO:0004383 | guanylate cyclase activity(GO:0004383) |
| 0.0 | 1.9 | GO:0005178 | integrin binding(GO:0005178) |
| 0.0 | 0.1 | GO:0050699 | WW domain binding(GO:0050699) |
| 0.0 | 2.1 | GO:0031490 | chromatin DNA binding(GO:0031490) |
| 0.0 | 0.3 | GO:0016175 | superoxide-generating NADPH oxidase activity(GO:0016175) |
| 0.0 | 1.6 | GO:0030215 | semaphorin receptor binding(GO:0030215) |
| 0.0 | 0.7 | GO:0022842 | leak channel activity(GO:0022840) potassium ion leak channel activity(GO:0022841) narrow pore channel activity(GO:0022842) |
| 0.0 | 0.3 | GO:0005522 | profilin binding(GO:0005522) |
| 0.0 | 0.7 | GO:0008574 | ATP-dependent microtubule motor activity, plus-end-directed(GO:0008574) |
| 0.0 | 0.5 | GO:0017080 | sodium channel regulator activity(GO:0017080) |
| 0.0 | 0.6 | GO:0005164 | tumor necrosis factor receptor binding(GO:0005164) |
| 0.0 | 0.7 | GO:0015175 | neutral amino acid transmembrane transporter activity(GO:0015175) |
| 0.0 | 0.1 | GO:0005093 | Rab GDP-dissociation inhibitor activity(GO:0005093) |
| 0.0 | 0.4 | GO:0017110 | nucleoside-diphosphatase activity(GO:0017110) |
| 0.0 | 0.7 | GO:0008013 | beta-catenin binding(GO:0008013) |
| 0.0 | 0.2 | GO:0005247 | voltage-gated chloride channel activity(GO:0005247) |
| 0.0 | 1.6 | GO:0004879 | RNA polymerase II transcription factor activity, ligand-activated sequence-specific DNA binding(GO:0004879) |
| 0.0 | 0.2 | GO:0048495 | Roundabout binding(GO:0048495) |
| 0.0 | 0.4 | GO:0005184 | neuropeptide hormone activity(GO:0005184) |
| 0.0 | 0.4 | GO:0004890 | GABA-A receptor activity(GO:0004890) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 2.0 | NABA PROTEOGLYCANS | Genes encoding proteoglycans |
| 0.0 | 0.6 | PID PDGFRA PATHWAY | PDGFR-alpha signaling pathway |
| 0.0 | 0.2 | PID CONE PATHWAY | Visual signal transduction: Cones |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 1.2 | REACTOME RECEPTOR LIGAND BINDING INITIATES THE SECOND PROTEOLYTIC CLEAVAGE OF NOTCH RECEPTOR | Genes involved in Receptor-ligand binding initiates the second proteolytic cleavage of Notch receptor |
| 0.1 | 1.4 | REACTOME KERATAN SULFATE DEGRADATION | Genes involved in Keratan sulfate degradation |
| 0.1 | 1.3 | REACTOME AMYLOIDS | Genes involved in Amyloids |
| 0.0 | 0.7 | REACTOME INHIBITION OF INSULIN SECRETION BY ADRENALINE NORADRENALINE | Genes involved in Inhibition of Insulin Secretion by Adrenaline/Noradrenaline |
| 0.0 | 0.5 | REACTOME ABCA TRANSPORTERS IN LIPID HOMEOSTASIS | Genes involved in ABCA transporters in lipid homeostasis |
| 0.0 | 0.3 | REACTOME NA CL DEPENDENT NEUROTRANSMITTER TRANSPORTERS | Genes involved in Na+/Cl- dependent neurotransmitter transporters |