Project
PRJNA195909:zebrafish embryo and larva development
Navigation
Downloads
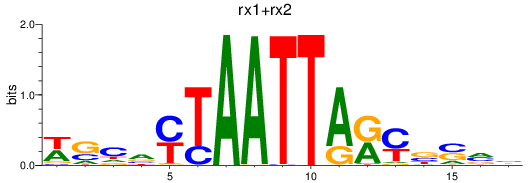
Results for rx1+rx2
Z-value: 0.34
Transcription factors associated with rx1+rx2
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
rx2
|
ENSDARG00000040321 | retinal homeobox gene 2 |
|
rx1
|
ENSDARG00000071684 | retinal homeobox gene 1 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| rx2 | dr11_v1_chr2_-_55861351_55861351 | 0.84 | 5.1e-03 | Click! |
| rx1 | dr11_v1_chr22_-_5006801_5006801 | 0.79 | 1.1e-02 | Click! |
Activity profile of rx1+rx2 motif
Sorted Z-values of rx1+rx2 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr4_-_9891874 | 0.69 |
ENSDART00000067193
|
adm2a
|
adrenomedullin 2a |
| chr18_-_5598958 | 0.54 |
ENSDART00000161538
|
cyp1a
|
cytochrome P450, family 1, subfamily A |
| chr21_+_6751760 | 0.49 |
ENSDART00000135914
|
olfm1b
|
olfactomedin 1b |
| chr5_+_32260502 | 0.47 |
ENSDART00000149020
|
si:ch211-158m24.12
|
si:ch211-158m24.12 |
| chr21_+_6751405 | 0.47 |
ENSDART00000037265
ENSDART00000146371 |
olfm1b
|
olfactomedin 1b |
| chr8_+_16025554 | 0.43 |
ENSDART00000110171
|
elavl4
|
ELAV like neuron-specific RNA binding protein 4 |
| chr22_-_10121880 | 0.42 |
ENSDART00000002348
|
rdh5
|
retinol dehydrogenase 5 (11-cis/9-cis) |
| chr16_+_23984179 | 0.41 |
ENSDART00000175879
|
apoc2
|
apolipoprotein C-II |
| chr21_-_43015383 | 0.40 |
ENSDART00000065097
|
dpysl3
|
dihydropyrimidinase-like 3 |
| chr23_+_43255328 | 0.38 |
ENSDART00000102712
|
tgm2a
|
transglutaminase 2, C polypeptide A |
| chr15_-_5815006 | 0.37 |
ENSDART00000102459
|
rbp2a
|
retinol binding protein 2a, cellular |
| chr8_-_16697615 | 0.36 |
ENSDART00000187929
|
rpe65b
|
retinal pigment epithelium-specific protein 65b |
| chr18_+_9171778 | 0.32 |
ENSDART00000101192
|
sema3d
|
sema domain, immunoglobulin domain (Ig), short basic domain, secreted, (semaphorin) 3D |
| chr6_-_7720332 | 0.31 |
ENSDART00000135945
|
rpsa
|
ribosomal protein SA |
| chr3_-_53533128 | 0.29 |
ENSDART00000183591
|
notch3
|
notch 3 |
| chr23_-_15216654 | 0.28 |
ENSDART00000131649
|
sulf2b
|
sulfatase 2b |
| chr3_-_26017831 | 0.28 |
ENSDART00000179982
|
hmox1a
|
heme oxygenase 1a |
| chr17_-_49978986 | 0.27 |
ENSDART00000154728
|
col12a1a
|
collagen, type XII, alpha 1a |
| chr15_+_23799461 | 0.26 |
ENSDART00000154885
|
si:ch211-167j9.4
|
si:ch211-167j9.4 |
| chr8_+_29986265 | 0.26 |
ENSDART00000148258
|
ptch1
|
patched 1 |
| chr4_+_21129752 | 0.25 |
ENSDART00000169764
|
syt1a
|
synaptotagmin Ia |
| chr3_+_28953274 | 0.25 |
ENSDART00000133528
ENSDART00000103602 |
lgals2a
|
lectin, galactoside-binding, soluble, 2a |
| chr8_+_25079470 | 0.23 |
ENSDART00000000744
|
sypl2b
|
synaptophysin-like 2b |
| chr19_+_42983613 | 0.23 |
ENSDART00000033724
|
fabp3
|
fatty acid binding protein 3, muscle and heart |
| chr25_+_11008419 | 0.23 |
ENSDART00000156589
|
mhc1lia
|
major histocompatibility complex class I LIA |
| chr7_+_6652967 | 0.22 |
ENSDART00000102681
|
pnp5a
|
purine nucleoside phosphorylase 5a |
| chr24_+_38301080 | 0.21 |
ENSDART00000105672
|
mybpc2b
|
myosin binding protein C, fast type b |
| chr6_+_57541776 | 0.21 |
ENSDART00000157330
|
necab3
|
N-terminal EF-hand calcium binding protein 3 |
| chr18_+_21408794 | 0.20 |
ENSDART00000140161
|
necab2
|
N-terminal EF-hand calcium binding protein 2 |
| chr5_-_12219572 | 0.20 |
ENSDART00000167834
|
nos1
|
nitric oxide synthase 1 (neuronal) |
| chr19_-_42391383 | 0.20 |
ENSDART00000110075
ENSDART00000087002 |
plekho1a
|
pleckstrin homology domain containing, family O member 1a |
| chr20_+_43925266 | 0.20 |
ENSDART00000037379
|
clic5b
|
chloride intracellular channel 5b |
| chr4_-_4387012 | 0.18 |
ENSDART00000191836
|
CU468826.3
|
Danio rerio U2 small nuclear ribonucleoprotein auxiliary factor 35 kDa subunit-related protein 1-like (LOC100331497), mRNA. |
| chr5_+_58550795 | 0.18 |
ENSDART00000192282
|
pou2f3
|
POU class 2 homeobox 3 |
| chr9_+_21165484 | 0.17 |
ENSDART00000177286
|
si:rp71-68n21.9
|
si:rp71-68n21.9 |
| chr14_-_33824329 | 0.17 |
ENSDART00000189271
ENSDART00000180458 |
vimr2
|
vimentin-related 2 |
| chr16_-_32013913 | 0.16 |
ENSDART00000030282
ENSDART00000138701 |
gstk1
|
glutathione S-transferase kappa 1 |
| chr4_+_9400012 | 0.16 |
ENSDART00000191960
|
tmtc1
|
transmembrane and tetratricopeptide repeat containing 1 |
| chr14_-_413273 | 0.16 |
ENSDART00000163976
ENSDART00000179907 |
FAT4
|
FAT atypical cadherin 4 |
| chr15_-_22074315 | 0.16 |
ENSDART00000149830
|
drd2a
|
dopamine receptor D2a |
| chr7_+_46368520 | 0.15 |
ENSDART00000192821
|
znf536
|
zinc finger protein 536 |
| chr6_+_7421898 | 0.15 |
ENSDART00000043946
|
ccdc65
|
coiled-coil domain containing 65 |
| chr1_+_21731382 | 0.15 |
ENSDART00000054395
|
pax5
|
paired box 5 |
| chr15_-_14552101 | 0.14 |
ENSDART00000171169
|
numbl
|
numb homolog (Drosophila)-like |
| chr24_-_31425799 | 0.14 |
ENSDART00000157998
|
cngb3.1
|
cyclic nucleotide gated channel beta 3, tandem duplicate 1 |
| chr24_-_36680261 | 0.14 |
ENSDART00000059507
|
ccr10
|
chemokine (C-C motif) receptor 10 |
| chr7_-_33960170 | 0.14 |
ENSDART00000180766
|
skor1a
|
SKI family transcriptional corepressor 1a |
| chr2_+_50608099 | 0.13 |
ENSDART00000185805
ENSDART00000111135 |
neurod6b
|
neuronal differentiation 6b |
| chr3_+_17537352 | 0.13 |
ENSDART00000104549
|
hcrt
|
hypocretin (orexin) neuropeptide precursor |
| chr16_-_25568512 | 0.13 |
ENSDART00000149411
|
atxn1b
|
ataxin 1b |
| chr13_+_4871886 | 0.12 |
ENSDART00000132301
|
micu1
|
mitochondrial calcium uptake 1 |
| chr11_-_21528056 | 0.12 |
ENSDART00000181626
|
srgap2
|
SLIT-ROBO Rho GTPase activating protein 2 |
| chr14_+_30795559 | 0.12 |
ENSDART00000006132
|
cfl1
|
cofilin 1 |
| chr11_+_7264457 | 0.11 |
ENSDART00000154182
|
reep6
|
receptor accessory protein 6 |
| chr19_-_5669122 | 0.10 |
ENSDART00000112211
|
si:ch211-264f5.2
|
si:ch211-264f5.2 |
| chr8_-_24332284 | 0.09 |
ENSDART00000163241
|
pimr142
|
Pim proto-oncogene, serine/threonine kinase, related 142 |
| chr18_+_28106139 | 0.09 |
ENSDART00000089615
|
kiaa1549lb
|
KIAA1549-like b |
| chr1_+_44127292 | 0.09 |
ENSDART00000160542
|
cabp2a
|
calcium binding protein 2a |
| chr9_-_52962521 | 0.09 |
ENSDART00000170419
|
CU855885.1
|
|
| chr5_+_1079423 | 0.09 |
ENSDART00000172231
|
si:zfos-128g4.2
|
si:zfos-128g4.2 |
| chr3_-_23407720 | 0.09 |
ENSDART00000155658
|
rapgefl1
|
Rap guanine nucleotide exchange factor (GEF)-like 1 |
| chr4_+_6032640 | 0.09 |
ENSDART00000157487
|
tfec
|
transcription factor EC |
| chr15_+_5360407 | 0.08 |
ENSDART00000110420
|
or112-1
|
odorant receptor, family A, subfamily 112, member 1 |
| chr7_+_54642005 | 0.08 |
ENSDART00000171864
|
fgf19
|
fibroblast growth factor 19 |
| chr1_-_44701313 | 0.08 |
ENSDART00000193926
|
si:dkey-28b4.8
|
si:dkey-28b4.8 |
| chr16_-_51288178 | 0.08 |
ENSDART00000079864
|
zgc:173729
|
zgc:173729 |
| chr8_+_18011522 | 0.08 |
ENSDART00000136756
|
ssbp3b
|
single stranded DNA binding protein 3b |
| chr10_-_5581487 | 0.07 |
ENSDART00000141943
|
syk
|
spleen tyrosine kinase |
| chr11_+_16153207 | 0.07 |
ENSDART00000192356
|
tada3l
|
transcriptional adaptor 3 (NGG1 homolog, yeast)-like |
| chr7_+_34620418 | 0.06 |
ENSDART00000081338
|
slc9a5
|
solute carrier family 9, subfamily A (NHE5, cation proton antiporter 5), member 5 |
| chr9_+_43799829 | 0.05 |
ENSDART00000186240
|
ube2e3
|
ubiquitin-conjugating enzyme E2E 3 (UBC4/5 homolog, yeast) |
| chr8_-_24502997 | 0.05 |
ENSDART00000184801
ENSDART00000179794 |
BX296549.1
|
|
| chr2_+_53204750 | 0.05 |
ENSDART00000163644
|
zgc:165603
|
zgc:165603 |
| chr6_+_24420523 | 0.05 |
ENSDART00000185461
|
tgfbr3
|
transforming growth factor, beta receptor III |
| chr20_-_13774826 | 0.05 |
ENSDART00000063831
|
opn8c
|
opsin 8, group member c |
| chr11_-_16152400 | 0.05 |
ENSDART00000123665
|
arpc4l
|
actin related protein 2/3 complex, subunit 4, like |
| chr16_+_13818500 | 0.05 |
ENSDART00000135245
|
flcn
|
folliculin |
| chr16_+_35379720 | 0.05 |
ENSDART00000170497
|
si:dkey-34d22.2
|
si:dkey-34d22.2 |
| chr8_-_50888806 | 0.05 |
ENSDART00000053750
|
acsl2
|
acyl-CoA synthetase long chain family member 2 |
| chr7_+_26173751 | 0.04 |
ENSDART00000065131
|
si:ch211-196f2.7
|
si:ch211-196f2.7 |
| chr22_+_7738966 | 0.04 |
ENSDART00000147073
|
si:ch73-44m9.5
|
si:ch73-44m9.5 |
| chr5_-_68074592 | 0.04 |
ENSDART00000165052
ENSDART00000018792 |
spag7
|
sperm associated antigen 7 |
| chr16_-_11798994 | 0.04 |
ENSDART00000135408
|
cnfn
|
cornifelin |
| chr8_-_24366726 | 0.04 |
ENSDART00000182351
ENSDART00000187099 ENSDART00000181534 |
CR450805.1
|
|
| chr8_+_28695914 | 0.04 |
ENSDART00000033386
|
ocstamp
|
osteoclast stimulatory transmembrane protein |
| chr15_+_45640906 | 0.04 |
ENSDART00000149361
ENSDART00000149079 |
sagb
|
S-antigen; retina and pineal gland (arrestin) b |
| chr14_-_4556896 | 0.04 |
ENSDART00000044678
ENSDART00000192863 |
GABRA2
|
gamma-aminobutyric acid type A receptor alpha2 subunit |
| chr4_+_47636303 | 0.03 |
ENSDART00000167272
ENSDART00000166961 |
BX324142.1
|
|
| chr10_+_35358675 | 0.03 |
ENSDART00000193263
|
si:dkey-259j3.5
|
si:dkey-259j3.5 |
| chr22_-_20011476 | 0.03 |
ENSDART00000093312
ENSDART00000093310 |
celf5a
|
cugbp, Elav-like family member 5a |
| chr1_-_19502322 | 0.03 |
ENSDART00000181888
ENSDART00000044030 |
kitb
|
v-kit Hardy-Zuckerman 4 feline sarcoma viral oncogene homolog b |
| chr8_-_53044089 | 0.03 |
ENSDART00000135982
|
nr6a1a
|
nuclear receptor subfamily 6, group A, member 1a |
| chr8_-_25034411 | 0.03 |
ENSDART00000135973
|
nfyal
|
nuclear transcription factor Y, alpha, like |
| chr16_-_27677930 | 0.03 |
ENSDART00000145991
|
tbrg4
|
transforming growth factor beta regulator 4 |
| chr4_-_56954002 | 0.03 |
ENSDART00000160934
|
si:dkey-269o24.1
|
si:dkey-269o24.1 |
| chr25_+_20715950 | 0.03 |
ENSDART00000180223
|
ergic2
|
ERGIC and golgi 2 |
| chr20_-_9095105 | 0.02 |
ENSDART00000140792
|
oma1
|
OMA1 zinc metallopeptidase |
| chr18_+_17827149 | 0.02 |
ENSDART00000190237
ENSDART00000189345 |
ZNF423
|
si:ch211-216l23.1 |
| chr12_-_41684729 | 0.02 |
ENSDART00000184461
|
jakmip3
|
Janus kinase and microtubule interacting protein 3 |
| chr1_-_17715493 | 0.01 |
ENSDART00000133027
|
si:dkey-256e7.8
|
si:dkey-256e7.8 |
| chr24_+_37640626 | 0.01 |
ENSDART00000008047
|
wdr24
|
WD repeat domain 24 |
| chr11_+_16152316 | 0.01 |
ENSDART00000081054
|
tada3l
|
transcriptional adaptor 3 (NGG1 homolog, yeast)-like |
| chr16_+_54263921 | 0.01 |
ENSDART00000002856
|
drd2l
|
dopamine receptor D2 like |
| chr13_-_37229245 | 0.01 |
ENSDART00000140923
|
si:dkeyp-77c8.5
|
si:dkeyp-77c8.5 |
| chr8_+_25034544 | 0.01 |
ENSDART00000123300
|
ngrn
|
neugrin, neurite outgrowth associated |
| chr8_-_12264486 | 0.00 |
ENSDART00000091612
ENSDART00000135812 |
dab2ipa
|
DAB2 interacting protein a |
Network of associatons between targets according to the STRING database.
First level regulatory network of rx1+rx2
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.3 | GO:0035019 | somatic stem cell population maintenance(GO:0035019) symmetric cell division(GO:0098725) |
| 0.1 | 0.3 | GO:0007585 | respiratory gaseous exchange(GO:0007585) regulation of respiratory gaseous exchange(GO:0043576) |
| 0.1 | 0.4 | GO:1901825 | tetraterpenoid metabolic process(GO:0016108) tetraterpenoid biosynthetic process(GO:0016109) carotenoid metabolic process(GO:0016116) carotenoid biosynthetic process(GO:0016117) xanthophyll metabolic process(GO:0016122) xanthophyll biosynthetic process(GO:0016123) zeaxanthin metabolic process(GO:1901825) zeaxanthin biosynthetic process(GO:1901827) |
| 0.1 | 0.4 | GO:0042362 | fat-soluble vitamin biosynthetic process(GO:0042362) |
| 0.1 | 0.2 | GO:0098924 | retrograde trans-synaptic signaling by soluble gas(GO:0098923) retrograde trans-synaptic signaling by nitric oxide(GO:0098924) |
| 0.1 | 0.3 | GO:0002164 | larval development(GO:0002164) larval heart development(GO:0007508) |
| 0.1 | 0.4 | GO:0042984 | amyloid precursor protein biosynthetic process(GO:0042983) regulation of amyloid precursor protein biosynthetic process(GO:0042984) |
| 0.0 | 0.4 | GO:0051491 | positive regulation of filopodium assembly(GO:0051491) |
| 0.0 | 0.1 | GO:0030043 | actin filament fragmentation(GO:0030043) |
| 0.0 | 0.1 | GO:0042755 | eating behavior(GO:0042755) |
| 0.0 | 0.3 | GO:0048635 | negative regulation of muscle organ development(GO:0048635) |
| 0.0 | 0.1 | GO:0045579 | positive regulation of B cell differentiation(GO:0045579) |
| 0.0 | 0.1 | GO:2001223 | negative regulation of neuron migration(GO:2001223) |
| 0.0 | 0.4 | GO:0042953 | lipoprotein transport(GO:0042953) lipoprotein localization(GO:0044872) |
| 0.0 | 0.1 | GO:0051561 | positive regulation of mitochondrial calcium ion concentration(GO:0051561) |
| 0.0 | 0.1 | GO:0003352 | regulation of cilium movement(GO:0003352) |
| 0.0 | 0.3 | GO:0090303 | positive regulation of wound healing(GO:0090303) |
| 0.0 | 0.4 | GO:0018149 | peptide cross-linking(GO:0018149) |
| 0.0 | 0.1 | GO:0060114 | reflex(GO:0060004) vestibular reflex(GO:0060005) vestibular receptor cell differentiation(GO:0060114) vestibular receptor cell development(GO:0060118) |
| 0.0 | 0.2 | GO:0014846 | esophagus smooth muscle contraction(GO:0014846) glial cell-derived neurotrophic factor receptor signaling pathway(GO:0035860) |
| 0.0 | 0.3 | GO:0000028 | ribosomal small subunit assembly(GO:0000028) |
| 0.0 | 0.3 | GO:0070570 | regulation of axon regeneration(GO:0048679) regulation of neuron projection regeneration(GO:0070570) |
| 0.0 | 0.2 | GO:0051967 | negative regulation of cytosolic calcium ion concentration(GO:0051481) negative regulation of synaptic transmission, glutamatergic(GO:0051967) |
| 0.0 | 0.3 | GO:0048791 | calcium ion-regulated exocytosis of neurotransmitter(GO:0048791) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.3 | GO:0042584 | chromaffin granule membrane(GO:0042584) |
| 0.0 | 0.4 | GO:0034362 | low-density lipoprotein particle(GO:0034362) |
| 0.0 | 0.8 | GO:0030426 | growth cone(GO:0030426) |
| 0.0 | 0.1 | GO:1990246 | uniplex complex(GO:1990246) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.4 | GO:0050251 | retinol isomerase activity(GO:0050251) all-trans-retinyl-ester hydrolase, 11-cis retinol forming activity(GO:0052885) |
| 0.1 | 0.5 | GO:0070330 | aromatase activity(GO:0070330) |
| 0.1 | 0.3 | GO:0097108 | hedgehog family protein binding(GO:0097108) |
| 0.0 | 0.3 | GO:0004392 | heme oxygenase (decyclizing) activity(GO:0004392) |
| 0.0 | 0.4 | GO:0004157 | dihydropyrimidinase activity(GO:0004157) |
| 0.0 | 0.2 | GO:0004517 | nitric-oxide synthase activity(GO:0004517) |
| 0.0 | 0.3 | GO:0016936 | galactoside binding(GO:0016936) |
| 0.0 | 0.2 | GO:0004731 | purine-nucleoside phosphorylase activity(GO:0004731) |
| 0.0 | 0.3 | GO:0038191 | neuropilin binding(GO:0038191) |
| 0.0 | 0.4 | GO:0003810 | protein-glutamine gamma-glutamyltransferase activity(GO:0003810) |
| 0.0 | 0.3 | GO:0043236 | laminin binding(GO:0043236) |
| 0.0 | 0.2 | GO:0004169 | dolichyl-phosphate-mannose-protein mannosyltransferase activity(GO:0004169) |
| 0.0 | 0.2 | GO:0001591 | dopamine neurotransmitter receptor activity, coupled via Gi/Go(GO:0001591) |
| 0.0 | 0.2 | GO:0008449 | N-acetylglucosamine-6-sulfatase activity(GO:0008449) |
| 0.0 | 0.3 | GO:0005112 | Notch binding(GO:0005112) |
| 0.0 | 0.1 | GO:0005223 | intracellular cyclic nucleotide activated cation channel activity(GO:0005221) intracellular cAMP activated cation channel activity(GO:0005222) intracellular cGMP activated cation channel activity(GO:0005223) cyclic nucleotide-gated ion channel activity(GO:0043855) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.4 | PID CONE PATHWAY | Visual signal transduction: Cones |
| 0.0 | 0.1 | SA FAS SIGNALING | The TNF-type receptor Fas induces apoptosis on ligand binding. |
| 0.0 | 0.3 | PID HEDGEHOG 2PATHWAY | Signaling events mediated by the Hedgehog family |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.4 | REACTOME CHYLOMICRON MEDIATED LIPID TRANSPORT | Genes involved in Chylomicron-mediated lipid transport |
| 0.0 | 0.4 | REACTOME CRMPS IN SEMA3A SIGNALING | Genes involved in CRMPs in Sema3A signaling |
| 0.0 | 0.3 | REACTOME RECEPTOR LIGAND BINDING INITIATES THE SECOND PROTEOLYTIC CLEAVAGE OF NOTCH RECEPTOR | Genes involved in Receptor-ligand binding initiates the second proteolytic cleavage of Notch receptor |
| 0.0 | 0.1 | REACTOME CHEMOKINE RECEPTORS BIND CHEMOKINES | Genes involved in Chemokine receptors bind chemokines |
| 0.0 | 0.1 | REACTOME SEMA3A PAK DEPENDENT AXON REPULSION | Genes involved in Sema3A PAK dependent Axon repulsion |
| 0.0 | 0.2 | REACTOME LATENT INFECTION OF HOMO SAPIENS WITH MYCOBACTERIUM TUBERCULOSIS | Genes involved in Latent infection of Homo sapiens with Mycobacterium tuberculosis |
| 0.0 | 0.1 | REACTOME FGFR4 LIGAND BINDING AND ACTIVATION | Genes involved in FGFR4 ligand binding and activation |