Project
PRJNA195909:zebrafish embryo and larva development
Navigation
Downloads
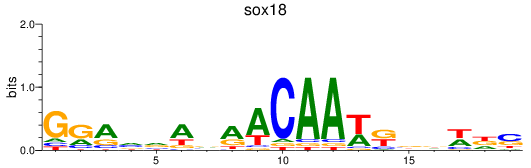
Results for sox18
Z-value: 0.73
Transcription factors associated with sox18
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
sox18
|
ENSDARG00000058598 | SRY-box transcription factor 18 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| sox18 | dr11_v1_chr23_+_8797143_8797143 | -0.73 | 2.5e-02 | Click! |
Activity profile of sox18 motif
Sorted Z-values of sox18 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr9_+_44994214 | 1.19 |
ENSDART00000141434
|
retsatl
|
retinol saturase (all-trans-retinol 13,14-reductase) like |
| chr8_+_23726708 | 1.10 |
ENSDART00000142395
|
mkrn4
|
makorin, ring finger protein, 4 |
| chr17_+_24597001 | 1.07 |
ENSDART00000191834
|
rlf
|
rearranged L-myc fusion |
| chr7_+_24814866 | 0.96 |
ENSDART00000173581
|
si:ch211-226l4.6
|
si:ch211-226l4.6 |
| chr7_-_24995631 | 0.84 |
ENSDART00000173955
ENSDART00000173791 |
rcor2
|
REST corepressor 2 |
| chr22_+_25352530 | 0.83 |
ENSDART00000176742
ENSDART00000113186 |
si:dkey-240e12.6
|
si:dkey-240e12.6 |
| chr5_-_5669879 | 0.76 |
ENSDART00000191963
|
CABZ01075628.1
|
|
| chr1_+_45707219 | 0.72 |
ENSDART00000143363
|
si:ch211-214c7.4
|
si:ch211-214c7.4 |
| chr14_+_30272891 | 0.66 |
ENSDART00000017122
|
asah1a
|
N-acylsphingosine amidohydrolase (acid ceramidase) 1a |
| chr4_-_8030583 | 0.63 |
ENSDART00000113628
|
si:ch211-240l19.8
|
si:ch211-240l19.8 |
| chr6_+_49052741 | 0.57 |
ENSDART00000011876
|
sycp1
|
synaptonemal complex protein 1 |
| chr19_+_15440841 | 0.57 |
ENSDART00000182329
|
lin28a
|
lin-28 homolog A (C. elegans) |
| chr22_+_24559947 | 0.56 |
ENSDART00000169847
|
wdr47b
|
WD repeat domain 47b |
| chr5_+_3892551 | 0.55 |
ENSDART00000134396
|
rpain
|
RPA interacting protein |
| chr17_-_43594864 | 0.53 |
ENSDART00000139980
|
zfyve28
|
zinc finger, FYVE domain containing 28 |
| chr21_+_21012210 | 0.53 |
ENSDART00000190317
|
ndr1
|
nodal-related 1 |
| chr17_+_10570906 | 0.52 |
ENSDART00000182371
ENSDART00000176097 ENSDART00000110593 ENSDART00000169356 |
mgaa
|
MGA, MAX dimerization protein a |
| chr14_-_35400671 | 0.51 |
ENSDART00000129676
|
lsm11
|
LSM11, U7 small nuclear RNA associated |
| chr16_-_9449712 | 0.50 |
ENSDART00000136522
|
prpf3
|
PRP3 pre-mRNA processing factor 3 homolog (yeast) |
| chr1_+_49435017 | 0.49 |
ENSDART00000124833
|
pdcd11
|
programmed cell death 11 |
| chr4_+_13412030 | 0.49 |
ENSDART00000003694
|
cand1
|
cullin-associated and neddylation-dissociated 1 |
| chr19_+_9113932 | 0.46 |
ENSDART00000060442
|
setdb1a
|
SET domain, bifurcated 1a |
| chr21_-_5066462 | 0.46 |
ENSDART00000067733
|
zgc:77838
|
zgc:77838 |
| chr23_-_10745288 | 0.46 |
ENSDART00000140745
ENSDART00000013768 |
eif4e3
|
eukaryotic translation initiation factor 4E family member 3 |
| chr11_+_17984354 | 0.46 |
ENSDART00000179986
|
rpusd4
|
RNA pseudouridylate synthase domain containing 4 |
| chr15_+_29727799 | 0.46 |
ENSDART00000182006
|
zgc:153372
|
zgc:153372 |
| chr10_+_15511885 | 0.46 |
ENSDART00000165663
|
erbin
|
erbb2 interacting protein |
| chr7_+_22853788 | 0.46 |
ENSDART00000148502
|
clcn5b
|
chloride channel, voltage-sensitive 5b |
| chr17_-_14780578 | 0.46 |
ENSDART00000154690
|
si:ch211-266o15.1
|
si:ch211-266o15.1 |
| chr8_-_25600888 | 0.46 |
ENSDART00000193860
|
stk38a
|
serine/threonine kinase 38a |
| chr2_-_9971609 | 0.44 |
ENSDART00000137924
ENSDART00000048655 ENSDART00000131613 |
zgc:55943
|
zgc:55943 |
| chr5_-_16475374 | 0.44 |
ENSDART00000134274
ENSDART00000136004 |
piwil2
|
piwi-like RNA-mediated gene silencing 2 |
| chr12_+_13348918 | 0.43 |
ENSDART00000181373
|
rnasen
|
ribonuclease type III, nuclear |
| chr3_+_22442445 | 0.42 |
ENSDART00000190921
|
wnk4b
|
WNK lysine deficient protein kinase 4b |
| chr11_+_17984167 | 0.41 |
ENSDART00000020283
ENSDART00000188329 |
rpusd4
|
RNA pseudouridylate synthase domain containing 4 |
| chr14_-_24277805 | 0.40 |
ENSDART00000054243
|
dpf2l
|
D4, zinc and double PHD fingers family 2, like |
| chr7_+_69459759 | 0.40 |
ENSDART00000160500
|
ctdnep1b
|
CTD nuclear envelope phosphatase 1b |
| chr5_-_68259747 | 0.39 |
ENSDART00000188756
|
CABZ01083944.1
|
|
| chr5_-_23517747 | 0.39 |
ENSDART00000137655
|
stag2a
|
stromal antigen 2a |
| chr16_-_21903083 | 0.37 |
ENSDART00000165849
|
setdb1b
|
SET domain, bifurcated 1b |
| chr10_-_42237304 | 0.37 |
ENSDART00000140341
|
tcf7l1a
|
transcription factor 7 like 1a |
| chr11_+_5565082 | 0.36 |
ENSDART00000112590
ENSDART00000183207 |
si:ch73-40i7.5
|
si:ch73-40i7.5 |
| chr5_+_22459087 | 0.36 |
ENSDART00000134781
|
BX546499.1
|
|
| chr3_+_34986837 | 0.34 |
ENSDART00000190341
|
smarce1
|
SWI/SNF related, matrix associated, actin dependent regulator of chromatin, subfamily e, member 1 |
| chr16_+_29586468 | 0.34 |
ENSDART00000148926
|
mcl1b
|
MCL1, BCL2 family apoptosis regulator b |
| chr2_+_31942390 | 0.33 |
ENSDART00000138684
ENSDART00000146758 ENSDART00000137921 |
otulinb
|
OTU deubiquitinase with linear linkage specificity b |
| chr7_+_22688781 | 0.32 |
ENSDART00000173509
|
ugt5g1
|
UDP glucuronosyltransferase 5 family, polypeptide G1 |
| chr3_-_31845816 | 0.31 |
ENSDART00000157028
|
map3k3
|
mitogen-activated protein kinase kinase kinase 3 |
| chr7_+_73827805 | 0.31 |
ENSDART00000109316
|
zgc:173587
|
zgc:173587 |
| chr3_+_34159192 | 0.30 |
ENSDART00000151119
|
carm1
|
coactivator-associated arginine methyltransferase 1 |
| chr18_-_1270220 | 0.29 |
ENSDART00000193754
|
ugt5f1
|
UDP glucuronosyltransferase 5 family, polypeptide F1 |
| chr25_-_35104083 | 0.29 |
ENSDART00000183252
ENSDART00000156727 |
si:dkey-108k21.21
|
si:dkey-108k21.21 |
| chr9_+_44832032 | 0.28 |
ENSDART00000002633
|
frzb
|
frizzled related protein |
| chr24_-_24724233 | 0.27 |
ENSDART00000127044
ENSDART00000012399 |
armc1
|
armadillo repeat containing 1 |
| chr22_+_1786230 | 0.27 |
ENSDART00000169318
ENSDART00000164948 |
znf1154
|
zinc finger protein 1154 |
| chr13_-_45063686 | 0.26 |
ENSDART00000130467
ENSDART00000136679 |
chp1
|
calcineurin-like EF-hand protein 1 |
| chr17_+_51682429 | 0.26 |
ENSDART00000004379
|
nol10
|
nucleolar protein 10 |
| chr22_+_2533514 | 0.26 |
ENSDART00000147967
|
si:ch73-92e7.4
|
si:ch73-92e7.4 |
| chr25_+_35061044 | 0.25 |
ENSDART00000111706
|
si:dkey-108k21.14
|
si:dkey-108k21.14 |
| chr5_+_61863996 | 0.25 |
ENSDART00000082879
|
sympk
|
symplekin |
| chr13_+_25397098 | 0.24 |
ENSDART00000132953
|
gsto2
|
glutathione S-transferase omega 2 |
| chr7_+_39011355 | 0.24 |
ENSDART00000173855
|
dgkza
|
diacylglycerol kinase, zeta a |
| chr2_+_33541928 | 0.23 |
ENSDART00000162852
|
BX548164.1
|
|
| chr11_+_43751263 | 0.22 |
ENSDART00000163843
|
zgc:153431
|
zgc:153431 |
| chr13_+_25396896 | 0.22 |
ENSDART00000041257
|
gsto2
|
glutathione S-transferase omega 2 |
| chr19_+_43753995 | 0.22 |
ENSDART00000058504
|
SLC17A3
|
si:ch1073-513e17.1 |
| chr11_+_29671661 | 0.22 |
ENSDART00000024318
ENSDART00000165024 |
rnf207a
|
ring finger protein 207a |
| chr13_-_31164749 | 0.22 |
ENSDART00000049180
|
mapk8a
|
mitogen-activated protein kinase 8a |
| chr1_-_32111882 | 0.22 |
ENSDART00000112639
ENSDART00000101958 |
pnpla4
|
patatin-like phospholipase domain containing 4 |
| chr9_+_32306862 | 0.21 |
ENSDART00000078513
|
mob4
|
MOB family member 4, phocein |
| chr14_+_6429399 | 0.20 |
ENSDART00000149783
ENSDART00000148461 |
abca1b
|
ATP-binding cassette, sub-family A (ABC1), member 1B |
| chr2_+_21185903 | 0.19 |
ENSDART00000131369
|
si:dkey-29d8.3
|
si:dkey-29d8.3 |
| chr15_+_17722054 | 0.18 |
ENSDART00000191390
ENSDART00000169550 |
si:ch211-213d14.1
|
si:ch211-213d14.1 |
| chr2_-_32262287 | 0.18 |
ENSDART00000056621
ENSDART00000039717 |
fam49ba
|
family with sequence similarity 49, member Ba |
| chr16_+_29586004 | 0.18 |
ENSDART00000149520
|
mcl1b
|
MCL1, BCL2 family apoptosis regulator b |
| chr17_+_16423721 | 0.18 |
ENSDART00000064233
|
efcab11
|
EF-hand calcium binding domain 11 |
| chr3_+_52145511 | 0.17 |
ENSDART00000078485
|
adgre14
|
adhesion G protein-coupled receptor E14 |
| chr9_+_45680802 | 0.17 |
ENSDART00000036369
|
asb1
|
ankyrin repeat and SOCS box containing 1 |
| chr12_-_35105670 | 0.16 |
ENSDART00000153034
|
si:ch73-127m5.2
|
si:ch73-127m5.2 |
| chr14_+_37545341 | 0.16 |
ENSDART00000105588
|
pcdh1b
|
protocadherin 1b |
| chr24_+_9475809 | 0.16 |
ENSDART00000132688
|
si:ch211-285f17.1
|
si:ch211-285f17.1 |
| chr20_-_42534153 | 0.16 |
ENSDART00000061122
|
rfx6
|
regulatory factor X, 6 |
| chr5_-_38107741 | 0.16 |
ENSDART00000156853
|
si:ch211-284e13.14
|
si:ch211-284e13.14 |
| chr10_-_8041948 | 0.14 |
ENSDART00000059017
|
nfkbil1
|
nuclear factor of kappa light polypeptide gene enhancer in B-cells inhibitor-like 1 |
| chr25_-_12982193 | 0.14 |
ENSDART00000159617
|
ccl39.5
|
chemokine (C-C motif) ligand 39, duplicate 5 |
| chr3_-_23470986 | 0.14 |
ENSDART00000113135
|
msl1a
|
male-specific lethal 1 homolog a (Drosophila) |
| chr3_-_33961589 | 0.13 |
ENSDART00000151533
|
ighd
|
immunoglobulin heavy constant delta |
| chr4_+_33012407 | 0.13 |
ENSDART00000151873
|
si:dkey-26h11.2
|
si:dkey-26h11.2 |
| chr5_-_62317496 | 0.13 |
ENSDART00000180089
|
zgc:85789
|
zgc:85789 |
| chr3_+_9588705 | 0.13 |
ENSDART00000172543
ENSDART00000104875 |
trap1
|
TNF receptor-associated protein 1 |
| chr5_-_7513082 | 0.12 |
ENSDART00000158913
|
bmpr1ba
|
bone morphogenetic protein receptor, type IBa |
| chr21_-_30166097 | 0.12 |
ENSDART00000130676
|
hbegfb
|
heparin-binding EGF-like growth factor b |
| chr22_+_29087308 | 0.12 |
ENSDART00000186776
ENSDART00000178615 ENSDART00000150811 ENSDART00000150489 |
pla2g6
|
phospholipase A2, group VI (cytosolic, calcium-independent) |
| chr15_-_20779624 | 0.10 |
ENSDART00000181936
|
tpst1
|
tyrosylprotein sulfotransferase 1 |
| chr3_-_40976463 | 0.10 |
ENSDART00000128450
ENSDART00000018676 |
cyp3c1
|
cytochrome P450, family 3, subfamily c, polypeptide 1 |
| chr21_+_43171013 | 0.10 |
ENSDART00000151362
|
zcchc10
|
zinc finger, CCHC domain containing 10 |
| chr21_-_1012269 | 0.09 |
ENSDART00000159835
|
CABZ01057159.1
|
|
| chr14_+_6962271 | 0.09 |
ENSDART00000148447
ENSDART00000149114 ENSDART00000149492 ENSDART00000148394 |
hnrnpaba
|
heterogeneous nuclear ribonucleoprotein A/Ba |
| chr25_+_10987817 | 0.09 |
ENSDART00000156529
|
si:dkey-52p2.5
|
si:dkey-52p2.5 |
| chr22_+_1853999 | 0.08 |
ENSDART00000163288
|
znf1174
|
zinc finger protein 1174 |
| chr13_+_40034176 | 0.08 |
ENSDART00000189797
|
golga7bb
|
golgin A7 family, member Bb |
| chr15_+_11683114 | 0.07 |
ENSDART00000168233
|
si:dkey-31c13.1
|
si:dkey-31c13.1 |
| chr8_-_13486258 | 0.07 |
ENSDART00000137459
|
pimr104
|
Pim proto-oncogene, serine/threonine kinase, related 104 |
| chr21_+_30662263 | 0.06 |
ENSDART00000154758
ENSDART00000138664 |
clcn5a
|
chloride channel, voltage-sensitive 5a |
| chr5_-_30704390 | 0.06 |
ENSDART00000016709
|
ift22
|
intraflagellar transport 22 homolog (Chlamydomonas) |
| chr20_+_34717403 | 0.06 |
ENSDART00000034252
|
pnocb
|
prepronociceptin b |
| chr18_-_40905901 | 0.06 |
ENSDART00000064848
|
pglyrp5
|
peptidoglycan recognition protein 5 |
| chr22_+_17118225 | 0.06 |
ENSDART00000135604
|
frem1b
|
Fras1 related extracellular matrix 1b |
| chr24_+_38658334 | 0.06 |
ENSDART00000155735
|
si:ch73-70c5.1
|
si:ch73-70c5.1 |
| chr21_+_14511685 | 0.05 |
ENSDART00000102099
|
CR392036.1
|
|
| chr7_+_4922104 | 0.05 |
ENSDART00000135154
|
si:dkey-28d5.11
|
si:dkey-28d5.11 |
| chr4_-_37479395 | 0.05 |
ENSDART00000168299
ENSDART00000157864 |
si:dkey-106c17.2
|
si:dkey-106c17.2 |
| chr1_-_50611031 | 0.05 |
ENSDART00000148285
|
ppm1k
|
protein phosphatase, Mg2+/Mn2+ dependent, 1K |
| chr2_-_51593835 | 0.04 |
ENSDART00000168072
|
si:ch211-9d9.7
|
si:ch211-9d9.7 |
| chr1_-_43987873 | 0.04 |
ENSDART00000108821
|
CR385050.1
|
|
| chr20_-_45062514 | 0.04 |
ENSDART00000183529
ENSDART00000182955 |
klhl29
|
kelch-like family member 29 |
| chr22_-_8499778 | 0.04 |
ENSDART00000142219
|
si:ch73-27e22.4
|
si:ch73-27e22.4 |
| chr25_+_12849609 | 0.04 |
ENSDART00000168144
|
ccl33.2
|
chemokine (C-C motif) ligand 33, duplicate 2 |
| chr8_-_13454281 | 0.04 |
ENSDART00000141959
|
CR354547.2
|
|
| chr16_+_4838808 | 0.03 |
ENSDART00000179363
|
ibtk
|
inhibitor of Bruton agammaglobulinemia tyrosine kinase |
| chr15_+_29728377 | 0.02 |
ENSDART00000099958
|
zgc:153372
|
zgc:153372 |
| chr10_-_29733194 | 0.02 |
ENSDART00000149252
|
si:ch73-261i21.2
|
si:ch73-261i21.2 |
| chr1_-_2061419 | 0.01 |
ENSDART00000058878
|
oxgr1a.3
|
oxoglutarate (alpha-ketoglutarate) receptor 1a, tandem duplicate 3 |
| chr1_-_11519934 | 0.01 |
ENSDART00000162060
|
sdk1b
|
sidekick cell adhesion molecule 1b |
| chr25_+_10416583 | 0.01 |
ENSDART00000073907
|
ehf
|
ets homologous factor |
| chr22_+_9110998 | 0.01 |
ENSDART00000141520
|
nlrp15
|
NACHT, LRR and PYD domains-containing protein 15 |
| chr18_+_7204378 | 0.00 |
ENSDART00000142905
|
vwf
|
von Willebrand factor |
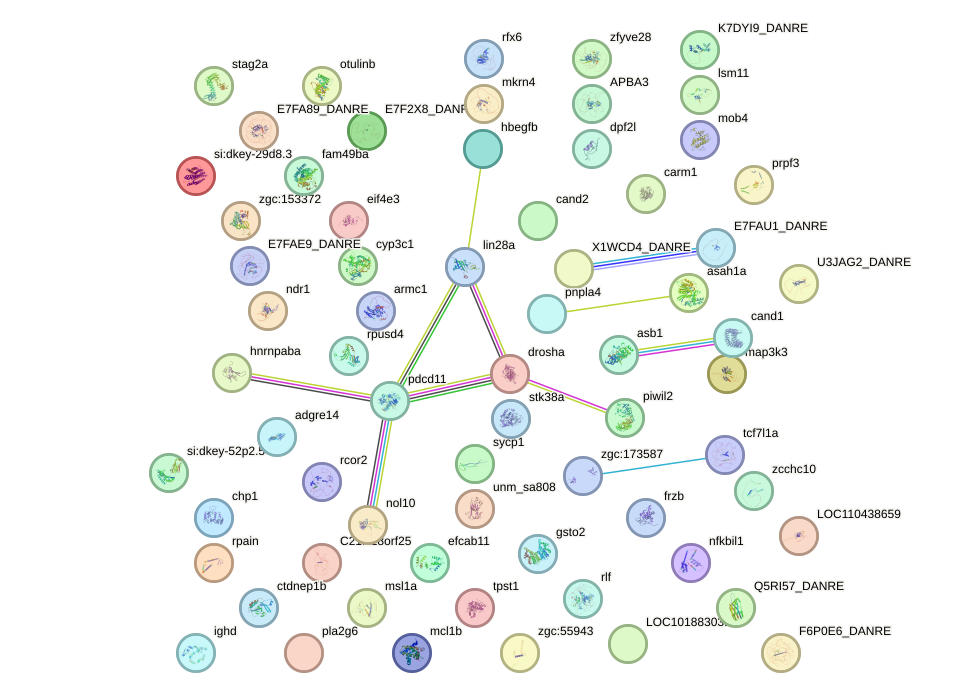
Network of associatons between targets according to the STRING database.
First level regulatory network of sox18
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.6 | GO:0051026 | meiotic DNA repair synthesis(GO:0000711) chiasma assembly(GO:0051026) |
| 0.2 | 0.5 | GO:0010265 | SCF complex assembly(GO:0010265) |
| 0.2 | 0.5 | GO:0018872 | arsonoacetate metabolic process(GO:0018872) |
| 0.1 | 0.9 | GO:0000455 | enzyme-directed rRNA pseudouridine synthesis(GO:0000455) |
| 0.1 | 0.4 | GO:0030718 | germ-line stem cell population maintenance(GO:0030718) |
| 0.1 | 0.4 | GO:0060829 | negative regulation of canonical Wnt signaling pathway involved in neural plate anterior/posterior pattern formation(GO:0060829) |
| 0.1 | 0.5 | GO:0007175 | negative regulation of epidermal growth factor-activated receptor activity(GO:0007175) |
| 0.1 | 0.4 | GO:0003096 | renal sodium ion transport(GO:0003096) renal sodium ion absorption(GO:0070294) |
| 0.1 | 0.4 | GO:0010866 | regulation of triglyceride biosynthetic process(GO:0010866) positive regulation of triglyceride biosynthetic process(GO:0010867) |
| 0.1 | 0.3 | GO:0045600 | positive regulation of fat cell differentiation(GO:0045600) |
| 0.1 | 0.5 | GO:0006398 | mRNA 3'-end processing by stem-loop binding and cleavage(GO:0006398) |
| 0.1 | 0.8 | GO:0090309 | methylation-dependent chromatin silencing(GO:0006346) positive regulation of chromatin silencing(GO:0031937) regulation of methylation-dependent chromatin silencing(GO:0090308) positive regulation of methylation-dependent chromatin silencing(GO:0090309) |
| 0.1 | 0.3 | GO:1990108 | protein linear deubiquitination(GO:1990108) |
| 0.1 | 0.3 | GO:0035988 | chondrocyte proliferation(GO:0035988) |
| 0.1 | 0.2 | GO:0035773 | insulin secretion involved in cellular response to glucose stimulus(GO:0035773) regulation of insulin secretion involved in cellular response to glucose stimulus(GO:0061178) |
| 0.0 | 0.5 | GO:0032088 | negative regulation of NF-kappaB transcription factor activity(GO:0032088) |
| 0.0 | 0.5 | GO:0043154 | negative regulation of cysteine-type endopeptidase activity involved in apoptotic process(GO:0043154) |
| 0.0 | 0.3 | GO:0003160 | endocardium morphogenesis(GO:0003160) |
| 0.0 | 0.5 | GO:0045743 | positive regulation of fibroblast growth factor receptor signaling pathway(GO:0045743) |
| 0.0 | 0.5 | GO:0016584 | nucleosome positioning(GO:0016584) |
| 0.0 | 0.1 | GO:0061400 | positive regulation of transcription from RNA polymerase II promoter in response to calcium ion(GO:0061400) |
| 0.0 | 0.2 | GO:0033700 | phospholipid efflux(GO:0033700) |
| 0.0 | 0.8 | GO:0016575 | histone deacetylation(GO:0016575) |
| 0.0 | 1.0 | GO:0006119 | oxidative phosphorylation(GO:0006119) |
| 0.0 | 0.1 | GO:0043984 | histone H4-K16 acetylation(GO:0043984) |
| 0.0 | 0.1 | GO:0006478 | protein sulfation(GO:0006477) peptidyl-tyrosine sulfation(GO:0006478) |
| 0.0 | 0.1 | GO:0007176 | regulation of epidermal growth factor-activated receptor activity(GO:0007176) |
| 0.0 | 0.2 | GO:0019433 | triglyceride catabolic process(GO:0019433) |
| 0.0 | 0.4 | GO:0048264 | determination of ventral identity(GO:0048264) |
| 0.0 | 0.2 | GO:0007205 | protein kinase C-activating G-protein coupled receptor signaling pathway(GO:0007205) |
| 0.0 | 0.2 | GO:0006378 | mRNA polyadenylation(GO:0006378) RNA polyadenylation(GO:0043631) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.6 | GO:0043073 | germ cell nucleus(GO:0043073) |
| 0.1 | 0.4 | GO:1990923 | PET complex(GO:1990923) |
| 0.1 | 0.4 | GO:0071595 | Nem1-Spo7 phosphatase complex(GO:0071595) |
| 0.1 | 0.5 | GO:0005845 | mRNA cap binding complex(GO:0005845) |
| 0.0 | 0.3 | GO:0071797 | LUBAC complex(GO:0071797) |
| 0.0 | 0.5 | GO:0046540 | U4/U6 x U5 tri-snRNP complex(GO:0046540) |
| 0.0 | 0.8 | GO:0032040 | small-subunit processome(GO:0032040) |
| 0.0 | 0.4 | GO:1990907 | beta-catenin-TCF complex(GO:1990907) |
| 0.0 | 0.7 | GO:0090544 | BAF-type complex(GO:0090544) |
| 0.0 | 0.1 | GO:0072487 | MSL complex(GO:0072487) |
| 0.0 | 0.4 | GO:0008278 | cohesin complex(GO:0008278) |
| 0.0 | 0.2 | GO:0005847 | mRNA cleavage and polyadenylation specificity factor complex(GO:0005847) |
| 0.0 | 0.5 | GO:0016605 | PML body(GO:0016605) |
| 0.0 | 0.3 | GO:0071141 | SMAD protein complex(GO:0071141) |
| 0.0 | 0.5 | GO:0030532 | small nuclear ribonucleoprotein complex(GO:0030532) |
| 0.0 | 0.8 | GO:0000118 | histone deacetylase complex(GO:0000118) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.7 | GO:0017064 | fatty acid amide hydrolase activity(GO:0017064) |
| 0.2 | 0.5 | GO:0030791 | arsenite methyltransferase activity(GO:0030791) methylarsonite methyltransferase activity(GO:0030792) |
| 0.2 | 0.5 | GO:0045174 | glutathione dehydrogenase (ascorbate) activity(GO:0045174) |
| 0.2 | 0.5 | GO:0005176 | ErbB-2 class receptor binding(GO:0005176) |
| 0.1 | 0.8 | GO:0046974 | histone methyltransferase activity (H3-K9 specific)(GO:0046974) |
| 0.1 | 0.5 | GO:0070698 | type I activin receptor binding(GO:0070698) |
| 0.1 | 0.2 | GO:0050253 | retinyl-palmitate esterase activity(GO:0050253) |
| 0.1 | 0.3 | GO:0008469 | histone-arginine N-methyltransferase activity(GO:0008469) |
| 0.1 | 0.4 | GO:0034584 | piRNA binding(GO:0034584) |
| 0.0 | 0.9 | GO:0009982 | pseudouridine synthase activity(GO:0009982) |
| 0.0 | 0.2 | GO:0090556 | phospholipid-translocating ATPase activity(GO:0004012) phosphatidylcholine-translocating ATPase activity(GO:0090554) phosphatidylserine-translocating ATPase activity(GO:0090556) |
| 0.0 | 0.3 | GO:0016922 | ligand-dependent nuclear receptor binding(GO:0016922) |
| 0.0 | 1.0 | GO:0004129 | cytochrome-c oxidase activity(GO:0004129) heme-copper terminal oxidase activity(GO:0015002) oxidoreductase activity, acting on a heme group of donors(GO:0016675) oxidoreductase activity, acting on a heme group of donors, oxygen as acceptor(GO:0016676) |
| 0.0 | 0.5 | GO:0005247 | voltage-gated chloride channel activity(GO:0005247) |
| 0.0 | 0.5 | GO:0000340 | RNA 7-methylguanosine cap binding(GO:0000340) |
| 0.0 | 0.4 | GO:0019870 | chloride channel inhibitor activity(GO:0019869) potassium channel inhibitor activity(GO:0019870) |
| 0.0 | 0.5 | GO:0031492 | nucleosomal DNA binding(GO:0031492) |
| 0.0 | 0.2 | GO:0004705 | JUN kinase activity(GO:0004705) SAP kinase activity(GO:0016909) |
| 0.0 | 0.5 | GO:0032266 | phosphatidylinositol-3-phosphate binding(GO:0032266) |
| 0.0 | 0.4 | GO:0001540 | beta-amyloid binding(GO:0001540) |
| 0.0 | 0.1 | GO:0008745 | N-acetylmuramoyl-L-alanine amidase activity(GO:0008745) |
| 0.0 | 0.5 | GO:0017069 | snRNA binding(GO:0017069) |
| 0.0 | 0.2 | GO:0030544 | Hsp70 protein binding(GO:0030544) |
| 0.0 | 0.1 | GO:0008476 | protein-tyrosine sulfotransferase activity(GO:0008476) |
| 0.0 | 0.2 | GO:0031726 | CCR1 chemokine receptor binding(GO:0031726) CCR4 chemokine receptor binding(GO:0031729) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.3 | PID ERB GENOMIC PATHWAY | Validated nuclear estrogen receptor beta network |
| 0.0 | 0.3 | PID P38 MKK3 6PATHWAY | p38 MAPK signaling pathway |
| 0.0 | 0.5 | PID ERBB1 INTERNALIZATION PATHWAY | Internalization of ErbB1 |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.5 | REACTOME SLBP DEPENDENT PROCESSING OF REPLICATION DEPENDENT HISTONE PRE MRNAS | Genes involved in SLBP Dependent Processing of Replication-Dependent Histone Pre-mRNAs |
| 0.0 | 0.5 | REACTOME GLUTATHIONE CONJUGATION | Genes involved in Glutathione conjugation |
| 0.0 | 0.6 | REACTOME MEIOTIC SYNAPSIS | Genes involved in Meiotic Synapsis |
| 0.0 | 0.1 | REACTOME ACYL CHAIN REMODELLING OF PE | Genes involved in Acyl chain remodelling of PE |