Project
PRJNA195909:zebrafish embryo and larva development
Navigation
Downloads
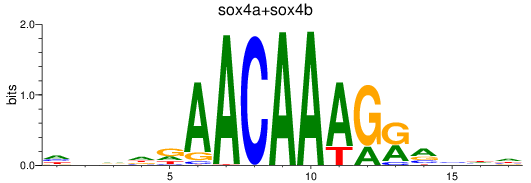
Results for sox4a+sox4b
Z-value: 0.53
Transcription factors associated with sox4a+sox4b
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
sox4a
|
ENSDARG00000004588 | SRY-box transcription factor 4a |
|
sox4b
|
ENSDARG00000098834 | SRY-box transcription factor 4b |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| sox4a | dr11_v1_chr19_-_28789404_28789409 | 0.54 | 1.3e-01 | Click! |
| sox4b | dr11_v1_chr16_+_68069_68124 | 0.17 | 6.6e-01 | Click! |
Activity profile of sox4a+sox4b motif
Sorted Z-values of sox4a+sox4b motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr24_-_37568359 | 0.77 |
ENSDART00000056286
|
h1f0
|
H1 histone family, member 0 |
| chr7_+_44713135 | 0.71 |
ENSDART00000170721
|
si:dkey-56m19.5
|
si:dkey-56m19.5 |
| chr13_+_22675802 | 0.67 |
ENSDART00000145538
ENSDART00000143312 |
zgc:193505
|
zgc:193505 |
| chr8_-_16697912 | 0.61 |
ENSDART00000076542
|
rpe65b
|
retinal pigment epithelium-specific protein 65b |
| chr23_+_35714574 | 0.61 |
ENSDART00000164616
|
tuba1c
|
tubulin, alpha 1c |
| chr24_-_33756003 | 0.54 |
ENSDART00000079283
|
tmeff1b
|
transmembrane protein with EGF-like and two follistatin-like domains 1b |
| chr21_+_26389391 | 0.53 |
ENSDART00000077197
|
tmsb
|
thymosin, beta |
| chr20_+_34770197 | 0.49 |
ENSDART00000018304
|
mcm3
|
minichromosome maintenance complex component 3 |
| chr1_+_17376922 | 0.48 |
ENSDART00000145068
|
fat1a
|
FAT atypical cadherin 1a |
| chr23_+_17865554 | 0.45 |
ENSDART00000181009
ENSDART00000162822 |
naca
|
nascent polypeptide-associated complex alpha subunit |
| chr6_+_12968101 | 0.44 |
ENSDART00000013781
|
mcm6
|
minichromosome maintenance complex component 6 |
| chr9_+_25330905 | 0.44 |
ENSDART00000101470
|
itm2bb
|
integral membrane protein 2Bb |
| chr23_-_3758637 | 0.42 |
ENSDART00000131536
ENSDART00000139408 ENSDART00000137826 |
hmga1a
|
high mobility group AT-hook 1a |
| chr20_+_31269778 | 0.42 |
ENSDART00000133353
|
apobb.1
|
apolipoprotein Bb, tandem duplicate 1 |
| chr14_-_8080416 | 0.42 |
ENSDART00000045109
|
zgc:92242
|
zgc:92242 |
| chr23_+_17865953 | 0.42 |
ENSDART00000014723
ENSDART00000140302 ENSDART00000144800 |
naca
|
nascent polypeptide-associated complex alpha subunit |
| chr15_+_35691783 | 0.41 |
ENSDART00000183994
|
CT573366.1
|
|
| chr8_-_16712111 | 0.40 |
ENSDART00000184147
ENSDART00000180419 ENSDART00000076600 |
rpe65c
|
retinal pigment epithelium-specific protein 65c |
| chr7_-_69636502 | 0.39 |
ENSDART00000126739
|
tspan5a
|
tetraspanin 5a |
| chr8_+_46217861 | 0.39 |
ENSDART00000038790
|
angptl7
|
angiopoietin-like 7 |
| chr5_-_46505691 | 0.39 |
ENSDART00000111589
ENSDART00000122966 ENSDART00000166907 |
hapln1a
|
hyaluronan and proteoglycan link protein 1a |
| chr9_+_54179306 | 0.39 |
ENSDART00000189829
|
tmsb4x
|
thymosin, beta 4 x |
| chr9_+_48007081 | 0.38 |
ENSDART00000060593
ENSDART00000099835 |
zgc:92380
|
zgc:92380 |
| chr5_-_30615901 | 0.37 |
ENSDART00000147769
|
si:ch211-117m20.5
|
si:ch211-117m20.5 |
| chr23_+_25428513 | 0.37 |
ENSDART00000144554
|
fmnl3
|
formin-like 3 |
| chr15_+_28368644 | 0.36 |
ENSDART00000168453
|
slc43a2a
|
solute carrier family 43 (amino acid system L transporter), member 2a |
| chr19_-_17774875 | 0.36 |
ENSDART00000151133
ENSDART00000130695 |
top2b
|
DNA topoisomerase II beta |
| chr25_+_31323978 | 0.36 |
ENSDART00000067030
|
lsp1
|
lymphocyte-specific protein 1 |
| chr7_-_26408472 | 0.36 |
ENSDART00000111494
|
gal3st4
|
galactose-3-O-sulfotransferase 4 |
| chr1_+_24076243 | 0.35 |
ENSDART00000014608
|
mab21l2
|
mab-21-like 2 |
| chr5_+_15203421 | 0.34 |
ENSDART00000040826
|
tbx1
|
T-box 1 |
| chr4_-_16354292 | 0.33 |
ENSDART00000139919
|
lum
|
lumican |
| chr19_-_15855427 | 0.33 |
ENSDART00000133059
|
cited4a
|
Cbp/p300-interacting transactivator, with Glu/Asp-rich carboxy-terminal domain, 4a |
| chr23_+_36144487 | 0.32 |
ENSDART00000082473
|
hoxc3a
|
homeobox C3a |
| chr8_+_23093155 | 0.32 |
ENSDART00000063075
|
zgc:100920
|
zgc:100920 |
| chr15_-_26549693 | 0.32 |
ENSDART00000186432
|
serpinf2b
|
serpin peptidase inhibitor, clade F (alpha-2 antiplasmin, pigment epithelium derived factor), member 2b |
| chr6_-_6976096 | 0.31 |
ENSDART00000151822
ENSDART00000039443 ENSDART00000177960 |
tuba8l4
|
tubulin, alpha 8 like 4 |
| chr25_+_34576067 | 0.30 |
ENSDART00000157519
|
trpm1b
|
transient receptor potential cation channel, subfamily M, member 1b |
| chr25_+_35502552 | 0.29 |
ENSDART00000189612
ENSDART00000058443 |
fibina
|
fin bud initiation factor a |
| chr15_-_15469079 | 0.29 |
ENSDART00000132637
ENSDART00000004220 |
rab34a
|
RAB34, member RAS oncogene family a |
| chr7_-_24149670 | 0.29 |
ENSDART00000005884
|
mmp14a
|
matrix metallopeptidase 14a (membrane-inserted) |
| chr23_+_36083529 | 0.29 |
ENSDART00000053295
ENSDART00000130260 |
hoxc10a
|
homeobox C10a |
| chr1_+_46194333 | 0.28 |
ENSDART00000010894
|
sox1b
|
SRY (sex determining region Y)-box 1b |
| chr10_+_18952271 | 0.28 |
ENSDART00000146517
|
dpysl2b
|
dihydropyrimidinase-like 2b |
| chr9_+_32978302 | 0.27 |
ENSDART00000007630
|
nhlh2
|
nescient helix loop helix 2 |
| chr14_-_1955257 | 0.26 |
ENSDART00000193254
|
pcdh2g5
|
protocadherin 2 gamma 5 |
| chr10_-_27196093 | 0.26 |
ENSDART00000185282
|
auts2a
|
autism susceptibility candidate 2a |
| chr2_-_20574193 | 0.26 |
ENSDART00000190448
|
si:ch211-267e7.3
|
si:ch211-267e7.3 |
| chr19_+_43969363 | 0.24 |
ENSDART00000051712
|
gatad1
|
GATA zinc finger domain containing 1 |
| chr16_-_26731928 | 0.24 |
ENSDART00000135860
|
rnf41l
|
ring finger protein 41, like |
| chr13_+_44857087 | 0.24 |
ENSDART00000017770
|
zbtb8os
|
zinc finger and BTB domain containing 8 opposite strand |
| chr16_-_9675982 | 0.24 |
ENSDART00000113724
|
mal2
|
mal, T cell differentiation protein 2 (gene/pseudogene) |
| chr7_+_65876335 | 0.24 |
ENSDART00000150143
|
tead1b
|
TEA domain family member 1b |
| chr15_-_15468326 | 0.24 |
ENSDART00000161192
|
rab34a
|
RAB34, member RAS oncogene family a |
| chr12_+_5081759 | 0.23 |
ENSDART00000164178
|
prrt2
|
proline-rich transmembrane protein 2 |
| chr20_-_49681850 | 0.23 |
ENSDART00000025926
|
col12a1b
|
collagen, type XII, alpha 1b |
| chr15_+_28368823 | 0.23 |
ENSDART00000142298
|
slc43a2a
|
solute carrier family 43 (amino acid system L transporter), member 2a |
| chr22_+_28446365 | 0.23 |
ENSDART00000189359
|
abi3bpb
|
ABI family, member 3 (NESH) binding protein b |
| chr21_-_35853245 | 0.23 |
ENSDART00000172245
|
sgcd
|
sarcoglycan, delta (dystrophin-associated glycoprotein) |
| chr21_-_2310064 | 0.22 |
ENSDART00000169520
|
si:ch211-241b2.1
|
si:ch211-241b2.1 |
| chr23_+_42819221 | 0.22 |
ENSDART00000180495
|
myl9a
|
myosin, light chain 9a, regulatory |
| chr2_+_26179096 | 0.22 |
ENSDART00000024662
|
plppr3a
|
phospholipid phosphatase related 3a |
| chr8_+_14158021 | 0.22 |
ENSDART00000080832
|
si:dkey-6n6.2
|
si:dkey-6n6.2 |
| chr14_-_1958994 | 0.21 |
ENSDART00000161783
|
pcdh2g5
|
protocadherin 2 gamma 5 |
| chr7_-_26308098 | 0.21 |
ENSDART00000146440
ENSDART00000146935 ENSDART00000164627 |
zgc:77439
|
zgc:77439 |
| chr17_-_5860222 | 0.21 |
ENSDART00000058894
|
si:ch73-340m8.2
|
si:ch73-340m8.2 |
| chr13_-_1151410 | 0.21 |
ENSDART00000007231
|
psmb1
|
proteasome subunit beta 1 |
| chr24_-_6345647 | 0.21 |
ENSDART00000108994
|
zgc:174877
|
zgc:174877 |
| chr25_+_28776562 | 0.21 |
ENSDART00000109702
|
slc41a2a
|
solute carrier family 41 (magnesium transporter), member 2a |
| chr4_+_15954293 | 0.20 |
ENSDART00000132695
|
si:dkey-117n7.4
|
si:dkey-117n7.4 |
| chr6_+_7249531 | 0.20 |
ENSDART00000125912
ENSDART00000083424 ENSDART00000049695 ENSDART00000136088 |
dzip1
|
DAZ interacting zinc finger protein 1 |
| chr23_-_3759345 | 0.20 |
ENSDART00000132205
ENSDART00000137707 ENSDART00000189382 |
hmga1a
|
high mobility group AT-hook 1a |
| chr5_+_64739762 | 0.20 |
ENSDART00000161112
ENSDART00000135610 ENSDART00000002908 |
olfm1a
|
olfactomedin 1a |
| chr10_+_31938796 | 0.20 |
ENSDART00000182579
|
lhfpl6
|
LHFPL tetraspan subfamily member 6 |
| chr11_+_30296332 | 0.19 |
ENSDART00000192843
|
ugt1b7
|
UDP glucuronosyltransferase 1 family, polypeptide B7 |
| chr6_-_54180699 | 0.19 |
ENSDART00000045901
|
rps10
|
ribosomal protein S10 |
| chr23_+_36122058 | 0.19 |
ENSDART00000184448
|
hoxc3a
|
homeobox C3a |
| chr21_-_15674802 | 0.19 |
ENSDART00000136666
|
mmp11b
|
matrix metallopeptidase 11b |
| chr16_-_24612871 | 0.19 |
ENSDART00000155614
ENSDART00000154787 ENSDART00000155983 ENSDART00000156519 |
fxyd6l
|
FXYD domain containing ion transport regulator 6 like |
| chr25_+_19149241 | 0.19 |
ENSDART00000184982
ENSDART00000067324 |
mfge8b
|
milk fat globule-EGF factor 8 protein b |
| chr19_+_43119014 | 0.19 |
ENSDART00000023156
|
eef1a1l1
|
eukaryotic translation elongation factor 1 alpha 1, like 1 |
| chr14_-_44773864 | 0.18 |
ENSDART00000158386
|
si:dkey-109l4.3
|
si:dkey-109l4.3 |
| chr1_+_54773877 | 0.18 |
ENSDART00000129831
|
nlrc6
|
NLR family CARD domain containing 6 |
| chr23_-_29824146 | 0.18 |
ENSDART00000020616
|
zgc:194189
|
zgc:194189 |
| chr8_-_28349859 | 0.18 |
ENSDART00000062671
|
tuba8l
|
tubulin, alpha 8 like |
| chr9_-_1939232 | 0.18 |
ENSDART00000146131
|
hoxd3a
|
homeobox D3a |
| chr8_+_48480529 | 0.18 |
ENSDART00000186631
|
PRDM16
|
si:ch211-263k4.2 |
| chr11_-_45185792 | 0.18 |
ENSDART00000171328
|
si:dkey-93h22.7
|
si:dkey-93h22.7 |
| chr3_+_56645710 | 0.18 |
ENSDART00000193978
|
CR759836.1
|
|
| chr25_-_3503164 | 0.17 |
ENSDART00000191477
ENSDART00000186345 ENSDART00000180199 |
PRKAR2B
|
si:ch211-272n13.7 |
| chr24_-_6345965 | 0.17 |
ENSDART00000178287
|
zgc:174877
|
zgc:174877 |
| chr20_+_26881600 | 0.16 |
ENSDART00000174799
|
serpinb1
|
serpin peptidase inhibitor, clade B (ovalbumin), member 1 |
| chr6_-_40098641 | 0.16 |
ENSDART00000017402
|
ip6k2b
|
inositol hexakisphosphate kinase 2b |
| chr12_-_30777540 | 0.16 |
ENSDART00000126466
|
entpd1
|
ectonucleoside triphosphate diphosphohydrolase 1 |
| chr11_+_3959495 | 0.16 |
ENSDART00000122953
|
gnl3
|
guanine nucleotide binding protein-like 3 (nucleolar) |
| chr7_+_20019125 | 0.16 |
ENSDART00000186391
|
bcl6b
|
B-cell CLL/lymphoma 6, member B |
| chr1_-_9652547 | 0.16 |
ENSDART00000016750
ENSDART00000127735 ENSDART00000160474 ENSDART00000080323 ENSDART00000126996 ENSDART00000126877 ENSDART00000123773 |
ugt5b3
ugt5b2
ugt5b3
ugt5b1
|
UDP glucuronosyltransferase 5 family, polypeptide B3 UDP glucuronosyltransferase 5 family, polypeptide B2 UDP glucuronosyltransferase 5 family, polypeptide B3 UDP glucuronosyltransferase 5 family, polypeptide B1 |
| chr12_-_30777743 | 0.15 |
ENSDART00000148888
|
entpd1
|
ectonucleoside triphosphate diphosphohydrolase 1 |
| chr5_+_43870389 | 0.15 |
ENSDART00000141002
|
zgc:112966
|
zgc:112966 |
| chr4_+_18843015 | 0.15 |
ENSDART00000152086
ENSDART00000066977 ENSDART00000132567 |
bik
|
BCL2 interacting killer |
| chr6_-_31224563 | 0.15 |
ENSDART00000104616
|
lepr
|
leptin receptor |
| chr5_+_22970617 | 0.15 |
ENSDART00000192859
|
hmgn7
|
high mobility group nucleosomal binding domain 7 |
| chr19_-_19339285 | 0.15 |
ENSDART00000158413
ENSDART00000170479 |
cspg5b
|
chondroitin sulfate proteoglycan 5b |
| chr6_-_19023468 | 0.15 |
ENSDART00000184729
|
sept9b
|
septin 9b |
| chr13_-_27916439 | 0.15 |
ENSDART00000139081
ENSDART00000087097 |
ogfrl1
|
opioid growth factor receptor-like 1 |
| chr13_-_43599898 | 0.14 |
ENSDART00000084416
ENSDART00000145705 |
ablim1a
|
actin binding LIM protein 1a |
| chr10_+_36650222 | 0.14 |
ENSDART00000126963
|
ucp3
|
uncoupling protein 3 |
| chr15_-_4094888 | 0.14 |
ENSDART00000166307
|
TM4SF19
|
si:dkey-83h2.3 |
| chr20_+_7584211 | 0.14 |
ENSDART00000132481
ENSDART00000127975 ENSDART00000144551 |
bloc1s2
|
biogenesis of lysosomal organelles complex-1, subunit 2 |
| chr21_-_4539899 | 0.14 |
ENSDART00000112460
|
dolk
|
dolichol kinase |
| chr9_+_13986427 | 0.14 |
ENSDART00000147200
|
cd28
|
CD28 molecule |
| chr13_+_28732101 | 0.14 |
ENSDART00000015773
|
ldb1a
|
LIM domain binding 1a |
| chr15_-_6976851 | 0.14 |
ENSDART00000158474
ENSDART00000168943 ENSDART00000169944 |
si:ch73-311h14.2
|
si:ch73-311h14.2 |
| chr25_+_7229046 | 0.14 |
ENSDART00000149965
ENSDART00000041820 |
lingo1a
|
leucine rich repeat and Ig domain containing 1a |
| chr5_-_12093618 | 0.14 |
ENSDART00000161542
|
lrrc74b
|
leucine rich repeat containing 74B |
| chr5_+_36900157 | 0.13 |
ENSDART00000183533
ENSDART00000051184 |
hnrnpl
|
heterogeneous nuclear ribonucleoprotein L |
| chr3_-_21348478 | 0.13 |
ENSDART00000114906
|
fam171a2a
|
family with sequence similarity 171, member A2a |
| chr9_-_42861080 | 0.13 |
ENSDART00000193688
|
ttn.1
|
titin, tandem duplicate 1 |
| chr22_-_10354381 | 0.13 |
ENSDART00000092050
|
stab1
|
stabilin 1 |
| chr23_-_26535875 | 0.13 |
ENSDART00000135988
|
si:dkey-205h13.2
|
si:dkey-205h13.2 |
| chr19_-_7540821 | 0.13 |
ENSDART00000143958
|
lix1l
|
limb and CNS expressed 1 like |
| chr15_-_20710 | 0.13 |
ENSDART00000161218
|
cyp2y3
|
cytochrome P450, family 2, subfamily Y, polypeptide 3 |
| chr12_-_30338779 | 0.13 |
ENSDART00000192511
|
vwa2
|
von Willebrand factor A domain containing 2 |
| chr2_-_3038904 | 0.13 |
ENSDART00000186795
|
guk1a
|
guanylate kinase 1a |
| chr14_-_2270973 | 0.13 |
ENSDART00000180729
|
pcdh2ab9
|
protocadherin 2 alpha b 9 |
| chr8_+_30797363 | 0.13 |
ENSDART00000077280
|
mmp11a
|
matrix metallopeptidase 11a |
| chr12_+_35654749 | 0.12 |
ENSDART00000169889
ENSDART00000167873 |
baiap2b
|
BAI1-associated protein 2b |
| chr24_+_21973929 | 0.12 |
ENSDART00000042495
|
sat1b
|
spermidine/spermine N1-acetyltransferase 1b |
| chr1_+_54835131 | 0.12 |
ENSDART00000145070
|
si:ch211-196h16.4
|
si:ch211-196h16.4 |
| chr2_-_7696287 | 0.12 |
ENSDART00000190769
|
CABZ01055306.1
|
|
| chr4_+_75310097 | 0.12 |
ENSDART00000187088
|
CABZ01041913.1
|
|
| chr17_-_24680965 | 0.12 |
ENSDART00000154880
|
arhgef33
|
Rho guanine nucleotide exchange factor (GEF) 33 |
| chr10_+_43189325 | 0.12 |
ENSDART00000185584
|
vcanb
|
versican b |
| chr11_+_807153 | 0.12 |
ENSDART00000173289
|
vgll4b
|
vestigial-like family member 4b |
| chr1_+_54834119 | 0.12 |
ENSDART00000140020
|
si:ch211-196h16.4
|
si:ch211-196h16.4 |
| chr23_-_44226556 | 0.12 |
ENSDART00000149115
|
zgc:158659
|
zgc:158659 |
| chr6_-_54126463 | 0.12 |
ENSDART00000161059
|
tusc2a
|
tumor suppressor candidate 2a |
| chr6_-_54180516 | 0.12 |
ENSDART00000149945
|
rps10
|
ribosomal protein S10 |
| chr1_+_14073891 | 0.12 |
ENSDART00000021693
|
ank2a
|
ankyrin 2a, neuronal |
| chr11_-_3629201 | 0.12 |
ENSDART00000136577
ENSDART00000132121 |
itih3a
|
inter-alpha-trypsin inhibitor heavy chain 3a |
| chr4_-_4932619 | 0.12 |
ENSDART00000103293
|
ndufa5
|
NADH dehydrogenase (ubiquinone) 1 alpha subcomplex, 5 |
| chr13_+_21779975 | 0.11 |
ENSDART00000021556
|
si:ch211-51a6.2
|
si:ch211-51a6.2 |
| chr10_+_20113830 | 0.11 |
ENSDART00000139722
|
dmtn
|
dematin actin binding protein |
| chr6_-_59505589 | 0.11 |
ENSDART00000170685
|
gli1
|
GLI family zinc finger 1 |
| chr17_+_51627209 | 0.11 |
ENSDART00000056886
|
zgc:113142
|
zgc:113142 |
| chr10_-_25591194 | 0.11 |
ENSDART00000131640
|
tiam1a
|
T cell lymphoma invasion and metastasis 1a |
| chr19_+_48018464 | 0.11 |
ENSDART00000172307
ENSDART00000163848 |
UBE2M
|
si:ch1073-205c8.3 |
| chr10_-_17284055 | 0.11 |
ENSDART00000167464
|
GNAZ
|
G protein subunit alpha z |
| chr7_-_57332915 | 0.11 |
ENSDART00000162653
|
BX470176.1
|
|
| chr2_+_24177006 | 0.11 |
ENSDART00000132582
|
map4l
|
microtubule associated protein 4 like |
| chr9_-_8670158 | 0.11 |
ENSDART00000077296
|
col4a1
|
collagen, type IV, alpha 1 |
| chr21_+_17110598 | 0.11 |
ENSDART00000101282
ENSDART00000191864 |
bcr
|
breakpoint cluster region |
| chr1_-_27743985 | 0.11 |
ENSDART00000165515
|
si:dkey-29d5.2
|
si:dkey-29d5.2 |
| chr23_-_30787932 | 0.11 |
ENSDART00000135771
|
myt1a
|
myelin transcription factor 1a |
| chr14_+_6962271 | 0.10 |
ENSDART00000148447
ENSDART00000149114 ENSDART00000149492 ENSDART00000148394 |
hnrnpaba
|
heterogeneous nuclear ribonucleoprotein A/Ba |
| chr3_-_27646070 | 0.10 |
ENSDART00000122031
ENSDART00000151027 |
si:ch211-157c3.4
|
si:ch211-157c3.4 |
| chr15_-_28677725 | 0.10 |
ENSDART00000060255
|
blmh
|
bleomycin hydrolase |
| chr1_+_5485799 | 0.10 |
ENSDART00000022307
|
atic
|
5-aminoimidazole-4-carboxamide ribonucleotide formyltransferase/IMP cyclohydrolase |
| chr2_+_47623202 | 0.10 |
ENSDART00000154465
|
si:ch211-165b10.3
|
si:ch211-165b10.3 |
| chr16_+_25095483 | 0.10 |
ENSDART00000155032
|
si:ch211-261d7.3
|
si:ch211-261d7.3 |
| chr14_-_2004291 | 0.10 |
ENSDART00000114039
|
pcdh2g5
|
protocadherin 2 gamma 5 |
| chr15_-_12360409 | 0.10 |
ENSDART00000164596
|
tmprss13a
|
transmembrane protease, serine 13a |
| chr7_-_38340674 | 0.10 |
ENSDART00000075782
|
slc7a10a
|
solute carrier family 7 (neutral amino acid transporter light chain, asc system), member 10a |
| chr24_-_31194847 | 0.10 |
ENSDART00000158808
|
cnn3a
|
calponin 3, acidic a |
| chr18_+_19972853 | 0.10 |
ENSDART00000180071
|
skor1b
|
SKI family transcriptional corepressor 1b |
| chr21_-_20733615 | 0.10 |
ENSDART00000145544
|
si:ch211-22d5.2
|
si:ch211-22d5.2 |
| chr25_+_12494079 | 0.10 |
ENSDART00000163508
|
map1lc3b
|
microtubule-associated protein 1 light chain 3 beta |
| chr23_+_36306539 | 0.09 |
ENSDART00000053267
|
hnrnpa1b
|
heterogeneous nuclear ribonucleoprotein A1b |
| chr21_+_36774542 | 0.09 |
ENSDART00000130790
|
prr7
|
proline rich 7 (synaptic) |
| chr20_+_3277620 | 0.09 |
ENSDART00000067397
ENSDART00000135194 |
ndufaf7
|
NADH dehydrogenase (ubiquinone) complex I, assembly factor 7 |
| chr21_-_22709251 | 0.09 |
ENSDART00000140032
|
si:dkeyp-69c1.9
|
si:dkeyp-69c1.9 |
| chr15_-_22147860 | 0.09 |
ENSDART00000149784
|
scn3b
|
sodium channel, voltage-gated, type III, beta |
| chr17_+_24632440 | 0.09 |
ENSDART00000157092
|
map4k3b
|
mitogen-activated protein kinase kinase kinase kinase 3b |
| chr2_-_32768951 | 0.09 |
ENSDART00000004712
|
bfsp2
|
beaded filament structural protein 2, phakinin |
| chr10_-_1827225 | 0.09 |
ENSDART00000058627
|
epb41l4a
|
erythrocyte membrane protein band 4.1 like 4A |
| chr7_-_41968171 | 0.09 |
ENSDART00000188027
|
neto2b
|
neuropilin (NRP) and tolloid (TLL)-like 2b |
| chr1_-_51720633 | 0.09 |
ENSDART00000045894
|
rnaseh2a
|
ribonuclease H2, subunit A |
| chr14_+_35806605 | 0.09 |
ENSDART00000173093
|
gria2b
|
glutamate receptor, ionotropic, AMPA 2b |
| chr3_+_17537352 | 0.08 |
ENSDART00000104549
|
hcrt
|
hypocretin (orexin) neuropeptide precursor |
| chr21_+_17768174 | 0.08 |
ENSDART00000141380
|
rxraa
|
retinoid X receptor, alpha a |
| chr14_-_2355833 | 0.08 |
ENSDART00000157677
|
PCDHGC3 (1 of many)
|
si:ch73-233f7.6 |
| chr25_+_26895394 | 0.08 |
ENSDART00000155820
|
si:dkey-42p14.3
|
si:dkey-42p14.3 |
| chr12_+_45677293 | 0.08 |
ENSDART00000152850
ENSDART00000153047 |
si:ch73-111m19.2
|
si:ch73-111m19.2 |
| chr23_+_9206350 | 0.08 |
ENSDART00000136236
|
si:dkey-66g10.2
|
si:dkey-66g10.2 |
| chr1_-_34335752 | 0.08 |
ENSDART00000140157
|
si:dkey-24h22.5
|
si:dkey-24h22.5 |
| chr20_+_30445971 | 0.08 |
ENSDART00000153150
|
myt1la
|
myelin transcription factor 1-like, a |
| chr4_+_64339299 | 0.08 |
ENSDART00000191590
|
si:ch211-223a21.1
|
si:ch211-223a21.1 |
| chr2_+_45068366 | 0.08 |
ENSDART00000142175
|
si:dkey-76d14.2
|
si:dkey-76d14.2 |
| chr2_+_18988407 | 0.08 |
ENSDART00000170216
|
glula
|
glutamate-ammonia ligase (glutamine synthase) a |
| chr7_+_30867008 | 0.08 |
ENSDART00000193106
|
apba2b
|
amyloid beta (A4) precursor protein-binding, family A, member 2b |
| chr23_-_36306337 | 0.08 |
ENSDART00000142760
ENSDART00000136929 ENSDART00000143340 |
cbx5
|
chromobox homolog 5 (HP1 alpha homolog, Drosophila) |
| chr14_-_31151148 | 0.08 |
ENSDART00000026569
|
gpc4
|
glypican 4 |
| chr7_-_28413224 | 0.08 |
ENSDART00000076502
|
rerglb
|
RERG/RAS-like b |
| chr6_+_8176486 | 0.08 |
ENSDART00000193308
|
nfil3-5
|
nuclear factor, interleukin 3 regulated, member 5 |
| chr14_+_6963312 | 0.07 |
ENSDART00000150050
|
hnrnpaba
|
heterogeneous nuclear ribonucleoprotein A/Ba |
| chr7_+_27290548 | 0.07 |
ENSDART00000100998
|
sox6
|
SRY (sex determining region Y)-box 6 |
| chr25_+_13620555 | 0.07 |
ENSDART00000163642
|
si:ch211-172l8.4
|
si:ch211-172l8.4 |
| chr5_-_38179220 | 0.07 |
ENSDART00000147701
|
si:ch211-284e13.11
|
si:ch211-284e13.11 |
| chr1_-_28607353 | 0.07 |
ENSDART00000191210
|
FO907124.1
|
|
| chr10_-_25628555 | 0.07 |
ENSDART00000143978
|
tiam1a
|
T cell lymphoma invasion and metastasis 1a |
Network of associatons between targets according to the STRING database.
First level regulatory network of sox4a+sox4b
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 1.0 | GO:0016116 | tetraterpenoid metabolic process(GO:0016108) tetraterpenoid biosynthetic process(GO:0016109) carotenoid metabolic process(GO:0016116) carotenoid biosynthetic process(GO:0016117) xanthophyll metabolic process(GO:0016122) xanthophyll biosynthetic process(GO:0016123) zeaxanthin metabolic process(GO:1901825) zeaxanthin biosynthetic process(GO:1901827) |
| 0.1 | 0.5 | GO:0043152 | induction of bacterial agglutination(GO:0043152) |
| 0.1 | 0.3 | GO:0003156 | regulation of organ formation(GO:0003156) |
| 0.1 | 0.4 | GO:0042985 | negative regulation of glycoprotein biosynthetic process(GO:0010561) negative regulation of amyloid precursor protein biosynthetic process(GO:0042985) negative regulation of glycoprotein metabolic process(GO:1903019) |
| 0.1 | 0.9 | GO:1902299 | pre-replicative complex assembly involved in nuclear cell cycle DNA replication(GO:0006267) pre-replicative complex assembly(GO:0036388) pre-replicative complex assembly involved in cell cycle DNA replication(GO:1902299) |
| 0.1 | 0.4 | GO:0006642 | triglyceride mobilization(GO:0006642) |
| 0.1 | 0.4 | GO:0007412 | axon target recognition(GO:0007412) |
| 0.1 | 0.5 | GO:0090385 | phagosome-lysosome fusion(GO:0090385) |
| 0.1 | 0.3 | GO:0033986 | response to methanol(GO:0033986) cellular response to methanol(GO:0071405) |
| 0.0 | 0.4 | GO:0042989 | sequestering of actin monomers(GO:0042989) |
| 0.0 | 0.2 | GO:0045056 | transcytosis(GO:0045056) |
| 0.0 | 0.4 | GO:0036075 | endochondral ossification(GO:0001958) replacement ossification(GO:0036075) |
| 0.0 | 0.1 | GO:0021563 | glossopharyngeal nerve development(GO:0021563) |
| 0.0 | 0.4 | GO:0061303 | cornea development in camera-type eye(GO:0061303) |
| 0.0 | 0.1 | GO:1901492 | positive regulation of lymphangiogenesis(GO:1901492) |
| 0.0 | 0.1 | GO:0042941 | D-amino acid transport(GO:0042940) D-alanine transport(GO:0042941) D-serine transport(GO:0042942) |
| 0.0 | 0.1 | GO:0009447 | polyamine catabolic process(GO:0006598) putrescine catabolic process(GO:0009447) |
| 0.0 | 0.1 | GO:0051881 | regulation of mitochondrial membrane potential(GO:0051881) |
| 0.0 | 0.1 | GO:0042745 | circadian sleep/wake cycle process(GO:0022410) circadian sleep/wake cycle(GO:0042745) |
| 0.0 | 0.1 | GO:0086005 | ventricular cardiac muscle cell action potential(GO:0086005) |
| 0.0 | 0.2 | GO:1902868 | positive regulation of neural retina development(GO:0061075) positive regulation of retina development in camera-type eye(GO:1902868) |
| 0.0 | 0.2 | GO:0007525 | somatic muscle development(GO:0007525) |
| 0.0 | 0.1 | GO:0051230 | mitotic spindle disassembly(GO:0051228) spindle disassembly(GO:0051230) |
| 0.0 | 0.1 | GO:1990845 | adaptive thermogenesis(GO:1990845) |
| 0.0 | 0.6 | GO:0015804 | neutral amino acid transport(GO:0015804) |
| 0.0 | 0.2 | GO:0006607 | NLS-bearing protein import into nucleus(GO:0006607) |
| 0.0 | 0.0 | GO:0021961 | posterior commissure morphogenesis(GO:0021961) |
| 0.0 | 0.2 | GO:0000459 | exonucleolytic trimming involved in rRNA processing(GO:0000459) exonucleolytic trimming to generate mature 3'-end of 5.8S rRNA from tricistronic rRNA transcript (SSU-rRNA, 5.8S rRNA, LSU-rRNA)(GO:0000467) |
| 0.0 | 0.1 | GO:1902038 | positive regulation of hematopoietic stem cell differentiation(GO:1902038) |
| 0.0 | 0.3 | GO:0006208 | pyrimidine nucleobase catabolic process(GO:0006208) |
| 0.0 | 0.1 | GO:0036072 | intramembranous ossification(GO:0001957) direct ossification(GO:0036072) |
| 0.0 | 0.2 | GO:0099550 | trans-synaptic signalling, modulating synaptic transmission(GO:0099550) |
| 0.0 | 0.1 | GO:0000098 | sulfur amino acid catabolic process(GO:0000098) |
| 0.0 | 0.1 | GO:0043137 | DNA replication, removal of RNA primer(GO:0043137) |
| 0.0 | 0.1 | GO:2000251 | positive regulation of actin cytoskeleton reorganization(GO:2000251) |
| 0.0 | 0.5 | GO:0016339 | calcium-dependent cell-cell adhesion via plasma membrane cell adhesion molecules(GO:0016339) |
| 0.0 | 0.3 | GO:0000028 | ribosomal small subunit assembly(GO:0000028) |
| 0.0 | 0.4 | GO:0035329 | hippo signaling(GO:0035329) |
| 0.0 | 0.3 | GO:0030032 | lamellipodium assembly(GO:0030032) |
| 0.0 | 0.1 | GO:0006542 | glutamine biosynthetic process(GO:0006542) |
| 0.0 | 0.4 | GO:0046847 | filopodium assembly(GO:0046847) |
| 0.0 | 0.2 | GO:0048679 | regulation of axon regeneration(GO:0048679) regulation of neuron projection regeneration(GO:0070570) |
| 0.0 | 0.3 | GO:0021884 | forebrain neuron development(GO:0021884) |
| 0.0 | 0.3 | GO:0044243 | collagen catabolic process(GO:0030574) multicellular organism catabolic process(GO:0044243) |
| 0.0 | 0.1 | GO:0006189 | 'de novo' IMP biosynthetic process(GO:0006189) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.9 | GO:0005854 | nascent polypeptide-associated complex(GO:0005854) |
| 0.1 | 0.4 | GO:0009330 | DNA topoisomerase complex (ATP-hydrolyzing)(GO:0009330) |
| 0.1 | 0.4 | GO:0034359 | mature chylomicron(GO:0034359) |
| 0.1 | 0.9 | GO:0042555 | MCM complex(GO:0042555) |
| 0.0 | 0.2 | GO:0072669 | tRNA-splicing ligase complex(GO:0072669) |
| 0.0 | 0.2 | GO:0016012 | sarcoglycan complex(GO:0016012) |
| 0.0 | 0.1 | GO:0035301 | Hedgehog signaling complex(GO:0035301) |
| 0.0 | 0.1 | GO:0032299 | ribonuclease H2 complex(GO:0032299) |
| 0.0 | 0.1 | GO:0034098 | VCP-NPL4-UFD1 AAA ATPase complex(GO:0034098) |
| 0.0 | 0.1 | GO:0000930 | gamma-tubulin complex(GO:0000930) |
| 0.0 | 0.1 | GO:0005943 | phosphatidylinositol 3-kinase complex, class IA(GO:0005943) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 1.0 | GO:0050251 | retinol isomerase activity(GO:0050251) all-trans-retinyl-ester hydrolase, 11-cis retinol forming activity(GO:0052885) |
| 0.1 | 0.4 | GO:0003918 | DNA topoisomerase type II (ATP-hydrolyzing) activity(GO:0003918) DNA topoisomerase II activity(GO:0061505) |
| 0.1 | 0.4 | GO:0043140 | ATP-dependent 3'-5' DNA helicase activity(GO:0043140) single-stranded DNA-dependent ATP-dependent 3'-5' DNA helicase activity(GO:1990518) |
| 0.0 | 0.4 | GO:0070325 | low-density lipoprotein particle receptor binding(GO:0050750) lipoprotein particle receptor binding(GO:0070325) |
| 0.0 | 0.9 | GO:0003785 | actin monomer binding(GO:0003785) |
| 0.0 | 0.4 | GO:0001733 | galactosylceramide sulfotransferase activity(GO:0001733) galactose 3-O-sulfotransferase activity(GO:0050694) |
| 0.0 | 0.1 | GO:0017077 | oxidative phosphorylation uncoupler activity(GO:0017077) |
| 0.0 | 0.1 | GO:0035243 | protein-arginine omega-N symmetric methyltransferase activity(GO:0035243) |
| 0.0 | 0.1 | GO:0001223 | transcription coactivator binding(GO:0001223) |
| 0.0 | 0.1 | GO:0004385 | guanylate kinase activity(GO:0004385) |
| 0.0 | 0.5 | GO:0003688 | DNA replication origin binding(GO:0003688) |
| 0.0 | 0.2 | GO:0004499 | N,N-dimethylaniline monooxygenase activity(GO:0004499) |
| 0.0 | 0.3 | GO:0004157 | dihydropyrimidinase activity(GO:0004157) |
| 0.0 | 0.5 | GO:0001540 | beta-amyloid binding(GO:0001540) |
| 0.0 | 0.1 | GO:0030274 | LIM domain binding(GO:0030274) |
| 0.0 | 0.3 | GO:0004382 | guanosine-diphosphatase activity(GO:0004382) uridine-diphosphatase activity(GO:0045134) |
| 0.0 | 0.1 | GO:0019809 | spermidine binding(GO:0019809) |
| 0.0 | 0.6 | GO:0005540 | hyaluronic acid binding(GO:0005540) |
| 0.0 | 1.0 | GO:0005200 | structural constituent of cytoskeleton(GO:0005200) |
| 0.0 | 0.7 | GO:0015175 | neutral amino acid transmembrane transporter activity(GO:0015175) |
| 0.0 | 0.1 | GO:0004985 | opioid receptor activity(GO:0004985) |
| 0.0 | 0.2 | GO:0019911 | structural constituent of myelin sheath(GO:0019911) |
| 0.0 | 0.1 | GO:0008349 | MAP kinase kinase kinase kinase activity(GO:0008349) |
| 0.0 | 0.1 | GO:0008046 | axon guidance receptor activity(GO:0008046) |
| 0.0 | 0.0 | GO:0047173 | phosphatidylcholine-retinol O-acyltransferase activity(GO:0047173) |
| 0.0 | 0.1 | GO:0004523 | RNA-DNA hybrid ribonuclease activity(GO:0004523) |
| 0.0 | 0.1 | GO:0019871 | sodium channel inhibitor activity(GO:0019871) |
| 0.0 | 0.1 | GO:0016211 | glutamate-ammonia ligase activity(GO:0004356) ammonia ligase activity(GO:0016211) acid-ammonia (or amide) ligase activity(GO:0016880) |
| 0.0 | 0.1 | GO:0016742 | hydroxymethyl-, formyl- and related transferase activity(GO:0016742) |
| 0.0 | 0.1 | GO:0031843 | neuropeptide Y receptor binding(GO:0031841) type 2 neuropeptide Y receptor binding(GO:0031843) |
| 0.0 | 0.2 | GO:0070003 | threonine-type endopeptidase activity(GO:0004298) threonine-type peptidase activity(GO:0070003) |
| 0.0 | 0.1 | GO:0046934 | phosphatidylinositol-4,5-bisphosphate 3-kinase activity(GO:0046934) phosphatidylinositol-3,4-bisphosphate 5-kinase activity(GO:0052812) |
| 0.0 | 0.2 | GO:0000828 | inositol hexakisphosphate kinase activity(GO:0000828) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.4 | PID P38 MK2 PATHWAY | p38 signaling mediated by MAPKAP kinases |
| 0.0 | 0.3 | NABA PROTEOGLYCANS | Genes encoding proteoglycans |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.8 | REACTOME APOPTOSIS INDUCED DNA FRAGMENTATION | Genes involved in Apoptosis induced DNA fragmentation |
| 0.1 | 0.9 | REACTOME UNWINDING OF DNA | Genes involved in Unwinding of DNA |
| 0.0 | 0.6 | REACTOME POST CHAPERONIN TUBULIN FOLDING PATHWAY | Genes involved in Post-chaperonin tubulin folding pathway |
| 0.0 | 0.3 | REACTOME KERATAN SULFATE DEGRADATION | Genes involved in Keratan sulfate degradation |
| 0.0 | 0.1 | REACTOME CD28 DEPENDENT VAV1 PATHWAY | Genes involved in CD28 dependent Vav1 pathway |
| 0.0 | 0.2 | REACTOME PKA MEDIATED PHOSPHORYLATION OF CREB | Genes involved in PKA-mediated phosphorylation of CREB |