Project
PRJNA195909:zebrafish embryo and larva development
Navigation
Downloads
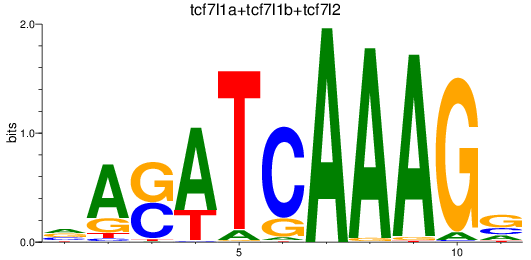
Results for tcf7l1a+tcf7l1b+tcf7l2
Z-value: 1.07
Transcription factors associated with tcf7l1a+tcf7l1b+tcf7l2
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
tcf7l2
|
ENSDARG00000004415 | transcription factor 7 like 2 |
|
tcf7l1b
|
ENSDARG00000007369 | transcription factor 7 like 1b |
|
tcf7l1a
|
ENSDARG00000038159 | transcription factor 7 like 1a |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| tcf7l2 | dr11_v1_chr12_-_31009315_31009315 | 0.98 | 7.0e-06 | Click! |
| tcf7l1b | dr11_v1_chr8_-_52230115_52230115 | -0.86 | 3.1e-03 | Click! |
| tcf7l1a | dr11_v1_chr10_-_42297889_42297889 | -0.71 | 3.2e-02 | Click! |
Activity profile of tcf7l1a+tcf7l1b+tcf7l2 motif
Sorted Z-values of tcf7l1a+tcf7l1b+tcf7l2 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr3_+_55131289 | 3.36 |
ENSDART00000111585
|
AL935210.1
|
|
| chr16_+_23913943 | 3.34 |
ENSDART00000175404
ENSDART00000129525 |
apoa4b.1
|
apolipoprotein A-IV b, tandem duplicate 1 |
| chr14_-_25949951 | 3.05 |
ENSDART00000141304
|
sparc
|
secreted protein, acidic, cysteine-rich (osteonectin) |
| chr3_+_55122662 | 2.54 |
ENSDART00000128380
|
hbbe1.2
|
hemoglobin beta embryonic-1.2 |
| chr3_+_55114097 | 2.31 |
ENSDART00000121686
|
hbbe1.1
|
hemoglobin beta embryonic-1.1 |
| chr16_-_45917322 | 2.27 |
ENSDART00000060822
|
afp4
|
antifreeze protein type IV |
| chr7_+_39446247 | 2.22 |
ENSDART00000033610
ENSDART00000099015 |
tnnt3b
|
troponin T type 3b (skeletal, fast) |
| chr10_-_39011514 | 1.92 |
ENSDART00000075123
|
pcp4a
|
Purkinje cell protein 4a |
| chr16_-_45917683 | 1.91 |
ENSDART00000184289
|
afp4
|
antifreeze protein type IV |
| chr3_+_27027781 | 1.65 |
ENSDART00000065495
|
emp2
|
epithelial membrane protein 2 |
| chr7_+_38750871 | 1.63 |
ENSDART00000114238
ENSDART00000052325 ENSDART00000137001 |
f2
|
coagulation factor II (thrombin) |
| chr20_-_40451115 | 1.57 |
ENSDART00000075092
|
pkib
|
protein kinase (cAMP-dependent, catalytic) inhibitor beta |
| chr25_+_21829777 | 1.56 |
ENSDART00000027393
|
ckmt1
|
creatine kinase, mitochondrial 1 |
| chr4_-_17409533 | 1.56 |
ENSDART00000011943
|
pah
|
phenylalanine hydroxylase |
| chr24_-_32408404 | 1.54 |
ENSDART00000144157
|
si:ch211-56a11.2
|
si:ch211-56a11.2 |
| chr13_-_31441042 | 1.51 |
ENSDART00000076571
|
rtn1a
|
reticulon 1a |
| chr15_+_32727848 | 1.49 |
ENSDART00000161361
|
postnb
|
periostin, osteoblast specific factor b |
| chr10_+_32104305 | 1.46 |
ENSDART00000099880
|
wnt11r
|
wingless-type MMTV integration site family, member 11, related |
| chr12_-_31103187 | 1.43 |
ENSDART00000005562
ENSDART00000031408 ENSDART00000125046 ENSDART00000009237 ENSDART00000122972 ENSDART00000153068 |
tcf7l2
|
transcription factor 7 like 2 |
| chr22_-_16042243 | 1.41 |
ENSDART00000062633
|
s1pr1
|
sphingosine-1-phosphate receptor 1 |
| chr21_-_41147818 | 1.38 |
ENSDART00000167339
ENSDART00000192730 |
msx2b
|
muscle segment homeobox 2b |
| chr11_-_42099645 | 1.36 |
ENSDART00000173312
|
abhd6a
|
abhydrolase domain containing 6a |
| chr9_+_7548533 | 1.34 |
ENSDART00000081543
|
ptprna
|
protein tyrosine phosphatase, receptor type, Na |
| chr6_-_26559921 | 1.34 |
ENSDART00000104532
|
sox14
|
SRY (sex determining region Y)-box 14 |
| chr20_-_48485354 | 1.33 |
ENSDART00000124040
ENSDART00000148437 |
insm1a
|
insulinoma-associated 1a |
| chr23_-_26077038 | 1.32 |
ENSDART00000126299
|
gdi1
|
GDP dissociation inhibitor 1 |
| chr6_+_49255706 | 1.28 |
ENSDART00000156866
|
si:dkey-183k8.2
|
si:dkey-183k8.2 |
| chr1_+_26356360 | 1.27 |
ENSDART00000113020
|
tet2
|
tet methylcytosine dioxygenase 2 |
| chr24_+_11334733 | 1.24 |
ENSDART00000147552
ENSDART00000143171 |
si:dkey-12l12.1
|
si:dkey-12l12.1 |
| chr20_+_22666548 | 1.21 |
ENSDART00000147520
|
lnx1
|
ligand of numb-protein X 1 |
| chr8_-_41228530 | 1.18 |
ENSDART00000165949
ENSDART00000173055 |
fahd2a
|
fumarylacetoacetate hydrolase domain containing 2A |
| chr1_-_29045426 | 1.16 |
ENSDART00000019770
|
gpm6ba
|
glycoprotein M6Ba |
| chr14_+_24277556 | 1.15 |
ENSDART00000122660
|
hnrnpa0a
|
heterogeneous nuclear ribonucleoprotein A0a |
| chr6_+_52804267 | 1.14 |
ENSDART00000065681
|
matn4
|
matrilin 4 |
| chr11_-_23219367 | 1.13 |
ENSDART00000003646
|
optc
|
opticin |
| chr1_-_36151377 | 1.12 |
ENSDART00000037516
|
znf827
|
zinc finger protein 827 |
| chr6_-_57938043 | 1.12 |
ENSDART00000171073
|
tox2
|
TOX high mobility group box family member 2 |
| chr5_+_42280372 | 1.10 |
ENSDART00000142855
|
tbx6l
|
T-box 6, like |
| chr1_-_40911332 | 1.07 |
ENSDART00000027463
|
hmx4
|
H6 family homeobox 4 |
| chr22_-_26100282 | 1.07 |
ENSDART00000166075
|
pdgfaa
|
platelet-derived growth factor alpha polypeptide a |
| chr23_-_29824146 | 1.06 |
ENSDART00000020616
|
zgc:194189
|
zgc:194189 |
| chr7_-_41554047 | 1.05 |
ENSDART00000174144
|
plxdc2
|
plexin domain containing 2 |
| chr6_-_40429411 | 1.05 |
ENSDART00000156005
ENSDART00000156357 |
si:dkey-28n18.9
|
si:dkey-28n18.9 |
| chr7_-_9803154 | 1.04 |
ENSDART00000055593
|
aldh1a3
|
aldehyde dehydrogenase 1 family, member A3 |
| chr8_+_13106760 | 1.04 |
ENSDART00000029308
|
itgb4
|
integrin, beta 4 |
| chr1_+_26356205 | 1.03 |
ENSDART00000190064
ENSDART00000176380 |
tet2
|
tet methylcytosine dioxygenase 2 |
| chr16_+_26774182 | 1.03 |
ENSDART00000042895
|
cdh17
|
cadherin 17, LI cadherin (liver-intestine) |
| chr25_-_22187397 | 1.03 |
ENSDART00000123211
ENSDART00000139110 |
pkp3a
|
plakophilin 3a |
| chr7_+_57836841 | 1.00 |
ENSDART00000136175
|
camk2d1
|
calcium/calmodulin-dependent protein kinase (CaM kinase) II delta 1 |
| chr22_-_26005894 | 0.99 |
ENSDART00000105088
|
pdgfaa
|
platelet-derived growth factor alpha polypeptide a |
| chr12_+_5129245 | 0.97 |
ENSDART00000169073
|
pde6c
|
phosphodiesterase 6C, cGMP-specific, cone, alpha prime |
| chr19_-_9522548 | 0.96 |
ENSDART00000045245
|
ing4
|
inhibitor of growth family, member 4 |
| chr21_+_19834072 | 0.96 |
ENSDART00000147555
|
ccdc80l2
|
coiled-coil domain containing 80 like 2 |
| chr16_+_32559821 | 0.96 |
ENSDART00000093250
|
pou3f2b
|
POU class 3 homeobox 2b |
| chr8_+_16407884 | 0.95 |
ENSDART00000133742
|
cdkn2c
|
cyclin-dependent kinase inhibitor 2C (p18, inhibits CDK4) |
| chr4_+_69619 | 0.94 |
ENSDART00000164425
|
mansc1
|
MANSC domain containing 1 |
| chr20_-_9436521 | 0.93 |
ENSDART00000133000
|
zgc:101840
|
zgc:101840 |
| chr5_-_24333684 | 0.91 |
ENSDART00000051553
|
znf703
|
zinc finger protein 703 |
| chr7_-_54677143 | 0.91 |
ENSDART00000163748
|
ccnd1
|
cyclin D1 |
| chr2_+_33368414 | 0.91 |
ENSDART00000077462
|
slc6a9
|
solute carrier family 6 (neurotransmitter transporter, glycine), member 9 |
| chr13_-_31647323 | 0.91 |
ENSDART00000135381
|
six4a
|
SIX homeobox 4a |
| chr15_-_2188332 | 0.90 |
ENSDART00000138941
ENSDART00000009564 |
shox2
|
short stature homeobox 2 |
| chr5_+_10015002 | 0.88 |
ENSDART00000140570
|
slc2a11b
|
solute carrier family 2 (facilitated glucose transporter), member 11b |
| chr11_+_14622379 | 0.87 |
ENSDART00000112589
|
efna2b
|
ephrin-A2b |
| chr7_-_37622370 | 0.85 |
ENSDART00000173523
|
nkd1
|
naked cuticle homolog 1 (Drosophila) |
| chr7_-_37622153 | 0.84 |
ENSDART00000052379
|
nkd1
|
naked cuticle homolog 1 (Drosophila) |
| chr19_+_48049202 | 0.83 |
ENSDART00000027158
|
psmd3
|
proteasome 26S subunit, non-ATPase 3 |
| chr20_+_34455645 | 0.83 |
ENSDART00000135789
|
mettl11b
|
methyltransferase like 11B |
| chr18_+_45796096 | 0.83 |
ENSDART00000087070
|
abcc5
|
ATP-binding cassette, sub-family C (CFTR/MRP), member 5 |
| chr2_+_22042745 | 0.82 |
ENSDART00000132039
|
tox
|
thymocyte selection-associated high mobility group box |
| chr5_-_29570141 | 0.81 |
ENSDART00000043259
|
entpd2a.2
|
ectonucleoside triphosphate diphosphohydrolase 2a, tandem duplicate 2 |
| chr1_-_42779075 | 0.80 |
ENSDART00000133917
|
lrrtm1
|
leucine rich repeat transmembrane neuronal 1 |
| chr24_+_6107901 | 0.80 |
ENSDART00000156419
|
si:ch211-37e10.2
|
si:ch211-37e10.2 |
| chr9_-_9992697 | 0.79 |
ENSDART00000123415
|
ugt1ab
|
UDP glucuronosyltransferase 1 family a, b |
| chr19_+_31771270 | 0.78 |
ENSDART00000147474
|
stmn2b
|
stathmin 2b |
| chr7_+_69653981 | 0.77 |
ENSDART00000090165
|
frem1a
|
Fras1 related extracellular matrix 1a |
| chr15_-_8309207 | 0.77 |
ENSDART00000143880
ENSDART00000061351 |
tnfrsf19
|
tumor necrosis factor receptor superfamily, member 19 |
| chr14_+_15064870 | 0.76 |
ENSDART00000167075
|
nkx1.2lb
|
NK1 transcription factor related 2-like,b |
| chr20_-_42702832 | 0.76 |
ENSDART00000134689
ENSDART00000045816 |
plg
|
plasminogen |
| chr20_+_25225112 | 0.76 |
ENSDART00000153088
ENSDART00000127291 ENSDART00000130494 |
moxd1
|
monooxygenase, DBH-like 1 |
| chr3_+_12484008 | 0.76 |
ENSDART00000182229
|
vasnb
|
vasorin b |
| chr17_-_20897250 | 0.74 |
ENSDART00000088106
|
ank3b
|
ankyrin 3b |
| chr17_-_14816465 | 0.73 |
ENSDART00000156326
ENSDART00000156287 |
nid2a
|
nidogen 2a (osteonidogen) |
| chr5_-_65000312 | 0.72 |
ENSDART00000192893
|
ANXA1 (1 of many)
|
zgc:110283 |
| chr1_+_2190714 | 0.72 |
ENSDART00000132126
|
mbnl2
|
muscleblind-like splicing regulator 2 |
| chr24_+_23742690 | 0.72 |
ENSDART00000130162
|
TCF24
|
transcription factor 24 |
| chr19_+_29798064 | 0.69 |
ENSDART00000167803
ENSDART00000051804 |
marcksl1b
|
MARCKS-like 1b |
| chr9_+_28693592 | 0.68 |
ENSDART00000110198
|
zgc:162780
|
zgc:162780 |
| chr1_-_8428736 | 0.68 |
ENSDART00000138435
ENSDART00000121823 |
syngr3b
|
synaptogyrin 3b |
| chr9_+_17306162 | 0.68 |
ENSDART00000075926
|
scel
|
sciellin |
| chr4_-_77432218 | 0.68 |
ENSDART00000158683
|
slco1d1
|
solute carrier organic anion transporter family, member 1D1 |
| chr23_-_20764227 | 0.68 |
ENSDART00000089750
|
znf362b
|
zinc finger protein 362b |
| chr19_+_42983613 | 0.68 |
ENSDART00000033724
|
fabp3
|
fatty acid binding protein 3, muscle and heart |
| chr18_-_44888375 | 0.67 |
ENSDART00000160506
|
si:ch211-71n6.4
|
si:ch211-71n6.4 |
| chr15_-_12545683 | 0.66 |
ENSDART00000162807
|
scn2b
|
sodium channel, voltage-gated, type II, beta |
| chr10_-_17466990 | 0.66 |
ENSDART00000147794
|
nt5c2l1
|
5'-nucleotidase, cytosolic II, like 1 |
| chr1_+_17376922 | 0.64 |
ENSDART00000145068
|
fat1a
|
FAT atypical cadherin 1a |
| chr8_-_38105053 | 0.64 |
ENSDART00000131546
|
adgra2
|
adhesion G protein-coupled receptor A2 |
| chr21_-_25522510 | 0.64 |
ENSDART00000162711
|
cnksr2b
|
connector enhancer of kinase suppressor of Ras 2b |
| chr21_-_17482465 | 0.63 |
ENSDART00000004548
|
barhl1b
|
BarH-like homeobox 1b |
| chr6_+_58915889 | 0.60 |
ENSDART00000083628
|
ddit3
|
DNA-damage-inducible transcript 3 |
| chr1_-_42778510 | 0.60 |
ENSDART00000190172
|
lrrtm1
|
leucine rich repeat transmembrane neuronal 1 |
| chr23_-_45705525 | 0.58 |
ENSDART00000148959
|
ednrab
|
endothelin receptor type Ab |
| chr13_-_18835254 | 0.58 |
ENSDART00000147579
ENSDART00000146795 |
lzts2a
|
leucine zipper, putative tumor suppressor 2a |
| chr15_+_7086327 | 0.57 |
ENSDART00000114560
|
pik3cb
|
phosphatidylinositol-4,5-bisphosphate 3-kinase, catalytic subunit beta |
| chr20_+_43942278 | 0.57 |
ENSDART00000100571
|
clic5b
|
chloride intracellular channel 5b |
| chr6_-_6976096 | 0.57 |
ENSDART00000151822
ENSDART00000039443 ENSDART00000177960 |
tuba8l4
|
tubulin, alpha 8 like 4 |
| chr17_+_27176243 | 0.56 |
ENSDART00000162527
|
si:ch211-160f23.7
|
si:ch211-160f23.7 |
| chr10_-_31562695 | 0.55 |
ENSDART00000186456
|
robo3
|
roundabout, axon guidance receptor, homolog 3 (Drosophila) |
| chr5_+_34407763 | 0.54 |
ENSDART00000188849
ENSDART00000145127 |
lamc3
|
laminin, gamma 3 |
| chr7_+_34492744 | 0.54 |
ENSDART00000109635
ENSDART00000173844 |
calml4a
|
calmodulin-like 4a |
| chr13_+_27073901 | 0.54 |
ENSDART00000146227
|
slc24a3
|
solute carrier family 24 (sodium/potassium/calcium exchanger), member 3 |
| chr16_-_383664 | 0.53 |
ENSDART00000051693
|
irx4a
|
iroquois homeobox 4a |
| chr16_+_16933002 | 0.53 |
ENSDART00000173163
|
myh14
|
myosin, heavy chain 14, non-muscle |
| chr14_+_22447662 | 0.53 |
ENSDART00000161776
|
sowahab
|
sosondowah ankyrin repeat domain family member Ab |
| chr9_-_9998087 | 0.53 |
ENSDART00000124423
|
ugt1ab
|
UDP glucuronosyltransferase 1 family a, b |
| chr20_+_43083745 | 0.52 |
ENSDART00000139014
ENSDART00000153438 |
moxd1l
|
monooxygenase, DBH-like 1, like |
| chr8_+_16408385 | 0.52 |
ENSDART00000177231
|
cdkn2c
|
cyclin-dependent kinase inhibitor 2C (p18, inhibits CDK4) |
| chr1_+_36674584 | 0.52 |
ENSDART00000186772
ENSDART00000192274 |
ednraa
|
endothelin receptor type Aa |
| chr20_+_30445971 | 0.50 |
ENSDART00000153150
|
myt1la
|
myelin transcription factor 1-like, a |
| chr22_+_35930526 | 0.49 |
ENSDART00000169242
|
LO016987.1
|
|
| chr2_+_30379650 | 0.49 |
ENSDART00000129542
|
crispld1b
|
cysteine-rich secretory protein LCCL domain containing 1b |
| chr23_+_18722915 | 0.48 |
ENSDART00000025057
|
myh7bb
|
myosin, heavy chain 7B, cardiac muscle, beta b |
| chr25_+_7229046 | 0.48 |
ENSDART00000149965
ENSDART00000041820 |
lingo1a
|
leucine rich repeat and Ig domain containing 1a |
| chr4_+_10366532 | 0.47 |
ENSDART00000189901
|
kcnd2
|
potassium voltage-gated channel, Shal-related subfamily, member 2 |
| chr8_-_34051548 | 0.46 |
ENSDART00000105204
|
pbx3b
|
pre-B-cell leukemia homeobox 3b |
| chr8_-_34052019 | 0.46 |
ENSDART00000040126
ENSDART00000159208 ENSDART00000048994 ENSDART00000098822 |
pbx3b
|
pre-B-cell leukemia homeobox 3b |
| chr5_+_27442247 | 0.46 |
ENSDART00000184129
|
loxl2b
|
lysyl oxidase-like 2b |
| chr16_+_19014886 | 0.46 |
ENSDART00000079298
|
si:ch211-254p10.2
|
si:ch211-254p10.2 |
| chr5_-_27867657 | 0.45 |
ENSDART00000112495
|
tcima
|
transcriptional and immune response regulator a |
| chr19_-_8880688 | 0.45 |
ENSDART00000039629
|
celf3a
|
cugbp, Elav-like family member 3a |
| chr8_-_25329967 | 0.44 |
ENSDART00000139682
|
eps8l3b
|
EPS8-like 3b |
| chr19_-_32487469 | 0.44 |
ENSDART00000050130
|
gmpr
|
guanosine monophosphate reductase |
| chr25_+_14092871 | 0.44 |
ENSDART00000067239
|
guca1g
|
guanylate cyclase activator 1g |
| chr22_+_13917311 | 0.43 |
ENSDART00000022654
|
sh3bp4a
|
SH3-domain binding protein 4a |
| chr13_+_3252950 | 0.43 |
ENSDART00000020671
|
prph2b
|
peripherin 2b (retinal degeneration, slow) |
| chr5_-_22082918 | 0.42 |
ENSDART00000020908
|
zc4h2
|
zinc finger, C4H2 domain containing |
| chr21_-_37889727 | 0.41 |
ENSDART00000163612
ENSDART00000180958 |
gabrb4
|
gamma-aminobutyric acid (GABA) A receptor, beta 4 |
| chr14_+_22591624 | 0.41 |
ENSDART00000108987
|
gfra4b
|
GDNF family receptor alpha 4b |
| chr17_-_20897407 | 0.40 |
ENSDART00000149481
|
ank3b
|
ankyrin 3b |
| chr7_+_38936132 | 0.40 |
ENSDART00000173945
|
creb3l1
|
cAMP responsive element binding protein 3-like 1 |
| chr15_-_163586 | 0.40 |
ENSDART00000163597
|
SEPT4
|
septin-4 |
| chr24_-_6546479 | 0.38 |
ENSDART00000160538
|
arhgap21a
|
Rho GTPase activating protein 21a |
| chr2_-_43083749 | 0.38 |
ENSDART00000084303
ENSDART00000145494 |
kcnq3
|
potassium voltage-gated channel, KQT-like subfamily, member 3 |
| chr20_+_37294112 | 0.38 |
ENSDART00000076293
ENSDART00000140450 |
cx23
|
connexin 23 |
| chr10_-_31563049 | 0.38 |
ENSDART00000023575
|
robo3
|
roundabout, axon guidance receptor, homolog 3 (Drosophila) |
| chr23_+_28077953 | 0.37 |
ENSDART00000186122
ENSDART00000111570 |
slc26a10
|
solute carrier family 26, member 10 |
| chr1_-_39976492 | 0.35 |
ENSDART00000181680
|
stox2a
|
storkhead box 2a |
| chr20_+_25597527 | 0.35 |
ENSDART00000130242
|
cyp2j20
|
cytochrome P450, family 2, subfamily J, polypeptide 20 |
| chr1_-_681116 | 0.35 |
ENSDART00000165894
|
adamts1
|
ADAM metallopeptidase with thrombospondin type 1 motif, 1 |
| chr5_+_45008418 | 0.35 |
ENSDART00000189882
|
dmrt2a
|
doublesex and mab-3 related transcription factor 2a |
| chr21_-_25522906 | 0.34 |
ENSDART00000110923
|
cnksr2b
|
connector enhancer of kinase suppressor of Ras 2b |
| chr13_+_22476742 | 0.33 |
ENSDART00000078759
ENSDART00000130101 ENSDART00000137220 ENSDART00000133065 ENSDART00000147348 |
ldb3a
|
LIM domain binding 3a |
| chr11_+_40043569 | 0.33 |
ENSDART00000130278
|
si:dkey-264d12.1
|
si:dkey-264d12.1 |
| chr3_-_32299892 | 0.32 |
ENSDART00000181472
|
si:dkey-16p21.7
|
si:dkey-16p21.7 |
| chr23_-_39784368 | 0.32 |
ENSDART00000110282
|
si:ch211-286f9.2
|
si:ch211-286f9.2 |
| chr19_+_43119698 | 0.31 |
ENSDART00000167847
ENSDART00000186962 ENSDART00000187305 |
eef1a1l1
|
eukaryotic translation elongation factor 1 alpha 1, like 1 |
| chr9_+_10014817 | 0.31 |
ENSDART00000132065
|
nxph2a
|
neurexophilin 2a |
| chr11_-_29818866 | 0.31 |
ENSDART00000176367
|
cybb
|
cytochrome b-245, beta polypeptide (chronic granulomatous disease) |
| chr20_+_10727022 | 0.31 |
ENSDART00000104185
|
si:ch211-182e10.4
|
si:ch211-182e10.4 |
| chr24_+_11105786 | 0.30 |
ENSDART00000175182
|
prlh2
|
prolactin releasing hormone 2 |
| chr9_-_3671911 | 0.30 |
ENSDART00000102900
|
sp5a
|
Sp5 transcription factor a |
| chr16_+_17389116 | 0.30 |
ENSDART00000103750
ENSDART00000173448 |
fam131bb
|
family with sequence similarity 131, member Bb |
| chr4_-_2036620 | 0.30 |
ENSDART00000150490
|
si:dkey-97m3.1
|
si:dkey-97m3.1 |
| chr13_-_31829786 | 0.30 |
ENSDART00000138667
|
sertad4
|
SERTA domain containing 4 |
| chr21_-_20328375 | 0.29 |
ENSDART00000079593
|
slc26a1
|
solute carrier family 26 (anion exchanger), member 1 |
| chr2_-_34483597 | 0.29 |
ENSDART00000133224
|
brinp2
|
bone morphogenetic protein/retinoic acid inducible neural-specific 2 |
| chr8_-_30242706 | 0.29 |
ENSDART00000139864
ENSDART00000143809 |
zgc:162939
|
zgc:162939 |
| chr9_+_29994439 | 0.29 |
ENSDART00000012447
|
tmem30c
|
transmembrane protein 30C |
| chr7_-_18554603 | 0.29 |
ENSDART00000108938
|
msantd1
|
Myb/SANT-like DNA-binding domain containing 1 |
| chr20_+_39283849 | 0.28 |
ENSDART00000002481
ENSDART00000146683 |
scara3
|
scavenger receptor class A, member 3 |
| chr5_-_52921532 | 0.28 |
ENSDART00000160566
|
CU695117.1
|
|
| chr18_-_34143189 | 0.28 |
ENSDART00000079341
|
plch1
|
phospholipase C, eta 1 |
| chr15_-_31588162 | 0.28 |
ENSDART00000153598
|
hsph1
|
heat shock 105/110 protein 1 |
| chr2_-_59376399 | 0.27 |
ENSDART00000137134
|
ftr38
|
finTRIM family, member 38 |
| chr2_-_59376735 | 0.27 |
ENSDART00000193624
|
ftr38
|
finTRIM family, member 38 |
| chr3_+_41714966 | 0.27 |
ENSDART00000155440
|
eif3ba
|
eukaryotic translation initiation factor 3, subunit Ba |
| chr4_-_880415 | 0.27 |
ENSDART00000149162
|
aim1b
|
crystallin beta-gamma domain containing 1b |
| chr9_+_10014514 | 0.26 |
ENSDART00000185590
|
nxph2a
|
neurexophilin 2a |
| chr11_+_6080528 | 0.24 |
ENSDART00000183099
|
ushbp1
|
Usher syndrome 1C binding protein 1 |
| chr3_-_23643751 | 0.24 |
ENSDART00000078425
ENSDART00000140264 |
eve1
|
even-skipped-like1 |
| chr23_+_33907899 | 0.24 |
ENSDART00000159445
|
cs
|
citrate synthase |
| chr20_+_36234335 | 0.24 |
ENSDART00000193484
ENSDART00000181664 |
cnih3
|
cornichon family AMPA receptor auxiliary protein 3 |
| chr15_+_24691088 | 0.24 |
ENSDART00000110618
|
LRRC75A
|
si:dkey-151p21.7 |
| chr22_+_21324398 | 0.24 |
ENSDART00000168509
|
shc2
|
SHC (Src homology 2 domain containing) transforming protein 2 |
| chr9_-_35155089 | 0.23 |
ENSDART00000077901
|
appb
|
amyloid beta (A4) precursor protein b |
| chr1_+_35862550 | 0.23 |
ENSDART00000132118
|
si:ch211-194g2.4
|
si:ch211-194g2.4 |
| chr13_+_28732101 | 0.22 |
ENSDART00000015773
|
ldb1a
|
LIM domain binding 1a |
| chr6_+_15762647 | 0.22 |
ENSDART00000127133
ENSDART00000128939 |
iqca1
|
IQ motif containing with AAA domain 1 |
| chr9_+_44034790 | 0.21 |
ENSDART00000166110
ENSDART00000176954 |
itga4
|
integrin alpha 4 |
| chr25_+_35020529 | 0.21 |
ENSDART00000158016
|
flnca
|
filamin C, gamma a (actin binding protein 280) |
| chr19_+_42469058 | 0.21 |
ENSDART00000076915
|
si:dkey-166k12.1
|
si:dkey-166k12.1 |
| chr6_-_29105727 | 0.20 |
ENSDART00000184355
|
fam69ab
|
family with sequence similarity 69, member Ab |
| chr12_+_48204891 | 0.20 |
ENSDART00000190534
ENSDART00000164427 |
ndr2
|
nodal-related 2 |
| chr19_+_43119014 | 0.20 |
ENSDART00000023156
|
eef1a1l1
|
eukaryotic translation elongation factor 1 alpha 1, like 1 |
| chr12_+_6465557 | 0.20 |
ENSDART00000066477
ENSDART00000122271 |
dkk1b
|
dickkopf WNT signaling pathway inhibitor 1b |
| chr21_-_40782393 | 0.19 |
ENSDART00000075808
|
apbb3
|
amyloid beta (A4) precursor protein-binding, family B, member 3 |
| chr8_+_36803415 | 0.19 |
ENSDART00000111680
|
iqsec2b
|
IQ motif and Sec7 domain 2b |
| chr17_+_5800286 | 0.19 |
ENSDART00000175504
|
rp1l1a
|
retinitis pigmentosa 1-like 1a |
| chr24_+_15897717 | 0.18 |
ENSDART00000105956
|
neto1l
|
neuropilin (NRP) and tolloid (TLL)-like 1, like |
Network of associatons between targets according to the STRING database.
First level regulatory network of tcf7l1a+tcf7l1b+tcf7l2
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.6 | 1.7 | GO:0001977 | renal system process involved in regulation of blood volume(GO:0001977) renal system process involved in regulation of systemic arterial blood pressure(GO:0003071) regulation of glomerular filtration(GO:0003093) |
| 0.5 | 1.4 | GO:0003376 | sphingosine-1-phosphate signaling pathway(GO:0003376) |
| 0.4 | 1.3 | GO:0046333 | octopamine biosynthetic process(GO:0006589) dopamine catabolic process(GO:0042420) norepinephrine biosynthetic process(GO:0042421) octopamine metabolic process(GO:0046333) |
| 0.4 | 1.7 | GO:0046823 | regulation of protein import into nucleus(GO:0042306) negative regulation of protein import into nucleus(GO:0042308) negative regulation of nucleocytoplasmic transport(GO:0046823) negative regulation of protein localization to nucleus(GO:1900181) regulation of protein import(GO:1904589) negative regulation of protein import(GO:1904590) |
| 0.4 | 2.1 | GO:0003261 | cardiac muscle progenitor cell migration to the midline involved in heart field formation(GO:0003261) |
| 0.4 | 1.2 | GO:0051580 | regulation of neurotransmitter uptake(GO:0051580) negative regulation of anion transport(GO:1903792) |
| 0.4 | 1.4 | GO:0010226 | response to lithium ion(GO:0010226) |
| 0.3 | 2.3 | GO:0019857 | 5-methylcytosine catabolic process(GO:0006211) 5-methylcytosine metabolic process(GO:0019857) |
| 0.3 | 0.6 | GO:0003319 | cardioblast migration to the midline involved in heart rudiment formation(GO:0003319) |
| 0.3 | 1.4 | GO:0014045 | establishment of endothelial blood-brain barrier(GO:0014045) |
| 0.3 | 0.8 | GO:0006480 | N-terminal protein amino acid methylation(GO:0006480) |
| 0.3 | 1.6 | GO:0006953 | acute-phase response(GO:0006953) |
| 0.3 | 0.8 | GO:0042730 | fibrinolysis(GO:0042730) |
| 0.2 | 0.9 | GO:0060092 | regulation of synaptic transmission, glycinergic(GO:0060092) |
| 0.2 | 1.3 | GO:0060221 | retinal rod cell differentiation(GO:0060221) |
| 0.2 | 0.9 | GO:0031571 | mitotic G1 DNA damage checkpoint(GO:0031571) |
| 0.2 | 0.9 | GO:2000480 | negative regulation of cAMP-dependent protein kinase activity(GO:2000480) |
| 0.1 | 0.6 | GO:2001244 | positive regulation of intrinsic apoptotic signaling pathway(GO:2001244) |
| 0.1 | 0.8 | GO:0033986 | response to methanol(GO:0033986) cellular response to methanol(GO:0071405) |
| 0.1 | 1.6 | GO:0006559 | L-phenylalanine metabolic process(GO:0006558) L-phenylalanine catabolic process(GO:0006559) erythrose 4-phosphate/phosphoenolpyruvate family amino acid metabolic process(GO:1902221) erythrose 4-phosphate/phosphoenolpyruvate family amino acid catabolic process(GO:1902222) |
| 0.1 | 1.6 | GO:0042396 | phosphagen metabolic process(GO:0006599) phosphocreatine metabolic process(GO:0006603) phosphagen biosynthetic process(GO:0042396) phosphocreatine biosynthetic process(GO:0046314) |
| 0.1 | 3.3 | GO:0060030 | dorsal convergence(GO:0060030) |
| 0.1 | 0.5 | GO:1902455 | negative regulation of stem cell population maintenance(GO:1902455) |
| 0.1 | 2.4 | GO:0015671 | oxygen transport(GO:0015671) |
| 0.1 | 0.9 | GO:0031063 | regulation of histone deacetylation(GO:0031063) |
| 0.1 | 0.4 | GO:0034260 | negative regulation of GTPase activity(GO:0034260) |
| 0.1 | 3.1 | GO:0032474 | otolith morphogenesis(GO:0032474) semicircular canal morphogenesis(GO:0048752) |
| 0.1 | 2.2 | GO:0006942 | regulation of striated muscle contraction(GO:0006942) |
| 0.1 | 0.5 | GO:0035176 | social behavior(GO:0035176) intraspecies interaction between organisms(GO:0051703) |
| 0.1 | 0.8 | GO:0015886 | heme transport(GO:0015886) iron coordination entity transport(GO:1901678) |
| 0.1 | 1.5 | GO:0016203 | muscle attachment(GO:0016203) |
| 0.1 | 0.9 | GO:0071679 | commissural neuron axon guidance(GO:0071679) |
| 0.1 | 0.4 | GO:1900029 | positive regulation of ruffle assembly(GO:1900029) |
| 0.1 | 0.3 | GO:0019532 | oxalate transport(GO:0019532) |
| 0.1 | 0.6 | GO:0051013 | microtubule severing(GO:0051013) |
| 0.1 | 0.3 | GO:0006570 | tyrosine metabolic process(GO:0006570) |
| 0.1 | 0.2 | GO:0002568 | somatic diversification of T cell receptor genes(GO:0002568) somatic recombination of T cell receptor gene segments(GO:0002681) T cell receptor V(D)J recombination(GO:0033153) |
| 0.1 | 0.5 | GO:0034755 | iron ion transmembrane transport(GO:0034755) |
| 0.1 | 1.0 | GO:0031581 | hemidesmosome assembly(GO:0031581) |
| 0.1 | 0.2 | GO:0055057 | neuroblast division(GO:0055057) |
| 0.0 | 0.9 | GO:0030316 | osteoclast differentiation(GO:0030316) |
| 0.0 | 0.2 | GO:0061011 | hepatic duct development(GO:0061011) |
| 0.0 | 1.5 | GO:0010921 | regulation of phosphatase activity(GO:0010921) |
| 0.0 | 0.3 | GO:0042554 | superoxide anion generation(GO:0042554) |
| 0.0 | 0.3 | GO:0003313 | heart rudiment development(GO:0003313) |
| 0.0 | 1.4 | GO:0061386 | closure of optic fissure(GO:0061386) |
| 0.0 | 0.7 | GO:0043252 | sodium-independent organic anion transport(GO:0043252) |
| 0.0 | 0.8 | GO:0008345 | larval locomotory behavior(GO:0008345) larval behavior(GO:0030537) |
| 0.0 | 1.0 | GO:0045494 | photoreceptor cell maintenance(GO:0045494) |
| 0.0 | 0.6 | GO:0036092 | phosphatidylinositol-3-phosphate biosynthetic process(GO:0036092) |
| 0.0 | 0.8 | GO:1903845 | negative regulation of transforming growth factor beta receptor signaling pathway(GO:0030512) negative regulation of cellular response to transforming growth factor beta stimulus(GO:1903845) |
| 0.0 | 0.3 | GO:0071300 | cellular response to retinoic acid(GO:0071300) |
| 0.0 | 0.4 | GO:0048923 | posterior lateral line neuromast hair cell differentiation(GO:0048923) |
| 0.0 | 0.2 | GO:0046619 | optic placode formation involved in camera-type eye formation(GO:0046619) |
| 0.0 | 0.2 | GO:0046716 | muscle cell cellular homeostasis(GO:0046716) |
| 0.0 | 0.5 | GO:0048026 | positive regulation of mRNA splicing, via spliceosome(GO:0048026) |
| 0.0 | 3.3 | GO:0042157 | lipoprotein metabolic process(GO:0042157) |
| 0.0 | 1.0 | GO:0001885 | endothelial cell development(GO:0001885) |
| 0.0 | 0.4 | GO:0006144 | purine nucleobase metabolic process(GO:0006144) |
| 0.0 | 0.4 | GO:0008272 | sulfate transport(GO:0008272) |
| 0.0 | 0.2 | GO:0045471 | response to ethanol(GO:0045471) |
| 0.0 | 0.4 | GO:0048665 | neuron fate specification(GO:0048665) |
| 0.0 | 0.1 | GO:0038063 | collagen-activated tyrosine kinase receptor signaling pathway(GO:0038063) collagen-activated signaling pathway(GO:0038065) |
| 0.0 | 0.2 | GO:0014823 | response to activity(GO:0014823) |
| 0.0 | 0.1 | GO:0015015 | heparan sulfate proteoglycan biosynthetic process, enzymatic modification(GO:0015015) |
| 0.0 | 0.9 | GO:0008016 | regulation of heart contraction(GO:0008016) |
| 0.0 | 0.5 | GO:0031103 | axon regeneration(GO:0031103) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.4 | 1.4 | GO:0071664 | catenin-TCF7L2 complex(GO:0071664) |
| 0.2 | 2.4 | GO:0031838 | haptoglobin-hemoglobin complex(GO:0031838) |
| 0.2 | 0.6 | GO:1990909 | Wnt signalosome(GO:1990909) |
| 0.1 | 0.4 | GO:1902560 | GMP reductase complex(GO:1902560) |
| 0.1 | 0.8 | GO:0008541 | proteasome regulatory particle, lid subcomplex(GO:0008541) |
| 0.1 | 2.2 | GO:0017053 | transcriptional repressor complex(GO:0017053) |
| 0.1 | 1.3 | GO:0030667 | secretory granule membrane(GO:0030667) |
| 0.1 | 1.0 | GO:0030057 | desmosome(GO:0030057) |
| 0.1 | 2.3 | GO:0005861 | troponin complex(GO:0005861) |
| 0.1 | 0.4 | GO:0043194 | axon initial segment(GO:0043194) |
| 0.1 | 0.9 | GO:0016363 | nuclear matrix(GO:0016363) |
| 0.0 | 0.3 | GO:0043020 | NADPH oxidase complex(GO:0043020) |
| 0.0 | 1.3 | GO:0008305 | integrin complex(GO:0008305) |
| 0.0 | 2.8 | GO:0045121 | membrane raft(GO:0045121) membrane microdomain(GO:0098857) |
| 0.0 | 0.7 | GO:0031594 | neuromuscular junction(GO:0031594) |
| 0.0 | 0.7 | GO:0034706 | voltage-gated sodium channel complex(GO:0001518) sodium channel complex(GO:0034706) |
| 0.0 | 1.0 | GO:0016342 | catenin complex(GO:0016342) |
| 0.0 | 1.3 | GO:0030141 | secretory granule(GO:0030141) |
| 0.0 | 0.1 | GO:0033268 | node of Ranvier(GO:0033268) |
| 0.0 | 1.8 | GO:0005604 | basement membrane(GO:0005604) |
| 0.0 | 4.8 | GO:0031012 | extracellular matrix(GO:0031012) |
| 0.0 | 1.0 | GO:0034707 | chloride channel complex(GO:0034707) |
| 0.0 | 0.9 | GO:0031225 | anchored component of membrane(GO:0031225) |
| 0.0 | 0.5 | GO:0043204 | perikaryon(GO:0043204) |
| 0.0 | 0.3 | GO:0016282 | eukaryotic 43S preinitiation complex(GO:0016282) |
| 0.0 | 0.4 | GO:0032587 | ruffle membrane(GO:0032587) |
| 0.0 | 0.6 | GO:0030496 | midbody(GO:0030496) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.4 | 1.3 | GO:0005093 | Rab GDP-dissociation inhibitor activity(GO:0005093) |
| 0.4 | 1.3 | GO:0004500 | dopamine beta-monooxygenase activity(GO:0004500) |
| 0.3 | 2.3 | GO:0070579 | methylcytosine dioxygenase activity(GO:0070579) |
| 0.3 | 0.8 | GO:0071885 | N-terminal protein N-methyltransferase activity(GO:0071885) |
| 0.2 | 2.1 | GO:0005161 | platelet-derived growth factor receptor binding(GO:0005161) |
| 0.2 | 2.2 | GO:0030172 | troponin C binding(GO:0030172) troponin I binding(GO:0031013) |
| 0.2 | 2.4 | GO:0031720 | haptoglobin binding(GO:0031720) |
| 0.2 | 1.4 | GO:0038036 | sphingosine-1-phosphate receptor activity(GO:0038036) |
| 0.2 | 1.6 | GO:0016714 | oxidoreductase activity, acting on paired donors, with incorporation or reduction of molecular oxygen, reduced pteridine as one donor, and incorporation of one atom of oxygen(GO:0016714) |
| 0.2 | 1.5 | GO:0004865 | protein serine/threonine phosphatase inhibitor activity(GO:0004865) |
| 0.2 | 0.7 | GO:0015349 | thyroid hormone transmembrane transporter activity(GO:0015349) |
| 0.2 | 0.9 | GO:0004862 | cAMP-dependent protein kinase inhibitor activity(GO:0004862) |
| 0.2 | 1.1 | GO:0004962 | endothelin receptor activity(GO:0004962) |
| 0.1 | 0.4 | GO:0016657 | GMP reductase activity(GO:0003920) oxidoreductase activity, acting on NAD(P)H, nitrogenous group as acceptor(GO:0016657) |
| 0.1 | 0.8 | GO:0050431 | transforming growth factor beta binding(GO:0050431) |
| 0.1 | 1.6 | GO:0016775 | creatine kinase activity(GO:0004111) phosphotransferase activity, nitrogenous group as acceptor(GO:0016775) |
| 0.1 | 3.1 | GO:0005518 | collagen binding(GO:0005518) |
| 0.1 | 0.5 | GO:0005250 | A-type (transient outward) potassium channel activity(GO:0005250) |
| 0.1 | 1.0 | GO:0004029 | aldehyde dehydrogenase (NAD) activity(GO:0004029) |
| 0.1 | 0.9 | GO:0015187 | glycine transmembrane transporter activity(GO:0015187) |
| 0.1 | 0.5 | GO:0008273 | calcium, potassium:sodium antiporter activity(GO:0008273) |
| 0.1 | 0.4 | GO:0008048 | calcium sensitive guanylate cyclase activator activity(GO:0008048) |
| 0.1 | 0.6 | GO:0052812 | phosphatidylinositol-4,5-bisphosphate 3-kinase activity(GO:0046934) phosphatidylinositol-3,4-bisphosphate 5-kinase activity(GO:0052812) |
| 0.1 | 0.5 | GO:0005381 | iron ion transmembrane transporter activity(GO:0005381) |
| 0.0 | 0.8 | GO:0045134 | guanosine-diphosphatase activity(GO:0004382) uridine-diphosphatase activity(GO:0045134) |
| 0.0 | 3.3 | GO:0031490 | chromatin DNA binding(GO:0031490) |
| 0.0 | 0.3 | GO:0080019 | fatty-acyl-CoA reductase (alcohol-forming) activity(GO:0080019) |
| 0.0 | 0.9 | GO:0046875 | ephrin receptor binding(GO:0046875) |
| 0.0 | 0.2 | GO:0039706 | co-receptor binding(GO:0039706) |
| 0.0 | 0.7 | GO:0019531 | oxalate transmembrane transporter activity(GO:0019531) |
| 0.0 | 0.3 | GO:0016175 | superoxide-generating NADPH oxidase activity(GO:0016175) |
| 0.0 | 0.1 | GO:0001607 | neuromedin U receptor activity(GO:0001607) |
| 0.0 | 0.2 | GO:0030274 | LIM domain binding(GO:0030274) |
| 0.0 | 0.7 | GO:0008253 | 5'-nucleotidase activity(GO:0008253) |
| 0.0 | 0.2 | GO:0046912 | transferase activity, transferring acyl groups, acyl groups converted into alkyl on transfer(GO:0046912) |
| 0.0 | 0.4 | GO:0005092 | GDP-dissociation inhibitor activity(GO:0005092) |
| 0.0 | 0.1 | GO:0033906 | hyaluronoglucuronidase activity(GO:0033906) |
| 0.0 | 0.9 | GO:0098632 | protein binding involved in cell adhesion(GO:0098631) protein binding involved in cell-cell adhesion(GO:0098632) |
| 0.0 | 0.5 | GO:0004720 | protein-lysine 6-oxidase activity(GO:0004720) |
| 0.0 | 1.0 | GO:0035064 | methylated histone binding(GO:0035064) |
| 0.0 | 0.7 | GO:0017080 | sodium channel regulator activity(GO:0017080) |
| 0.0 | 1.3 | GO:0015020 | glucuronosyltransferase activity(GO:0015020) |
| 0.0 | 1.0 | GO:0004683 | calmodulin-dependent protein kinase activity(GO:0004683) |
| 0.0 | 2.1 | GO:0045296 | cadherin binding(GO:0045296) |
| 0.0 | 2.9 | GO:0005516 | calmodulin binding(GO:0005516) |
| 0.0 | 0.5 | GO:0005154 | epidermal growth factor receptor binding(GO:0005154) |
| 0.0 | 0.1 | GO:0017095 | heparan sulfate 6-O-sulfotransferase activity(GO:0017095) |
| 0.0 | 0.3 | GO:0035497 | cAMP response element binding(GO:0035497) |
| 0.0 | 0.1 | GO:0004332 | fructose-bisphosphate aldolase activity(GO:0004332) |
| 0.0 | 0.9 | GO:0042826 | histone deacetylase binding(GO:0042826) |
| 0.0 | 0.8 | GO:0008013 | beta-catenin binding(GO:0008013) |
| 0.0 | 0.3 | GO:0031369 | translation initiation factor binding(GO:0031369) |
| 0.0 | 0.3 | GO:0051371 | muscle alpha-actinin binding(GO:0051371) alpha-actinin binding(GO:0051393) |
| 0.0 | 0.1 | GO:0001642 | group III metabotropic glutamate receptor activity(GO:0001642) |
| 0.0 | 0.2 | GO:0003691 | double-stranded telomeric DNA binding(GO:0003691) |
| 0.0 | 0.9 | GO:0016538 | cyclin-dependent protein serine/threonine kinase regulator activity(GO:0016538) |
| 0.0 | 0.1 | GO:0032052 | bile acid binding(GO:0032052) |
| 0.0 | 0.4 | GO:0004890 | GABA-A receptor activity(GO:0004890) |
| 0.0 | 0.9 | GO:0016836 | hydro-lyase activity(GO:0016836) |
| 0.0 | 0.5 | GO:0003746 | translation elongation factor activity(GO:0003746) |
| 0.0 | 0.2 | GO:0010181 | FMN binding(GO:0010181) |
| 0.0 | 2.3 | GO:0004252 | serine-type endopeptidase activity(GO:0004252) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 1.6 | PID THROMBIN PAR4 PATHWAY | PAR4-mediated thrombin signaling events |
| 0.1 | 1.4 | PID S1P META PATHWAY | Sphingosine 1-phosphate (S1P) pathway |
| 0.1 | 1.0 | PID INTEGRIN4 PATHWAY | Alpha6 beta4 integrin-ligand interactions |
| 0.1 | 0.9 | PID P38 GAMMA DELTA PATHWAY | Signaling mediated by p38-gamma and p38-delta |
| 0.1 | 1.0 | PID CONE PATHWAY | Visual signal transduction: Cones |
| 0.1 | 1.3 | SIG REGULATION OF THE ACTIN CYTOSKELETON BY RHO GTPASES | Genes related to regulation of the actin cytoskeleton |
| 0.1 | 0.8 | PID SYNDECAN 4 PATHWAY | Syndecan-4-mediated signaling events |
| 0.0 | 1.1 | NABA PROTEOGLYCANS | Genes encoding proteoglycans |
| 0.0 | 0.3 | PID INTEGRIN5 PATHWAY | Beta5 beta6 beta7 and beta8 integrin cell surface interactions |
| 0.0 | 4.4 | NABA ECM GLYCOPROTEINS | Genes encoding structural ECM glycoproteins |
| 0.0 | 0.2 | PID VEGFR1 PATHWAY | VEGFR1 specific signals |
| 0.0 | 1.2 | PID PS1 PATHWAY | Presenilin action in Notch and Wnt signaling |
| 0.0 | 1.2 | PID NOTCH PATHWAY | Notch signaling pathway |
| 0.0 | 1.3 | PID AP1 PATHWAY | AP-1 transcription factor network |
| 0.0 | 0.6 | PID TRAIL PATHWAY | TRAIL signaling pathway |
| 0.0 | 0.5 | PID P38 ALPHA BETA DOWNSTREAM PATHWAY | Signaling mediated by p38-alpha and p38-beta |
| 0.0 | 1.5 | PID E2F PATHWAY | E2F transcription factor network |
| 0.0 | 0.0 | PID TCR JNK PATHWAY | JNK signaling in the CD4+ TCR pathway |
| 0.0 | 1.1 | NABA ECM AFFILIATED | Genes encoding proteins affiliated structurally or functionally to extracellular matrix proteins |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 2.4 | REACTOME REGULATION OF INSULIN LIKE GROWTH FACTOR IGF ACTIVITY BY INSULIN LIKE GROWTH FACTOR BINDING PROTEINS IGFBPS | Genes involved in Regulation of Insulin-like Growth Factor (IGF) Activity by Insulin-like Growth Factor Binding Proteins (IGFBPs) |
| 0.1 | 1.1 | REACTOME INTERACTION BETWEEN L1 AND ANKYRINS | Genes involved in Interaction between L1 and Ankyrins |
| 0.1 | 0.8 | REACTOME HYALURONAN METABOLISM | Genes involved in Hyaluronan metabolism |
| 0.1 | 0.9 | REACTOME NA CL DEPENDENT NEUROTRANSMITTER TRANSPORTERS | Genes involved in Na+/Cl- dependent neurotransmitter transporters |
| 0.1 | 2.4 | REACTOME G1 PHASE | Genes involved in G1 Phase |
| 0.0 | 0.4 | REACTOME PURINE SALVAGE | Genes involved in Purine salvage |
| 0.0 | 3.1 | REACTOME RESPONSE TO ELEVATED PLATELET CYTOSOLIC CA2 | Genes involved in Response to elevated platelet cytosolic Ca2+ |
| 0.0 | 0.6 | REACTOME NEPHRIN INTERACTIONS | Genes involved in Nephrin interactions |
| 0.0 | 0.5 | REACTOME SEMA4D INDUCED CELL MIGRATION AND GROWTH CONE COLLAPSE | Genes involved in Sema4D induced cell migration and growth-cone collapse |
| 0.0 | 0.9 | REACTOME SIGNALING BY ROBO RECEPTOR | Genes involved in Signaling by Robo receptor |
| 0.0 | 0.5 | REACTOME ACTIVATION OF GENES BY ATF4 | Genes involved in Activation of Genes by ATF4 |
| 0.0 | 0.2 | REACTOME SHC MEDIATED SIGNALLING | Genes involved in SHC-mediated signalling |
| 0.0 | 0.8 | REACTOME CROSS PRESENTATION OF SOLUBLE EXOGENOUS ANTIGENS ENDOSOMES | Genes involved in Cross-presentation of soluble exogenous antigens (endosomes) |
| 0.0 | 0.3 | REACTOME LATENT INFECTION OF HOMO SAPIENS WITH MYCOBACTERIUM TUBERCULOSIS | Genes involved in Latent infection of Homo sapiens with Mycobacterium tuberculosis |
| 0.0 | 0.5 | REACTOME VOLTAGE GATED POTASSIUM CHANNELS | Genes involved in Voltage gated Potassium channels |
| 0.0 | 0.8 | REACTOME CELL JUNCTION ORGANIZATION | Genes involved in Cell junction organization |
| 0.0 | 0.2 | REACTOME CITRIC ACID CYCLE TCA CYCLE | Genes involved in Citric acid cycle (TCA cycle) |
| 0.0 | 0.8 | REACTOME TRANSPORT OF INORGANIC CATIONS ANIONS AND AMINO ACIDS OLIGOPEPTIDES | Genes involved in Transport of inorganic cations/anions and amino acids/oligopeptides |