Project
PRJNA195909:zebrafish embryo and larva development
Navigation
Downloads
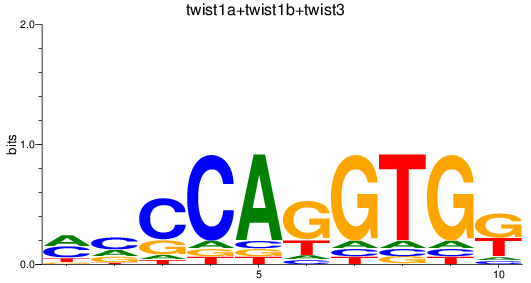
Results for twist1a+twist1b+twist3
Z-value: 1.12
Transcription factors associated with twist1a+twist1b+twist3
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
twist3
|
ENSDARG00000019646 | twist3 |
|
twist1a
|
ENSDARG00000030402 | twist family bHLH transcription factor 1a |
|
twist1b
|
ENSDARG00000076010 | twist family bHLH transcription factor 1b |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| twist1b | dr11_v1_chr16_+_19732543_19732543 | -0.97 | 1.8e-05 | Click! |
| twist3 | dr11_v1_chr23_+_2669_2669 | -0.94 | 2.0e-04 | Click! |
| twist1a | dr11_v1_chr19_-_2231146_2231188 | -0.62 | 7.5e-02 | Click! |
Activity profile of twist1a+twist1b+twist3 motif
Sorted Z-values of twist1a+twist1b+twist3 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr15_-_1590858 | 2.85 |
ENSDART00000081875
|
nnr
|
nanor |
| chr23_-_44574059 | 2.21 |
ENSDART00000123007
|
si:ch73-160p18.3
|
si:ch73-160p18.3 |
| chr21_-_43666420 | 2.12 |
ENSDART00000139008
ENSDART00000183996 ENSDART00000183395 |
si:dkey-229d11.3
si:dkey-229d11.5
|
si:dkey-229d11.3 si:dkey-229d11.5 |
| chr17_+_1360192 | 1.61 |
ENSDART00000184561
|
siva1
|
SIVA1, apoptosis-inducing factor |
| chr5_+_6670945 | 1.30 |
ENSDART00000185686
|
pxna
|
paxillin a |
| chr3_+_58472305 | 1.05 |
ENSDART00000154122
|
si:ch211-165g14.1
|
si:ch211-165g14.1 |
| chr5_-_30080332 | 1.03 |
ENSDART00000140049
|
bco2a
|
beta-carotene oxygenase 2a |
| chr11_-_40257225 | 1.00 |
ENSDART00000139009
|
TRIM62 (1 of many)
|
si:ch211-193i15.2 |
| chr15_-_766015 | 0.97 |
ENSDART00000190648
|
si:dkey-7i4.15
|
si:dkey-7i4.15 |
| chr8_-_14067517 | 0.97 |
ENSDART00000140948
|
dedd
|
death effector domain containing |
| chr13_+_1381953 | 0.94 |
ENSDART00000019983
|
rab23
|
RAB23, member RAS oncogene family |
| chr7_-_9674073 | 0.90 |
ENSDART00000187902
|
lrrk1
|
leucine-rich repeat kinase 1 |
| chr5_-_11809404 | 0.83 |
ENSDART00000132564
|
nf2a
|
neurofibromin 2a (merlin) |
| chr20_-_44575103 | 0.82 |
ENSDART00000192573
|
ubxn2a
|
UBX domain protein 2A |
| chr17_-_40876680 | 0.81 |
ENSDART00000127200
|
supt7l
|
SPT7-like STAGA complex gamma subunit |
| chr5_-_23696926 | 0.80 |
ENSDART00000021462
|
rnf128a
|
ring finger protein 128a |
| chr6_-_55423220 | 0.78 |
ENSDART00000158929
|
ctsa
|
cathepsin A |
| chr23_+_2914577 | 0.74 |
ENSDART00000184897
|
DHX35
|
zgc:158828 |
| chr20_+_2134816 | 0.73 |
ENSDART00000039249
|
l3mbtl3
|
l(3)mbt-like 3 (Drosophila) |
| chr5_-_15321451 | 0.72 |
ENSDART00000139203
|
txnrd2.1
|
thioredoxin reductase 2, tandem duplicate 1 |
| chr3_+_22935183 | 0.69 |
ENSDART00000157378
|
hdac5
|
histone deacetylase 5 |
| chr2_+_38002717 | 0.65 |
ENSDART00000139564
|
dcaf8
|
DDB1 and CUL4 associated factor 8 |
| chr22_-_26251563 | 0.60 |
ENSDART00000060888
ENSDART00000142821 |
ccdc130
|
coiled-coil domain containing 130 |
| chr11_-_34232906 | 0.59 |
ENSDART00000162150
|
lsg1
|
large 60S subunit nuclear export GTPase 1 |
| chr1_+_45925365 | 0.59 |
ENSDART00000144245
|
eif5b
|
eukaryotic translation initiation factor 5B |
| chr3_+_16663373 | 0.58 |
ENSDART00000100961
|
zgc:55558
|
zgc:55558 |
| chr18_+_45666489 | 0.58 |
ENSDART00000180147
ENSDART00000151351 |
prrg4
|
proline rich Gla (G-carboxyglutamic acid) 4 (transmembrane) |
| chr4_+_77973876 | 0.56 |
ENSDART00000057423
|
terfa
|
telomeric repeat binding factor a |
| chr8_+_247163 | 0.52 |
ENSDART00000122378
|
cep120
|
centrosomal protein 120 |
| chr6_-_10728057 | 0.50 |
ENSDART00000002247
|
sp3b
|
Sp3b transcription factor |
| chr1_+_45925150 | 0.47 |
ENSDART00000074689
|
eif5b
|
eukaryotic translation initiation factor 5B |
| chr13_+_35690023 | 0.45 |
ENSDART00000128865
ENSDART00000130050 |
psme4a
|
proteasome activator subunit 4a |
| chr24_+_39660124 | 0.45 |
ENSDART00000066500
|
stub1
|
STIP1 homology and U-Box containing protein 1 |
| chr21_-_26520629 | 0.41 |
ENSDART00000142731
|
rce1b
|
Ras converting CAAX endopeptidase 1b |
| chr9_-_40014339 | 0.41 |
ENSDART00000166918
|
IKZF2
|
si:zfos-1425h8.1 |
| chr8_+_20140321 | 0.41 |
ENSDART00000012120
|
acsbg2
|
acyl-CoA synthetase bubblegum family member 2 |
| chr21_-_35325466 | 0.40 |
ENSDART00000134780
ENSDART00000145930 ENSDART00000076715 ENSDART00000065341 ENSDART00000162189 |
ublcp1
|
ubiquitin-like domain containing CTD phosphatase 1 |
| chr6_+_42693288 | 0.39 |
ENSDART00000155010
|
si:ch211-207d10.2
|
si:ch211-207d10.2 |
| chr3_+_59411956 | 0.39 |
ENSDART00000166982
|
sec14l1
|
SEC14-like lipid binding 1 |
| chr1_-_55058795 | 0.38 |
ENSDART00000187293
|
peli1a
|
pellino E3 ubiquitin protein ligase 1a |
| chr6_+_42693114 | 0.36 |
ENSDART00000154353
|
si:ch211-207d10.2
|
si:ch211-207d10.2 |
| chr5_-_32505109 | 0.36 |
ENSDART00000188219
|
ntmt1
|
N-terminal Xaa-Pro-Lys N-methyltransferase 1 |
| chr4_+_33830944 | 0.32 |
ENSDART00000170238
|
znf1026
|
zinc finger protein 1026 |
| chr6_-_10728921 | 0.30 |
ENSDART00000151484
|
sp3b
|
Sp3b transcription factor |
| chr20_-_22193190 | 0.28 |
ENSDART00000047624
|
tmem165
|
transmembrane protein 165 |
| chr16_-_31791165 | 0.28 |
ENSDART00000148389
|
chd4b
|
chromodomain helicase DNA binding protein 4b |
| chr13_-_9119867 | 0.28 |
ENSDART00000137255
|
si:dkey-112g5.15
|
si:dkey-112g5.15 |
| chr16_-_21047483 | 0.27 |
ENSDART00000136235
|
cbx3b
|
chromobox homolog 3b |
| chr7_-_12596727 | 0.26 |
ENSDART00000186413
|
adamtsl3
|
ADAMTS-like 3 |
| chr3_+_49021079 | 0.25 |
ENSDART00000162012
|
zgc:163083
|
zgc:163083 |
| chr13_+_35689749 | 0.24 |
ENSDART00000158726
|
psme4a
|
proteasome activator subunit 4a |
| chr14_-_31087830 | 0.22 |
ENSDART00000002250
|
hs6st2
|
heparan sulfate 6-O-sulfotransferase 2 |
| chr7_+_17485782 | 0.20 |
ENSDART00000098083
|
CU179759.1
|
Danio rerio novel immune-type receptor 1d (nitr1d), mRNA. |
| chr22_-_5752009 | 0.19 |
ENSDART00000190052
|
bcdin3d
|
BCDIN3 domain containing |
| chr10_+_40737540 | 0.18 |
ENSDART00000125577
|
taar19a
|
trace amine associated receptor 19a |
| chr6_+_103361 | 0.16 |
ENSDART00000151899
|
ldlrb
|
low density lipoprotein receptor b |
| chr23_-_24047054 | 0.16 |
ENSDART00000184308
ENSDART00000185902 |
CR925720.1
|
|
| chr20_-_46046758 | 0.15 |
ENSDART00000127455
|
taar12j
|
trace amine associated receptor 12j |
| chr6_+_16982613 | 0.15 |
ENSDART00000156104
|
pimr8
|
Pim proto-oncogene, serine/threonine kinase, related 8 |
| chr16_-_19568795 | 0.11 |
ENSDART00000185141
|
abcb5
|
ATP-binding cassette, sub-family B (MDR/TAP), member 5 |
| chr10_+_1052591 | 0.09 |
ENSDART00000123405
|
unc5c
|
unc-5 netrin receptor C |
| chr15_-_46736432 | 0.09 |
ENSDART00000124567
|
ilkap
|
integrin-linked kinase-associated serine/threonine phosphatase |
| chr14_+_46216703 | 0.09 |
ENSDART00000136045
ENSDART00000142317 |
mgst2
|
microsomal glutathione S-transferase 2 |
| chr13_-_12602920 | 0.09 |
ENSDART00000102311
|
lrit3b
|
leucine-rich repeat, immunoglobulin-like and transmembrane domains 3b |
| chr20_-_2134620 | 0.06 |
ENSDART00000064375
|
tmem244
|
transmembrane protein 244 |
| chr18_-_35842554 | 0.03 |
ENSDART00000088488
|
opa3
|
optic atrophy 3 |
| chr21_-_35419486 | 0.03 |
ENSDART00000138529
|
si:dkeyp-23e4.3
|
si:dkeyp-23e4.3 |
| chr16_-_21047872 | 0.02 |
ENSDART00000131582
|
cbx3b
|
chromobox homolog 3b |
| chr5_-_2752573 | 0.02 |
ENSDART00000168121
|
lzts3a
|
leucine zipper, putative tumor suppressor family member 3a |
Network of associatons between targets according to the STRING database.
First level regulatory network of twist1a+twist1b+twist3
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.6 | GO:0032207 | regulation of telomere maintenance via recombination(GO:0032207) negative regulation of telomere maintenance via recombination(GO:0032208) |
| 0.1 | 0.4 | GO:0080120 | CAAX-box protein processing(GO:0071586) CAAX-box protein maturation(GO:0080120) |
| 0.1 | 0.5 | GO:0010825 | positive regulation of centrosome duplication(GO:0010825) |
| 0.1 | 0.4 | GO:0006480 | N-terminal protein amino acid methylation(GO:0006480) |
| 0.1 | 0.7 | GO:0035093 | spermatid nucleus differentiation(GO:0007289) sperm chromatin condensation(GO:0035092) spermatogenesis, exchange of chromosomal proteins(GO:0035093) |
| 0.1 | 1.0 | GO:0016121 | carotene metabolic process(GO:0016119) carotene catabolic process(GO:0016121) terpene metabolic process(GO:0042214) terpene catabolic process(GO:0046247) |
| 0.1 | 0.3 | GO:0071421 | manganese ion transmembrane transport(GO:0071421) |
| 0.1 | 1.3 | GO:0061055 | myotome development(GO:0061055) |
| 0.1 | 0.4 | GO:0008592 | regulation of Toll signaling pathway(GO:0008592) |
| 0.1 | 0.4 | GO:0030579 | ubiquitin-dependent SMAD protein catabolic process(GO:0030579) |
| 0.0 | 1.0 | GO:0008625 | extrinsic apoptotic signaling pathway via death domain receptors(GO:0008625) |
| 0.0 | 0.2 | GO:0070920 | regulation of production of small RNA involved in gene silencing by RNA(GO:0070920) regulation of production of miRNAs involved in gene silencing by miRNA(GO:1903798) |
| 0.0 | 0.8 | GO:0031468 | nuclear envelope reassembly(GO:0031468) |
| 0.0 | 0.1 | GO:0046689 | response to mercury ion(GO:0046689) detoxification of mercury ion(GO:0050787) |
| 0.0 | 0.9 | GO:0035469 | determination of pancreatic left/right asymmetry(GO:0035469) |
| 0.0 | 0.6 | GO:0033753 | ribosomal subunit export from nucleus(GO:0000054) ribosome localization(GO:0033750) establishment of ribosome localization(GO:0033753) rRNA-containing ribonucleoprotein complex export from nucleus(GO:0071428) |
| 0.0 | 0.7 | GO:0045454 | cell redox homeostasis(GO:0045454) |
| 0.0 | 0.2 | GO:0015015 | heparan sulfate proteoglycan biosynthetic process, enzymatic modification(GO:0015015) |
| 0.0 | 1.0 | GO:0032922 | circadian regulation of gene expression(GO:0032922) |
| 0.0 | 0.8 | GO:0043966 | histone H3 acetylation(GO:0043966) |
| 0.0 | 0.7 | GO:0072114 | pronephros morphogenesis(GO:0072114) |
| 0.0 | 0.4 | GO:0042759 | long-chain fatty acid biosynthetic process(GO:0042759) |
| 0.0 | 0.1 | GO:0019370 | leukotriene metabolic process(GO:0006691) leukotriene biosynthetic process(GO:0019370) |
| 0.0 | 0.7 | GO:0097191 | extrinsic apoptotic signaling pathway(GO:0097191) |
| 0.0 | 0.9 | GO:0090263 | positive regulation of canonical Wnt signaling pathway(GO:0090263) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.7 | GO:1990111 | spermatoproteasome complex(GO:1990111) |
| 0.1 | 0.8 | GO:0030914 | STAGA complex(GO:0030914) |
| 0.0 | 0.6 | GO:0071014 | post-mRNA release spliceosomal complex(GO:0071014) |
| 0.0 | 0.6 | GO:0070187 | telosome(GO:0070187) |
| 0.0 | 0.6 | GO:0015030 | Cajal body(GO:0015030) |
| 0.0 | 0.7 | GO:0080008 | Cul4-RING E3 ubiquitin ligase complex(GO:0080008) |
| 0.0 | 0.9 | GO:0005776 | autophagosome(GO:0005776) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.8 | GO:0004185 | serine-type carboxypeptidase activity(GO:0004185) |
| 0.1 | 0.4 | GO:0071885 | N-terminal protein N-methyltransferase activity(GO:0071885) |
| 0.1 | 1.0 | GO:0003834 | beta-carotene 15,15'-monooxygenase activity(GO:0003834) carotenoid dioxygenase activity(GO:0010436) |
| 0.1 | 0.7 | GO:0004791 | thioredoxin-disulfide reductase activity(GO:0004791) |
| 0.1 | 0.6 | GO:0098505 | G-rich strand telomeric DNA binding(GO:0098505) |
| 0.1 | 0.7 | GO:0016504 | peptidase activator activity(GO:0016504) |
| 0.0 | 0.7 | GO:0031078 | histone deacetylase activity (H3-K14 specific)(GO:0031078) NAD-dependent histone deacetylase activity (H3-K14 specific)(GO:0032041) |
| 0.0 | 0.3 | GO:0005384 | manganese ion transmembrane transporter activity(GO:0005384) |
| 0.0 | 0.2 | GO:0017095 | heparan sulfate 6-O-sulfotransferase activity(GO:0017095) |
| 0.0 | 1.6 | GO:0032813 | tumor necrosis factor receptor superfamily binding(GO:0032813) |
| 0.0 | 0.4 | GO:0016405 | CoA-ligase activity(GO:0016405) |
| 0.0 | 0.4 | GO:0051879 | Hsp70 protein binding(GO:0030544) Hsp90 protein binding(GO:0051879) |
| 0.0 | 0.1 | GO:0004464 | leukotriene-C4 synthase activity(GO:0004464) |
| 0.0 | 0.6 | GO:0019003 | GDP binding(GO:0019003) |
| 0.0 | 0.8 | GO:0004402 | histone acetyltransferase activity(GO:0004402) |
| 0.0 | 1.4 | GO:0004722 | protein serine/threonine phosphatase activity(GO:0004722) |
| 0.0 | 1.1 | GO:0003743 | translation initiation factor activity(GO:0003743) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.9 | PID HEDGEHOG GLI PATHWAY | Hedgehog signaling events mediated by Gli proteins |
| 0.0 | 0.8 | PID MYC PATHWAY | C-MYC pathway |
| 0.0 | 0.7 | PID HDAC CLASSII PATHWAY | Signaling events mediated by HDAC Class II |
| 0.0 | 0.4 | PID ALPHA SYNUCLEIN PATHWAY | Alpha-synuclein signaling |
| 0.0 | 1.0 | ST FAS SIGNALING PATHWAY | Fas Signaling Pathway |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.4 | REACTOME TGF BETA RECEPTOR SIGNALING ACTIVATES SMADS | Genes involved in TGF-beta receptor signaling activates SMADs |
| 0.0 | 0.8 | REACTOME GLYCOSPHINGOLIPID METABOLISM | Genes involved in Glycosphingolipid metabolism |
| 0.0 | 0.7 | REACTOME NOTCH1 INTRACELLULAR DOMAIN REGULATES TRANSCRIPTION | Genes involved in NOTCH1 Intracellular Domain Regulates Transcription |
| 0.0 | 0.2 | REACTOME HS GAG BIOSYNTHESIS | Genes involved in HS-GAG biosynthesis |