Project
PRJNA207719: Tissue specific transcriptome profiling
Navigation
Downloads
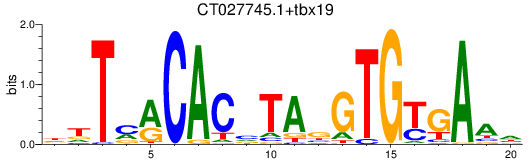
Results for CT027745.1+tbx19
Z-value: 0.80
Transcription factors associated with CT027745.1+tbx19
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
tbx19
|
ENSDARG00000079187 | T-box transcription factor 19 |
|
CT027745.1
|
ENSDARG00000112623 | ENSDARG00000112623 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| tbx19 | dr11_v1_chr6_+_9204099_9204099 | -0.86 | 6.0e-02 | Click! |
Activity profile of CT027745.1+tbx19 motif
Sorted Z-values of CT027745.1+tbx19 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr25_+_35020529 | 0.94 |
ENSDART00000158016
|
flnca
|
filamin C, gamma a (actin binding protein 280) |
| chr3_-_55104310 | 0.83 |
ENSDART00000101713
|
hbba1
|
hemoglobin, beta adult 1 |
| chr3_+_55105066 | 0.82 |
ENSDART00000110848
|
si:ch211-5k11.8
|
si:ch211-5k11.8 |
| chr23_+_32039386 | 0.63 |
ENSDART00000133801
|
mylk2
|
myosin light chain kinase 2 |
| chr9_-_54304684 | 0.54 |
ENSDART00000109512
|
il13
|
interleukin 13 |
| chr4_+_76722754 | 0.53 |
ENSDART00000153867
|
ms4a17a.3
|
membrane-spanning 4-domains, subfamily A, member 17A.3 |
| chr8_+_44783424 | 0.51 |
ENSDART00000025875
|
si:ch1073-459j12.1
|
si:ch1073-459j12.1 |
| chr22_+_4443689 | 0.47 |
ENSDART00000185490
|
ticam1
|
toll-like receptor adaptor molecule 1 |
| chr4_+_26357221 | 0.45 |
ENSDART00000187684
ENSDART00000101545 ENSDART00000148296 ENSDART00000171433 |
tnni1d
|
troponin I, skeletal, slow d |
| chr3_+_32416948 | 0.44 |
ENSDART00000157324
ENSDART00000154267 ENSDART00000186094 ENSDART00000155860 ENSDART00000156986 |
prrg2
|
proline rich Gla (G-carboxyglutamic acid) 2 |
| chr20_-_19864131 | 0.43 |
ENSDART00000057819
|
ptk2bb
|
protein tyrosine kinase 2 beta, b |
| chr1_-_7930679 | 0.43 |
ENSDART00000146090
|
si:dkey-79f11.10
|
si:dkey-79f11.10 |
| chr1_+_55662491 | 0.42 |
ENSDART00000152386
|
adgre8
|
adhesion G protein-coupled receptor E8 |
| chr25_-_12998202 | 0.40 |
ENSDART00000171661
|
ccl39.3
|
chemokine (C-C motif) ligand 39, duplicate 3 |
| chr5_+_1877464 | 0.40 |
ENSDART00000050658
|
zgc:101699
|
zgc:101699 |
| chr1_+_40802454 | 0.38 |
ENSDART00000193568
|
cpz
|
carboxypeptidase Z |
| chr7_-_20158380 | 0.37 |
ENSDART00000142891
|
acap1
|
ArfGAP with coiled-coil, ankyrin repeat and PH domains 1 |
| chr4_+_76787274 | 0.37 |
ENSDART00000156344
|
si:ch73-56d11.3
|
si:ch73-56d11.3 |
| chr17_-_8899323 | 0.37 |
ENSDART00000081590
|
nkl.1
|
NK-lysin tandem duplicate 1 |
| chr7_+_33172066 | 0.35 |
ENSDART00000174013
|
si:ch211-194p6.12
|
si:ch211-194p6.12 |
| chr3_-_20106233 | 0.34 |
ENSDART00000145285
|
selenow2b
|
selenoprotein W, 2b |
| chr18_+_7150144 | 0.33 |
ENSDART00000114690
|
vwf
|
von Willebrand factor |
| chr8_-_46486009 | 0.33 |
ENSDART00000140431
|
sult1st9
|
sulfotransferase family 1, cytosolic sulfotransferase 9 |
| chr8_-_28304534 | 0.32 |
ENSDART00000078545
|
lmod1a
|
leiomodin 1a (smooth muscle) |
| chr9_-_21918963 | 0.32 |
ENSDART00000090782
|
lmo7a
|
LIM domain 7a |
| chr15_+_6652396 | 0.30 |
ENSDART00000192813
ENSDART00000157678 |
nop53
|
NOP53 ribosome biogenesis factor |
| chr1_+_9966384 | 0.29 |
ENSDART00000132607
|
si:dkeyp-75b4.8
|
si:dkeyp-75b4.8 |
| chr12_+_38929663 | 0.29 |
ENSDART00000156334
|
si:dkey-239b22.1
|
si:dkey-239b22.1 |
| chr22_-_31020690 | 0.28 |
ENSDART00000130604
|
ssuh2.4
|
ssu-2 homolog, tandem duplicate 4 |
| chr13_+_3252950 | 0.28 |
ENSDART00000020671
|
prph2b
|
peripherin 2b (retinal degeneration, slow) |
| chr7_+_53152108 | 0.27 |
ENSDART00000171350
|
cdh29
|
cadherin 29 |
| chr14_-_6527010 | 0.27 |
ENSDART00000125058
|
nipsnap3a
|
nipsnap homolog 3A (C. elegans) |
| chr20_+_48739154 | 0.26 |
ENSDART00000100262
|
zgc:110348
|
zgc:110348 |
| chr4_-_33525219 | 0.26 |
ENSDART00000150491
|
CT027801.2
|
|
| chr9_+_343062 | 0.26 |
ENSDART00000164012
ENSDART00000164365 |
bpifcl
|
BPI fold containing family C, like |
| chr12_+_48683208 | 0.25 |
ENSDART00000188582
|
mmp21
|
matrix metallopeptidase 21 |
| chr3_-_11008532 | 0.25 |
ENSDART00000165086
|
CR382337.3
|
|
| chr15_+_29025090 | 0.25 |
ENSDART00000131755
|
si:ch211-137a8.2
|
si:ch211-137a8.2 |
| chr3_-_60401826 | 0.25 |
ENSDART00000144030
ENSDART00000160821 |
si:ch211-214b16.4
|
si:ch211-214b16.4 |
| chr13_+_40686133 | 0.24 |
ENSDART00000146112
|
hps1
|
Hermansky-Pudlak syndrome 1 |
| chr6_-_40756138 | 0.23 |
ENSDART00000156295
|
myhb
|
myosin, heavy chain b |
| chr21_-_31290582 | 0.23 |
ENSDART00000065362
|
ca4c
|
carbonic anhydrase IV c |
| chr17_+_23311377 | 0.23 |
ENSDART00000128073
|
ppp1r3ca
|
protein phosphatase 1, regulatory subunit 3Ca |
| chr5_+_29652513 | 0.22 |
ENSDART00000035400
|
tsc1a
|
TSC complex subunit 1a |
| chr17_-_20167206 | 0.22 |
ENSDART00000104874
ENSDART00000191995 |
p4ha1b
|
prolyl 4-hydroxylase, alpha polypeptide I b |
| chr14_+_34501245 | 0.22 |
ENSDART00000131424
|
lcp2b
|
lymphocyte cytosolic protein 2b |
| chr14_-_49859747 | 0.22 |
ENSDART00000169456
ENSDART00000164967 |
nfkb1
|
nuclear factor of kappa light polypeptide gene enhancer in B-cells 1 |
| chr6_+_598669 | 0.22 |
ENSDART00000151009
|
si:ch73-379f7.4
|
si:ch73-379f7.4 |
| chr20_-_54259780 | 0.21 |
ENSDART00000172631
|
fkbp3
|
FK506 binding protein 3 |
| chr5_+_29652198 | 0.21 |
ENSDART00000184083
|
tsc1a
|
TSC complex subunit 1a |
| chr8_-_44363596 | 0.21 |
ENSDART00000123658
|
si:busm1-228j01.4
|
si:busm1-228j01.4 |
| chr21_+_26071874 | 0.21 |
ENSDART00000003001
ENSDART00000146573 |
rpl23a
|
ribosomal protein L23a |
| chr20_-_40729364 | 0.21 |
ENSDART00000101014
|
cx32.2
|
connexin 32.2 |
| chr9_+_8365398 | 0.21 |
ENSDART00000138713
ENSDART00000136847 |
si:dkey-90l23.2
|
si:dkey-90l23.2 |
| chr6_-_10698125 | 0.21 |
ENSDART00000151636
|
dnah9l
|
dynein, axonemal, heavy polypeptide 9 like |
| chr22_-_8883097 | 0.20 |
ENSDART00000078237
|
CABZ01046432.1
|
|
| chr5_+_69856153 | 0.20 |
ENSDART00000124128
ENSDART00000073663 |
ugt2a6
|
UDP glucuronosyltransferase 2 family, polypeptide A6 |
| chr7_+_33151731 | 0.20 |
ENSDART00000029106
|
si:ch211-194p6.10
|
si:ch211-194p6.10 |
| chr5_-_20195350 | 0.20 |
ENSDART00000139675
|
dao.1
|
D-amino-acid oxidase, tandem duplicate 1 |
| chr9_+_20853894 | 0.20 |
ENSDART00000003648
|
wdr3
|
WD repeat domain 3 |
| chr16_+_33902006 | 0.19 |
ENSDART00000161807
ENSDART00000159474 |
gnl2
|
guanine nucleotide binding protein-like 2 (nucleolar) |
| chr5_-_1999417 | 0.19 |
ENSDART00000155437
ENSDART00000145781 |
si:ch211-160e1.5
|
si:ch211-160e1.5 |
| chr13_-_44808783 | 0.19 |
ENSDART00000099984
|
glo1
|
glyoxalase 1 |
| chr2_-_37140423 | 0.19 |
ENSDART00000144220
|
tspan37
|
tetraspanin 37 |
| chr24_-_40744672 | 0.19 |
ENSDART00000160672
|
CU633479.1
|
|
| chr2_-_3419890 | 0.18 |
ENSDART00000055618
|
iba57
|
IBA57, iron-sulfur cluster assembly |
| chr19_-_24745317 | 0.18 |
ENSDART00000142774
|
MYO1G
|
si:dkeyp-92c9.3 |
| chr5_-_66160415 | 0.18 |
ENSDART00000073895
|
mboat4
|
membrane bound O-acyltransferase domain containing 4 |
| chr4_+_52356485 | 0.18 |
ENSDART00000170639
|
zgc:173705
|
zgc:173705 |
| chr17_+_34186632 | 0.18 |
ENSDART00000014306
|
mpp5a
|
membrane protein, palmitoylated 5a (MAGUK p55 subfamily member 5) |
| chr9_-_41024438 | 0.17 |
ENSDART00000157398
|
pms1
|
PMS1 homolog 1, mismatch repair system component |
| chr23_+_32066168 | 0.17 |
ENSDART00000031047
|
cd63
|
CD63 molecule |
| chr7_-_15185811 | 0.17 |
ENSDART00000031049
|
si:dkey-172h23.2
|
si:dkey-172h23.2 |
| chr7_-_26571994 | 0.17 |
ENSDART00000128801
|
si:dkey-62k3.6
|
si:dkey-62k3.6 |
| chr23_-_31913231 | 0.17 |
ENSDART00000146852
ENSDART00000085054 |
mtfr2
|
mitochondrial fission regulator 2 |
| chr7_-_11383933 | 0.17 |
ENSDART00000163949
|
mesd
|
mesoderm development LRP chaperone |
| chr13_+_35635672 | 0.17 |
ENSDART00000148481
|
thbs2a
|
thrombospondin 2a |
| chr19_-_29832876 | 0.17 |
ENSDART00000005119
|
eif3i
|
eukaryotic translation initiation factor 3, subunit I |
| chr25_-_29080063 | 0.17 |
ENSDART00000181911
ENSDART00000138087 |
cox5aa
|
cytochrome c oxidase subunit Vaa |
| chr2_-_21349425 | 0.16 |
ENSDART00000171699
|
hhatla
|
hedgehog acyltransferase like, a |
| chr17_-_24890843 | 0.16 |
ENSDART00000184984
ENSDART00000135569 ENSDART00000193661 |
gale
|
UDP-galactose-4-epimerase |
| chr22_-_28624879 | 0.16 |
ENSDART00000188437
|
adgrg7.2
|
adhesion G protein-coupled receptor G7, tandem duplicate 2 |
| chr16_+_35595312 | 0.16 |
ENSDART00000170438
|
si:ch211-1i11.3
|
si:ch211-1i11.3 |
| chr15_+_12030367 | 0.16 |
ENSDART00000176178
|
FO904901.1
|
|
| chr3_+_36972298 | 0.16 |
ENSDART00000150917
|
si:ch211-18i17.2
|
si:ch211-18i17.2 |
| chr4_-_55436639 | 0.16 |
ENSDART00000163802
|
si:dkey-191j3.2
|
si:dkey-191j3.2 |
| chr10_+_8481660 | 0.15 |
ENSDART00000109559
|
BX682234.1
|
|
| chr22_-_7700179 | 0.15 |
ENSDART00000145047
|
si:ch211-59h6.1
|
si:ch211-59h6.1 |
| chr17_+_50389823 | 0.15 |
ENSDART00000154246
ENSDART00000153566 ENSDART00000153878 |
znf982
|
zinc finger protein 982 |
| chr23_-_32162810 | 0.15 |
ENSDART00000155905
|
nr4a1
|
nuclear receptor subfamily 4, group A, member 1 |
| chr15_+_29024895 | 0.14 |
ENSDART00000141164
ENSDART00000144126 |
si:ch211-137a8.2
|
si:ch211-137a8.2 |
| chr22_-_18544701 | 0.14 |
ENSDART00000132156
|
cirbpb
|
cold inducible RNA binding protein b |
| chr1_-_43963441 | 0.14 |
ENSDART00000137646
|
odam
|
odontogenic, ameloblast associated |
| chr20_-_48700370 | 0.14 |
ENSDART00000185195
ENSDART00000184898 |
pax1b
|
paired box 1b |
| chr1_+_11736420 | 0.14 |
ENSDART00000140329
|
chst12b
|
carbohydrate (chondroitin 4) sulfotransferase 12b |
| chr4_+_69863019 | 0.14 |
ENSDART00000168400
|
znf1117
|
zinc finger protein 1117 |
| chr25_-_10610961 | 0.14 |
ENSDART00000153474
|
ppp6r3
|
protein phosphatase 6, regulatory subunit 3 |
| chr3_+_9077907 | 0.14 |
ENSDART00000163056
|
znf985
|
zinc finger protein 985 |
| chr22_-_23067859 | 0.14 |
ENSDART00000137873
|
ptprc
|
protein tyrosine phosphatase, receptor type, C |
| chr4_-_71284106 | 0.13 |
ENSDART00000167459
|
si:ch211-205a14.1
|
si:ch211-205a14.1 |
| chr5_-_23715861 | 0.13 |
ENSDART00000019992
|
gbgt1l1
|
globoside alpha-1,3-N-acetylgalactosaminyltransferase 1, like 1 |
| chr9_-_41025062 | 0.13 |
ENSDART00000002053
|
pms1
|
PMS1 homolog 1, mismatch repair system component |
| chr7_+_24628546 | 0.13 |
ENSDART00000173745
|
MSMP
|
si:ch211-69e13.6 |
| chr2_-_39090258 | 0.13 |
ENSDART00000134270
ENSDART00000145268 |
NMNAT3
|
si:ch73-170l17.1 |
| chr4_+_65057234 | 0.13 |
ENSDART00000162985
|
si:dkey-14o6.2
|
si:dkey-14o6.2 |
| chr4_-_18954001 | 0.13 |
ENSDART00000144814
|
si:dkey-31f5.8
|
si:dkey-31f5.8 |
| chr4_-_63980140 | 0.13 |
ENSDART00000160818
|
znf1091
|
zinc finger protein 1091 |
| chr4_+_54480250 | 0.13 |
ENSDART00000158834
|
si:ch211-227e10.6
|
si:ch211-227e10.6 |
| chr13_+_281214 | 0.13 |
ENSDART00000137572
|
mpc1
|
mitochondrial pyruvate carrier 1 |
| chr23_+_38957738 | 0.12 |
ENSDART00000193480
|
ATP9A
|
ATPase phospholipid transporting 9A (putative) |
| chr19_-_31035155 | 0.12 |
ENSDART00000161882
|
bzw2
|
basic leucine zipper and W2 domains 2 |
| chr4_+_70015788 | 0.12 |
ENSDART00000161133
|
si:dkey-3h2.4
|
si:dkey-3h2.4 |
| chr2_-_26563978 | 0.12 |
ENSDART00000150016
|
glis1a
|
GLIS family zinc finger 1a |
| chr10_-_25343521 | 0.12 |
ENSDART00000166348
ENSDART00000009477 |
cct8
|
chaperonin containing TCP1, subunit 8 (theta) |
| chr10_+_15603082 | 0.12 |
ENSDART00000024450
|
zfand5b
|
zinc finger, AN1-type domain 5b |
| chr13_+_8696825 | 0.11 |
ENSDART00000109059
|
ttc7a
|
tetratricopeptide repeat domain 7A |
| chr16_+_7991274 | 0.11 |
ENSDART00000179704
|
ano10a
|
anoctamin 10a |
| chr5_-_42060121 | 0.11 |
ENSDART00000148021
ENSDART00000147407 |
cenpv
|
centromere protein V |
| chr25_+_27410352 | 0.11 |
ENSDART00000154362
|
pot1
|
protection of telomeres 1 homolog |
| chr6_+_46258866 | 0.10 |
ENSDART00000134584
|
zgc:162324
|
zgc:162324 |
| chr4_-_49987980 | 0.10 |
ENSDART00000150428
|
si:dkey-156k2.4
|
si:dkey-156k2.4 |
| chr5_-_3960161 | 0.10 |
ENSDART00000111453
|
myo19
|
myosin XIX |
| chr16_+_54829574 | 0.10 |
ENSDART00000148392
|
pabpc1a
|
poly(A) binding protein, cytoplasmic 1a |
| chr1_+_218524 | 0.10 |
ENSDART00000109529
|
tmco3
|
transmembrane and coiled-coil domains 3 |
| chr22_+_19522982 | 0.10 |
ENSDART00000192428
ENSDART00000190812 |
si:dkey-78l4.13
|
si:dkey-78l4.13 |
| chr4_+_69863750 | 0.10 |
ENSDART00000170800
|
znf1117
|
zinc finger protein 1117 |
| chr5_-_42059869 | 0.09 |
ENSDART00000193984
|
cenpv
|
centromere protein V |
| chr9_-_55128839 | 0.09 |
ENSDART00000085135
|
tbl1x
|
transducin beta like 1 X-linked |
| chr6_-_40651944 | 0.09 |
ENSDART00000187423
|
ppil1
|
peptidylprolyl isomerase (cyclophilin)-like 1 |
| chr14_+_22132896 | 0.09 |
ENSDART00000138274
|
ccng1
|
cyclin G1 |
| chr11_-_36446655 | 0.09 |
ENSDART00000179025
|
zbtb40
|
zinc finger and BTB domain containing 40 |
| chr4_-_37707809 | 0.09 |
ENSDART00000162606
|
znf1139
|
zinc finger protein 1139 |
| chr5_+_1624359 | 0.09 |
ENSDART00000165431
|
PPP1CC
|
protein phosphatase 1 catalytic subunit gamma |
| chr4_+_59425539 | 0.08 |
ENSDART00000127914
|
si:ch211-241n15.3
|
si:ch211-241n15.3 |
| chr13_-_280652 | 0.08 |
ENSDART00000193627
|
slc30a6
|
solute carrier family 30 (zinc transporter), member 6 |
| chr22_-_35006554 | 0.08 |
ENSDART00000138639
|
kcnip2
|
Kv channel interacting protein 2 |
| chr5_-_32890807 | 0.08 |
ENSDART00000007512
|
pole3
|
polymerase (DNA directed), epsilon 3 (p17 subunit) |
| chr4_+_63928858 | 0.08 |
ENSDART00000161958
|
si:dkey-179k24.2
|
si:dkey-179k24.2 |
| chr2_-_27315438 | 0.07 |
ENSDART00000087687
|
dsela
|
dermatan sulfate epimerase-like a |
| chr4_+_69231190 | 0.07 |
ENSDART00000158735
|
zgc:173615
|
zgc:173615 |
| chr17_-_33707589 | 0.07 |
ENSDART00000124788
|
si:dkey-84k17.2
|
si:dkey-84k17.2 |
| chr13_-_9024004 | 0.07 |
ENSDART00000169564
|
si:dkey-112g5.11
|
si:dkey-112g5.11 |
| chr16_+_11573471 | 0.07 |
ENSDART00000145858
|
si:dkey-11o1.2
|
si:dkey-11o1.2 |
| chr13_-_44831072 | 0.07 |
ENSDART00000137275
ENSDART00000188781 |
si:dkeyp-2e4.6
|
si:dkeyp-2e4.6 |
| chr3_-_25119839 | 0.07 |
ENSDART00000154724
|
chadla
|
chondroadherin-like a |
| chr23_+_31913292 | 0.07 |
ENSDART00000136910
|
armc1l
|
armadillo repeat containing 1, like |
| chr4_+_76353418 | 0.07 |
ENSDART00000171289
|
si:ch73-158p21.2
|
si:ch73-158p21.2 |
| chr23_+_38957472 | 0.07 |
ENSDART00000193836
|
ATP9A
|
ATPase phospholipid transporting 9A (putative) |
| chr5_-_69180587 | 0.07 |
ENSDART00000156681
ENSDART00000160753 |
zgc:171967
|
zgc:171967 |
| chr4_+_50891961 | 0.07 |
ENSDART00000150271
|
si:ch211-208f21.2
|
si:ch211-208f21.2 |
| chr2_-_27315604 | 0.06 |
ENSDART00000185540
|
dsela
|
dermatan sulfate epimerase-like a |
| chr5_+_56277866 | 0.06 |
ENSDART00000170610
ENSDART00000028854 ENSDART00000148749 |
aatf
|
apoptosis antagonizing transcription factor |
| chr25_+_4751879 | 0.06 |
ENSDART00000169465
|
si:zfos-2372e4.1
|
si:zfos-2372e4.1 |
| chr4_-_59094319 | 0.06 |
ENSDART00000162560
|
si:dkey-28i19.1
|
si:dkey-28i19.1 |
| chr2_-_37532772 | 0.06 |
ENSDART00000133951
|
arhgef18a
|
rho/rac guanine nucleotide exchange factor (GEF) 18a |
| chr2_+_1988036 | 0.06 |
ENSDART00000155956
|
ssx2ipa
|
synovial sarcoma, X breakpoint 2 interacting protein a |
| chr5_-_24543526 | 0.06 |
ENSDART00000046384
|
trmt2a
|
tRNA methyltransferase 2 homolog A |
| chr7_+_17063761 | 0.06 |
ENSDART00000182880
|
htatip2
|
HIV-1 Tat interactive protein 2 |
| chr4_-_54109066 | 0.06 |
ENSDART00000129685
ENSDART00000165283 ENSDART00000169701 |
si:dkey-199m13.7
|
si:dkey-199m13.7 |
| chr5_+_53482597 | 0.05 |
ENSDART00000180333
|
BX323994.1
|
|
| chr14_-_34830615 | 0.05 |
ENSDART00000167787
|
adrb2a
|
adrenoceptor beta 2, surface a |
| chr9_-_11560427 | 0.05 |
ENSDART00000127942
ENSDART00000061442 |
cryba2b
|
crystallin, beta A2b |
| chr4_-_1914228 | 0.05 |
ENSDART00000087835
|
ano6
|
anoctamin 6 |
| chr22_-_8648144 | 0.05 |
ENSDART00000078241
ENSDART00000191246 |
zmp:0000001175
|
zmp:0000001175 |
| chr24_-_25184553 | 0.05 |
ENSDART00000166917
|
plcxd2
|
phosphatidylinositol-specific phospholipase C, X domain containing 2 |
| chr9_+_41024973 | 0.05 |
ENSDART00000014660
ENSDART00000144467 |
ormdl1
|
ORMDL sphingolipid biosynthesis regulator 1 |
| chr7_+_23943597 | 0.04 |
ENSDART00000157408
|
si:dkey-183c6.7
|
si:dkey-183c6.7 |
| chr1_+_57752724 | 0.04 |
ENSDART00000169976
|
si:dkey-1c7.1
|
si:dkey-1c7.1 |
| chr21_-_45382112 | 0.04 |
ENSDART00000151029
ENSDART00000151335 ENSDART00000151687 ENSDART00000075438 |
cdkn2aipnl
|
CDKN2A interacting protein N-terminal like |
| chr12_-_27031060 | 0.04 |
ENSDART00000076103
|
chmp2a
|
charged multivesicular body protein 2A |
| chr4_-_55321457 | 0.03 |
ENSDART00000190996
|
znf998
|
zinc finger protein 998 |
| chr4_+_33819759 | 0.03 |
ENSDART00000188892
ENSDART00000182514 ENSDART00000133511 ENSDART00000160632 ENSDART00000169630 ENSDART00000146157 ENSDART00000182964 |
znf1026
|
zinc finger protein 1026 |
| chr4_+_35167118 | 0.03 |
ENSDART00000133667
|
si:dkey-269p2.1
|
si:dkey-269p2.1 |
| chr18_+_16133595 | 0.03 |
ENSDART00000080423
|
ctsd
|
cathepsin D |
| chr4_-_37236494 | 0.03 |
ENSDART00000165241
|
si:dkey-3p4.7
|
si:dkey-3p4.7 |
| chr2_-_31800521 | 0.03 |
ENSDART00000112763
|
retreg1
|
reticulophagy regulator 1 |
| chr4_-_65036768 | 0.03 |
ENSDART00000184455
|
si:ch211-283l16.1
|
si:ch211-283l16.1 |
| chr1_+_40263671 | 0.03 |
ENSDART00000045697
|
zgc:56493
|
zgc:56493 |
| chr23_+_4777348 | 0.03 |
ENSDART00000171971
|
vgll4a
|
vestigial-like family member 4a |
| chr4_+_60296027 | 0.03 |
ENSDART00000172479
|
si:dkey-248e17.5
|
si:dkey-248e17.5 |
| chr4_+_39055027 | 0.02 |
ENSDART00000150397
|
znf977
|
zinc finger protein 977 |
| chr6_+_21992820 | 0.02 |
ENSDART00000147507
|
thumpd3
|
THUMP domain containing 3 |
| chr5_-_24542726 | 0.02 |
ENSDART00000182975
|
trmt2a
|
tRNA methyltransferase 2 homolog A |
| chr20_+_27096430 | 0.02 |
ENSDART00000153358
|
ubr7
|
ubiquitin protein ligase E3 component n-recognin 7 |
| chr15_-_2973368 | 0.02 |
ENSDART00000163131
|
zgc:153184
|
zgc:153184 |
| chr15_-_20710 | 0.02 |
ENSDART00000161218
|
cyp2y3
|
cytochrome P450, family 2, subfamily Y, polypeptide 3 |
| chr2_-_24013260 | 0.02 |
ENSDART00000137065
|
col15a1b
|
collagen, type XV, alpha 1b |
| chr23_-_45682136 | 0.02 |
ENSDART00000164646
|
fam160a1b
|
family with sequence similarity 160, member A1b |
| chr18_-_33979693 | 0.02 |
ENSDART00000021215
|
si:ch211-203b20.7
|
si:ch211-203b20.7 |
| chr4_-_45136781 | 0.02 |
ENSDART00000141609
|
znf1046
|
zinc finger protein 1046 |
| chr21_-_40040178 | 0.02 |
ENSDART00000159741
|
BX248108.1
|
Danio rerio multidrug and toxin extrusion protein 1-like (mate4), mRNA. |
| chr11_-_24510995 | 0.01 |
ENSDART00000163489
|
plekhg5a
|
pleckstrin homology domain containing, family G (with RhoGef domain) member 5a |
| chr5_-_8765428 | 0.01 |
ENSDART00000167793
|
mybbp1a
|
MYB binding protein (P160) 1a |
| chr20_-_51831816 | 0.01 |
ENSDART00000060505
|
mia3
|
melanoma inhibitory activity family, member 3 |
| chr17_-_45164291 | 0.01 |
ENSDART00000062109
|
noxred1
|
NADP-dependent oxidoreductase domain containing 1 |
| chr17_-_24439672 | 0.00 |
ENSDART00000155020
|
wdpcp
|
WD repeat containing planar cell polarity effector |
| chr20_+_18994059 | 0.00 |
ENSDART00000152380
|
tdh
|
L-threonine dehydrogenase |
| chr8_+_10862353 | 0.00 |
ENSDART00000140717
|
brpf3b
|
bromodomain and PHD finger containing, 3b |
| chr14_+_18785727 | 0.00 |
ENSDART00000184452
|
si:ch211-111e20.1
|
si:ch211-111e20.1 |
| chr4_-_33371634 | 0.00 |
ENSDART00000150712
|
si:dkeyp-4f2.3
|
si:dkeyp-4f2.3 |
Network of associatons between targets according to the STRING database.
First level regulatory network of CT027745.1+tbx19
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.5 | GO:0032816 | regulation of natural killer cell activation(GO:0032814) positive regulation of natural killer cell activation(GO:0032816) TRIF-dependent toll-like receptor signaling pathway(GO:0035666) |
| 0.1 | 0.9 | GO:0046716 | muscle cell cellular homeostasis(GO:0046716) |
| 0.1 | 1.7 | GO:0015671 | oxygen transport(GO:0015671) |
| 0.1 | 0.2 | GO:1903232 | melanosome assembly(GO:1903232) |
| 0.1 | 0.3 | GO:0010269 | response to selenium ion(GO:0010269) |
| 0.1 | 0.2 | GO:0060262 | N-terminal protein palmitoylation(GO:0006500) negative regulation of lipoprotein metabolic process(GO:0050748) regulation of N-terminal protein palmitoylation(GO:0060254) negative regulation of N-terminal protein palmitoylation(GO:0060262) negative regulation of protein lipidation(GO:1903060) |
| 0.0 | 0.2 | GO:0006524 | alanine metabolic process(GO:0006522) alanine catabolic process(GO:0006524) pyruvate family amino acid metabolic process(GO:0009078) pyruvate family amino acid catabolic process(GO:0009080) D-alanine family amino acid metabolic process(GO:0046144) D-alanine metabolic process(GO:0046436) D-alanine catabolic process(GO:0055130) |
| 0.0 | 0.2 | GO:0000448 | cleavage in ITS2 between 5.8S rRNA and LSU-rRNA of tricistronic rRNA transcript (SSU-rRNA, 5.8S rRNA, LSU-rRNA)(GO:0000448) |
| 0.0 | 0.2 | GO:0016332 | establishment or maintenance of polarity of embryonic epithelium(GO:0016332) |
| 0.0 | 0.3 | GO:0051694 | pointed-end actin filament capping(GO:0051694) |
| 0.0 | 0.2 | GO:1904398 | positive regulation of neuromuscular junction development(GO:1904398) |
| 0.0 | 0.4 | GO:0007172 | signal complex assembly(GO:0007172) |
| 0.0 | 0.2 | GO:0019388 | galactose catabolic process(GO:0019388) galactose catabolic process via UDP-galactose(GO:0033499) |
| 0.0 | 0.4 | GO:1900077 | negative regulation of insulin receptor signaling pathway(GO:0046627) negative regulation of cellular response to insulin stimulus(GO:1900077) |
| 0.0 | 0.1 | GO:0035095 | behavioral response to nicotine(GO:0035095) |
| 0.0 | 0.1 | GO:0030205 | dermatan sulfate metabolic process(GO:0030205) |
| 0.0 | 0.2 | GO:0035188 | hatching(GO:0035188) organism emergence from protective structure(GO:0071684) |
| 0.0 | 0.5 | GO:0000027 | ribosomal large subunit assembly(GO:0000027) |
| 0.0 | 0.1 | GO:0051972 | regulation of telomerase activity(GO:0051972) |
| 0.0 | 0.1 | GO:0006272 | leading strand elongation(GO:0006272) |
| 0.0 | 0.4 | GO:0046475 | glycerophospholipid catabolic process(GO:0046475) |
| 0.0 | 0.0 | GO:0002407 | dendritic cell chemotaxis(GO:0002407) dendritic cell migration(GO:0036336) |
| 0.0 | 0.1 | GO:0071376 | response to corticotropin-releasing hormone(GO:0043435) cellular response to corticotropin-releasing hormone stimulus(GO:0071376) |
| 0.0 | 0.2 | GO:0000186 | activation of MAPKK activity(GO:0000186) |
| 0.0 | 0.0 | GO:0071918 | urea transmembrane transport(GO:0071918) |
| 0.0 | 0.5 | GO:0071219 | cellular response to molecule of bacterial origin(GO:0071219) cellular response to lipopolysaccharide(GO:0071222) |
| 0.0 | 0.2 | GO:0010962 | regulation of glycogen biosynthetic process(GO:0005979) regulation of glucan biosynthetic process(GO:0010962) |
| 0.0 | 0.3 | GO:0006298 | mismatch repair(GO:0006298) |
| 0.0 | 0.2 | GO:0032094 | response to food(GO:0032094) |
| 0.0 | 0.2 | GO:0006123 | mitochondrial electron transport, cytochrome c to oxygen(GO:0006123) |
| 0.0 | 0.2 | GO:0018401 | peptidyl-proline hydroxylation to 4-hydroxy-L-proline(GO:0018401) 4-hydroxyproline metabolic process(GO:0019471) |
| 0.0 | 0.3 | GO:0051923 | sulfation(GO:0051923) |
| 0.0 | 0.0 | GO:1905038 | regulation of sphingolipid biosynthetic process(GO:0090153) negative regulation of sphingolipid biosynthetic process(GO:0090155) negative regulation of ceramide biosynthetic process(GO:1900060) regulation of membrane lipid metabolic process(GO:1905038) regulation of ceramide biosynthetic process(GO:2000303) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 1.7 | GO:0005833 | hemoglobin complex(GO:0005833) |
| 0.1 | 0.4 | GO:0033596 | TSC1-TSC2 complex(GO:0033596) |
| 0.1 | 0.2 | GO:0031085 | BLOC-3 complex(GO:0031085) |
| 0.0 | 0.2 | GO:0071541 | eukaryotic translation initiation factor 3 complex, eIF3m(GO:0071541) |
| 0.0 | 0.3 | GO:0032300 | mismatch repair complex(GO:0032300) |
| 0.0 | 0.2 | GO:0034388 | Pwp2p-containing subcomplex of 90S preribosome(GO:0034388) |
| 0.0 | 0.4 | GO:0034992 | microtubule organizing center attachment site(GO:0034992) LINC complex(GO:0034993) |
| 0.0 | 0.1 | GO:0008622 | epsilon DNA polymerase complex(GO:0008622) |
| 0.0 | 0.2 | GO:0000164 | protein phosphatase type 1 complex(GO:0000164) |
| 0.0 | 0.8 | GO:0036379 | striated muscle thin filament(GO:0005865) myofilament(GO:0036379) |
| 0.0 | 0.2 | GO:0005751 | mitochondrial respiratory chain complex IV(GO:0005751) |
| 0.0 | 0.0 | GO:0035339 | SPOTS complex(GO:0035339) |
| 0.0 | 0.1 | GO:0005832 | chaperonin-containing T-complex(GO:0005832) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 1.7 | GO:0019825 | oxygen transporter activity(GO:0005344) oxygen binding(GO:0019825) |
| 0.1 | 0.3 | GO:0008097 | 5S rRNA binding(GO:0008097) |
| 0.1 | 0.2 | GO:0003978 | UDP-glucose 4-epimerase activity(GO:0003978) |
| 0.0 | 0.2 | GO:0003884 | D-amino-acid oxidase activity(GO:0003884) |
| 0.0 | 0.6 | GO:0004687 | myosin light chain kinase activity(GO:0004687) |
| 0.0 | 0.3 | GO:0004062 | aryl sulfotransferase activity(GO:0004062) |
| 0.0 | 0.4 | GO:0047498 | calcium-dependent phospholipase A2 activity(GO:0047498) |
| 0.0 | 0.1 | GO:0047757 | chondroitin-glucuronate 5-epimerase activity(GO:0047757) |
| 0.0 | 0.1 | GO:0010521 | telomerase inhibitor activity(GO:0010521) |
| 0.0 | 0.4 | GO:0031726 | CCR1 chemokine receptor binding(GO:0031726) CCR4 chemokine receptor binding(GO:0031729) |
| 0.0 | 0.5 | GO:0070492 | oligosaccharide binding(GO:0070492) |
| 0.0 | 0.2 | GO:0004691 | cAMP-dependent protein kinase activity(GO:0004691) |
| 0.0 | 0.4 | GO:0016846 | carbon-sulfur lyase activity(GO:0016846) |
| 0.0 | 0.2 | GO:0004656 | procollagen-proline 4-dioxygenase activity(GO:0004656) |
| 0.0 | 0.3 | GO:0030983 | mismatched DNA binding(GO:0030983) |
| 0.0 | 0.2 | GO:2001069 | glycogen binding(GO:2001069) |
| 0.0 | 0.3 | GO:0005523 | tropomyosin binding(GO:0005523) |
| 0.0 | 0.2 | GO:0017128 | phospholipid scramblase activity(GO:0017128) |
| 0.0 | 0.1 | GO:0035259 | glucocorticoid receptor binding(GO:0035259) |
| 0.0 | 0.0 | GO:0051380 | norepinephrine binding(GO:0051380) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.5 | ST IL 13 PATHWAY | Interleukin 13 (IL-13) Pathway |
| 0.0 | 0.6 | SIG REGULATION OF THE ACTIN CYTOSKELETON BY RHO GTPASES | Genes related to regulation of the actin cytoskeleton |
| 0.0 | 0.5 | PID TOLL ENDOGENOUS PATHWAY | Endogenous TLR signaling |
| 0.0 | 0.4 | PID ARF6 PATHWAY | Arf6 signaling events |
| 0.0 | 0.2 | PID IL23 PATHWAY | IL23-mediated signaling events |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.5 | REACTOME IKK COMPLEX RECRUITMENT MEDIATED BY RIP1 | Genes involved in IKK complex recruitment mediated by RIP1 |
| 0.0 | 0.3 | REACTOME PLATELET ADHESION TO EXPOSED COLLAGEN | Genes involved in Platelet Adhesion to exposed collagen |
| 0.0 | 0.1 | REACTOME PHOSPHORYLATION OF CD3 AND TCR ZETA CHAINS | Genes involved in Phosphorylation of CD3 and TCR zeta chains |
| 0.0 | 0.2 | REACTOME SYNTHESIS SECRETION AND DEACYLATION OF GHRELIN | Genes involved in Synthesis, Secretion, and Deacylation of Ghrelin |
| 0.0 | 0.1 | REACTOME HORMONE SENSITIVE LIPASE HSL MEDIATED TRIACYLGLYCEROL HYDROLYSIS | Genes involved in Hormone-sensitive lipase (HSL)-mediated triacylglycerol hydrolysis |