Project
PRJNA207719: Tissue specific transcriptome profiling
Navigation
Downloads
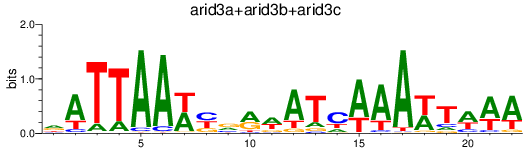
Results for arid3a+arid3b+arid3c
Z-value: 1.21
Transcription factors associated with arid3a+arid3b+arid3c
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
arid3c
|
ENSDARG00000067729 | AT rich interactive domain 3C (BRIGHT-like) |
|
arid3a
|
ENSDARG00000070843 | AT rich interactive domain 3A (BRIGHT-like) |
|
arid3b
|
ENSDARG00000104034 | AT rich interactive domain 3B (BRIGHT-like) |
|
arid3c
|
ENSDARG00000114447 | AT rich interactive domain 3C (BRIGHT-like) |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| arid3c | dr11_v1_chr5_+_41322783_41322783 | 0.89 | 4.3e-02 | Click! |
| arid3b | dr11_v1_chr25_-_29072162_29072162 | -0.66 | 2.3e-01 | Click! |
| arid3a | dr11_v1_chr11_+_5681762_5681762 | 0.65 | 2.4e-01 | Click! |
Activity profile of arid3a+arid3b+arid3c motif
Sorted Z-values of arid3a+arid3b+arid3c motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr19_-_3167729 | 2.39 |
ENSDART00000110763
ENSDART00000145710 ENSDART00000074620 ENSDART00000105174 |
stm
|
starmaker |
| chr8_+_3820134 | 2.35 |
ENSDART00000122454
|
citb
|
citron rho-interacting serine/threonine kinase b |
| chr5_+_70155935 | 1.93 |
ENSDART00000165570
|
rgs3a
|
regulator of G protein signaling 3a |
| chr18_-_47662696 | 1.90 |
ENSDART00000184260
|
CABZ01073963.1
|
|
| chr2_-_9489611 | 1.81 |
ENSDART00000146715
|
st6galnac3
|
ST6 (alpha-N-acetyl-neuraminyl-2,3-beta-galactosyl-1,3)-N-acetylgalactosaminide alpha-2,6-sialyltransferase 3 |
| chr1_+_7517454 | 1.67 |
ENSDART00000016139
|
lancl1
|
LanC antibiotic synthetase component C-like 1 (bacterial) |
| chr1_+_31112436 | 1.66 |
ENSDART00000075340
|
eef1a1b
|
eukaryotic translation elongation factor 1 alpha 1b |
| chr2_-_9059955 | 1.26 |
ENSDART00000022768
|
ak5
|
adenylate kinase 5 |
| chr19_+_4139065 | 1.09 |
ENSDART00000172524
|
si:dkey-218f9.10
|
si:dkey-218f9.10 |
| chr15_+_39096736 | 1.05 |
ENSDART00000129511
ENSDART00000014877 |
robo2
|
roundabout, axon guidance receptor, homolog 2 (Drosophila) |
| chr2_+_31492662 | 0.91 |
ENSDART00000123495
|
cacnb2b
|
calcium channel, voltage-dependent, beta 2b |
| chr23_-_24343363 | 0.89 |
ENSDART00000166392
|
fam131c
|
family with sequence similarity 131, member C |
| chr19_-_7406933 | 0.85 |
ENSDART00000151137
|
oxr1b
|
oxidation resistance 1b |
| chr15_+_45994123 | 0.77 |
ENSDART00000124704
|
lrfn1
|
leucine rich repeat and fibronectin type III domain containing 1 |
| chr8_+_7144066 | 0.75 |
ENSDART00000146306
|
slc6a6a
|
solute carrier family 6 (neurotransmitter transporter), member 6a |
| chr12_+_7445595 | 0.72 |
ENSDART00000103536
ENSDART00000152524 |
phyhiplb
|
phytanoyl-CoA 2-hydroxylase interacting protein-like b |
| chr20_+_52389858 | 0.72 |
ENSDART00000185863
ENSDART00000166651 |
arhgap39
|
Rho GTPase activating protein 39 |
| chr4_+_14660769 | 0.72 |
ENSDART00000168152
ENSDART00000013990 ENSDART00000079987 |
abcc9
|
ATP-binding cassette, sub-family C (CFTR/MRP), member 9 |
| chr12_+_8168272 | 0.72 |
ENSDART00000054092
|
arid5b
|
AT-rich interaction domain 5B |
| chr5_-_12407194 | 0.72 |
ENSDART00000125291
|
ksr2
|
kinase suppressor of ras 2 |
| chr10_-_2522588 | 0.69 |
ENSDART00000081926
|
CU856539.1
|
|
| chr22_-_15562933 | 0.68 |
ENSDART00000141528
|
ankmy1
|
ankyrin repeat and MYND domain containing 1 |
| chr4_+_9669717 | 0.68 |
ENSDART00000004604
|
si:dkey-153k10.9
|
si:dkey-153k10.9 |
| chr16_-_13730152 | 0.67 |
ENSDART00000138772
|
ttyh1
|
tweety family member 1 |
| chr19_-_11208782 | 0.66 |
ENSDART00000044426
ENSDART00000189754 |
si:dkey-240h12.4
|
si:dkey-240h12.4 |
| chr19_-_26869103 | 0.65 |
ENSDART00000089699
|
prrt1
|
proline-rich transmembrane protein 1 |
| chr21_+_13861589 | 0.62 |
ENSDART00000015629
ENSDART00000171306 |
stxbp1a
|
syntaxin binding protein 1a |
| chr21_+_3093419 | 0.61 |
ENSDART00000162520
|
SHC3
|
SHC adaptor protein 3 |
| chr18_+_6126506 | 0.61 |
ENSDART00000125725
|
si:ch1073-390k14.1
|
si:ch1073-390k14.1 |
| chr10_+_45089820 | 0.58 |
ENSDART00000175481
|
camk2b2
|
calcium/calmodulin-dependent protein kinase (CaM kinase) II beta 2 |
| chr8_+_31717175 | 0.57 |
ENSDART00000013434
|
oxct1a
|
3-oxoacid CoA transferase 1a |
| chr16_-_55028740 | 0.56 |
ENSDART00000156368
ENSDART00000161704 |
zgc:114181
|
zgc:114181 |
| chr19_+_24872159 | 0.56 |
ENSDART00000158490
|
si:ch211-195b13.1
|
si:ch211-195b13.1 |
| chr17_-_16965809 | 0.55 |
ENSDART00000153697
|
nrxn3a
|
neurexin 3a |
| chr16_-_8927425 | 0.55 |
ENSDART00000000382
|
triob
|
trio Rho guanine nucleotide exchange factor b |
| chr8_+_31716872 | 0.54 |
ENSDART00000161121
|
oxct1a
|
3-oxoacid CoA transferase 1a |
| chr5_+_58492699 | 0.54 |
ENSDART00000181584
|
FQ378016.1
|
|
| chr13_+_27951688 | 0.53 |
ENSDART00000050303
|
b3gat2
|
beta-1,3-glucuronyltransferase 2 (glucuronosyltransferase S) |
| chr22_+_12366516 | 0.53 |
ENSDART00000157802
|
r3hdm1
|
R3H domain containing 1 |
| chr11_+_39672874 | 0.53 |
ENSDART00000046663
ENSDART00000157659 |
camta1b
|
calmodulin binding transcription activator 1b |
| chr17_+_22311413 | 0.51 |
ENSDART00000151929
|
slc8a1b
|
solute carrier family 8 (sodium/calcium exchanger), member 1b |
| chr20_+_50852356 | 0.50 |
ENSDART00000167517
ENSDART00000168396 |
gphnb
|
gephyrin b |
| chr11_-_3343463 | 0.50 |
ENSDART00000066177
|
tuba2
|
tubulin, alpha 2 |
| chr12_+_3022882 | 0.47 |
ENSDART00000122905
|
rac3b
|
Rac family small GTPase 3b |
| chr19_-_3240605 | 0.46 |
ENSDART00000105168
|
si:ch211-133n4.4
|
si:ch211-133n4.4 |
| chr6_-_12275836 | 0.46 |
ENSDART00000189980
|
pkp4
|
plakophilin 4 |
| chr12_+_3871452 | 0.46 |
ENSDART00000066546
|
nif3l1
|
NIF3 NGG1 interacting factor 3-like 1 (S. cerevisiae) |
| chr25_+_14165447 | 0.45 |
ENSDART00000145387
|
shank2
|
SH3 and multiple ankyrin repeat domains 2 |
| chr8_-_17067364 | 0.44 |
ENSDART00000132687
|
rab3c
|
RAB3C, member RAS oncogene family |
| chr10_+_44584614 | 0.43 |
ENSDART00000163523
|
sez6l
|
seizure related 6 homolog (mouse)-like |
| chr25_-_6420800 | 0.43 |
ENSDART00000153768
|
ptpn9a
|
protein tyrosine phosphatase, non-receptor type 9, a |
| chr2_+_34967022 | 0.42 |
ENSDART00000134926
|
astn1
|
astrotactin 1 |
| chr12_+_47698356 | 0.42 |
ENSDART00000112010
|
lzts2b
|
leucine zipper, putative tumor suppressor 2b |
| chr18_+_39416357 | 0.42 |
ENSDART00000183174
ENSDART00000127955 ENSDART00000171303 |
lysmd2
|
LysM, putative peptidoglycan-binding, domain containing 2 |
| chr3_-_18711288 | 0.41 |
ENSDART00000183885
|
grid2ipa
|
glutamate receptor, ionotropic, delta 2 (Grid2) interacting protein, a |
| chr2_+_3472832 | 0.41 |
ENSDART00000115278
|
cx47.1
|
connexin 47.1 |
| chr3_-_21280373 | 0.41 |
ENSDART00000003939
|
syngr1a
|
synaptogyrin 1a |
| chr17_-_36936649 | 0.40 |
ENSDART00000145236
|
dpysl5a
|
dihydropyrimidinase-like 5a |
| chr17_+_2063693 | 0.40 |
ENSDART00000182349
|
zgc:162989
|
zgc:162989 |
| chr7_+_60551133 | 0.40 |
ENSDART00000148038
|
lrfn4b
|
leucine rich repeat and fibronectin type III domain containing 4b |
| chr22_-_26945493 | 0.40 |
ENSDART00000077411
|
cxcl12b
|
chemokine (C-X-C motif) ligand 12b (stromal cell-derived factor 1) |
| chr25_+_37126921 | 0.39 |
ENSDART00000124331
|
si:ch1073-174d20.1
|
si:ch1073-174d20.1 |
| chr8_-_34052019 | 0.38 |
ENSDART00000040126
ENSDART00000159208 ENSDART00000048994 ENSDART00000098822 |
pbx3b
|
pre-B-cell leukemia homeobox 3b |
| chr6_+_54221654 | 0.38 |
ENSDART00000128456
|
pacsin1b
|
protein kinase C and casein kinase substrate in neurons 1b |
| chr10_-_34870667 | 0.37 |
ENSDART00000161272
|
dclk1a
|
doublecortin-like kinase 1a |
| chr6_-_10780698 | 0.37 |
ENSDART00000151714
|
gpr155b
|
G protein-coupled receptor 155b |
| chr16_-_29458806 | 0.36 |
ENSDART00000047931
|
lingo4b
|
leucine rich repeat and Ig domain containing 4b |
| chr6_+_58698475 | 0.36 |
ENSDART00000056138
|
igsf8
|
immunoglobulin superfamily, member 8 |
| chr14_+_28281744 | 0.35 |
ENSDART00000173292
|
mid2
|
midline 2 |
| chr20_+_54383838 | 0.35 |
ENSDART00000157737
|
lrfn5b
|
leucine rich repeat and fibronectin type III domain containing 5b |
| chr9_+_2574122 | 0.35 |
ENSDART00000166326
ENSDART00000191822 |
CIR1
|
si:ch73-167c12.2 |
| chr19_-_34063567 | 0.34 |
ENSDART00000157815
ENSDART00000183907 |
elmo1
|
engulfment and cell motility 1 (ced-12 homolog, C. elegans) |
| chr2_-_1548330 | 0.34 |
ENSDART00000082155
ENSDART00000108481 ENSDART00000111272 |
dab1b
|
Dab, reelin signal transducer, homolog 1b (Drosophila) |
| chr16_+_36126310 | 0.34 |
ENSDART00000166040
ENSDART00000189802 |
sh3bp5b
|
SH3-domain binding protein 5b (BTK-associated) |
| chr15_+_37197494 | 0.33 |
ENSDART00000166203
|
aplp1
|
amyloid beta (A4) precursor-like protein 1 |
| chr22_+_5103349 | 0.33 |
ENSDART00000083474
|
atcaya
|
ataxia, cerebellar, Cayman type a |
| chr19_-_12145765 | 0.33 |
ENSDART00000032474
|
pabpc1b
|
poly A binding protein, cytoplasmic 1 b |
| chr14_+_46342882 | 0.33 |
ENSDART00000193707
ENSDART00000060577 |
tmem33
|
transmembrane protein 33 |
| chr3_-_21037840 | 0.32 |
ENSDART00000002393
|
rundc3aa
|
RUN domain containing 3Aa |
| chr5_+_69650148 | 0.32 |
ENSDART00000097244
|
gtf2h3
|
general transcription factor IIH, polypeptide 3 |
| chr22_+_27090136 | 0.31 |
ENSDART00000136770
|
si:dkey-246e1.3
|
si:dkey-246e1.3 |
| chr7_+_9189547 | 0.31 |
ENSDART00000169783
|
pcsk6
|
proprotein convertase subtilisin/kexin type 6 |
| chr9_+_34641237 | 0.31 |
ENSDART00000133996
|
shox
|
short stature homeobox |
| chr16_+_14812585 | 0.30 |
ENSDART00000134087
|
col14a1a
|
collagen, type XIV, alpha 1a |
| chr4_-_20027571 | 0.29 |
ENSDART00000003420
|
drd4-rs
|
dopamine receptor D4 related sequence |
| chr11_+_41459408 | 0.29 |
ENSDART00000182285
|
park7
|
parkinson protein 7 |
| chr10_-_31104983 | 0.29 |
ENSDART00000186941
|
pknox2
|
pbx/knotted 1 homeobox 2 |
| chr22_+_18389271 | 0.29 |
ENSDART00000088270
|
yjefn3
|
YjeF N-terminal domain containing 3 |
| chr5_-_42180205 | 0.29 |
ENSDART00000145247
|
fam222ba
|
family with sequence similarity 222, member Ba |
| chr14_+_45028062 | 0.28 |
ENSDART00000184717
ENSDART00000185481 |
ATP8A1
|
ATPase phospholipid transporting 8A1 |
| chr24_-_37484123 | 0.28 |
ENSDART00000111623
|
cluap1
|
clusterin associated protein 1 |
| chr19_+_23932259 | 0.28 |
ENSDART00000139040
|
si:dkey-222b8.1
|
si:dkey-222b8.1 |
| chr1_+_9886991 | 0.28 |
ENSDART00000135702
|
rgs11
|
regulator of G protein signaling 11 |
| chr25_+_34407529 | 0.28 |
ENSDART00000156751
|
OTUD7A
|
si:dkey-37f18.2 |
| chr18_-_19103929 | 0.28 |
ENSDART00000188370
ENSDART00000177621 |
dennd4a
|
DENN/MADD domain containing 4A |
| chr19_-_12145390 | 0.28 |
ENSDART00000143087
|
pabpc1b
|
poly A binding protein, cytoplasmic 1 b |
| chr2_+_49232559 | 0.27 |
ENSDART00000154285
ENSDART00000175147 |
sema6ba
|
sema domain, transmembrane domain (TM), and cytoplasmic domain, (semaphorin) 6Ba |
| chr16_+_32014552 | 0.27 |
ENSDART00000047570
|
mboat7
|
membrane bound O-acyltransferase domain containing 7 |
| chr10_-_20706109 | 0.27 |
ENSDART00000124751
|
kcnip3b
|
Kv channel interacting protein 3b, calsenilin |
| chr9_+_34380299 | 0.27 |
ENSDART00000131705
|
lamp1
|
lysosomal-associated membrane protein 1 |
| chr14_-_12051610 | 0.27 |
ENSDART00000193741
ENSDART00000029366 |
zgc:66447
|
zgc:66447 |
| chr6_-_1865323 | 0.27 |
ENSDART00000155775
|
dlgap4b
|
discs, large (Drosophila) homolog-associated protein 4b |
| chr7_+_57866292 | 0.26 |
ENSDART00000138757
|
camk2d1
|
calcium/calmodulin-dependent protein kinase (CaM kinase) II delta 1 |
| chr8_+_1651821 | 0.26 |
ENSDART00000060865
ENSDART00000186304 |
rasal1b
|
RAS protein activator like 1b (GAP1 like) |
| chr20_+_18943406 | 0.25 |
ENSDART00000193590
|
mtmr9
|
myotubularin related protein 9 |
| chr18_+_39074139 | 0.25 |
ENSDART00000142390
|
gnb5b
|
guanine nucleotide binding protein (G protein), beta 5b |
| chr21_+_10794914 | 0.25 |
ENSDART00000084035
|
znf532
|
zinc finger protein 532 |
| chr13_-_41546779 | 0.25 |
ENSDART00000163331
|
pcdh15a
|
protocadherin-related 15a |
| chr3_+_34919810 | 0.24 |
ENSDART00000055264
|
ca10b
|
carbonic anhydrase Xb |
| chr8_+_104114 | 0.24 |
ENSDART00000172101
|
sncaip
|
synuclein, alpha interacting protein |
| chr6_-_33871055 | 0.24 |
ENSDART00000115409
|
mast2
|
microtubule associated serine/threonine kinase 2 |
| chr18_+_11858397 | 0.24 |
ENSDART00000133762
|
tmtc2b
|
transmembrane and tetratricopeptide repeat containing 2b |
| chr3_-_41995321 | 0.23 |
ENSDART00000192277
|
ttyh3a
|
tweety family member 3a |
| chr11_-_43200994 | 0.23 |
ENSDART00000164700
|
sptbn1
|
spectrin, beta, non-erythrocytic 1 |
| chr14_+_5936996 | 0.23 |
ENSDART00000097144
ENSDART00000126777 |
kctd8
|
potassium channel tetramerization domain containing 8 |
| chr2_+_33457310 | 0.23 |
ENSDART00000056657
|
zgc:113531
|
zgc:113531 |
| chr25_-_518656 | 0.23 |
ENSDART00000156421
|
myo9ab
|
myosin IXAb |
| chr12_+_38774860 | 0.23 |
ENSDART00000130371
|
kif19
|
kinesin family member 19 |
| chr1_+_59293873 | 0.23 |
ENSDART00000168036
|
rdh8b
|
retinol dehydrogenase 8b |
| chr20_-_28638871 | 0.22 |
ENSDART00000184779
|
rgs6
|
regulator of G protein signaling 6 |
| chr13_+_18371208 | 0.22 |
ENSDART00000138172
|
ccar1
|
cell division cycle and apoptosis regulator 1 |
| chr6_+_11989537 | 0.22 |
ENSDART00000190817
|
baz2ba
|
bromodomain adjacent to zinc finger domain, 2Ba |
| chr15_+_42285643 | 0.21 |
ENSDART00000152731
|
scaf4b
|
SR-related CTD-associated factor 4b |
| chr21_+_40695345 | 0.21 |
ENSDART00000143594
|
ccdc82
|
coiled-coil domain containing 82 |
| chr10_+_22510280 | 0.21 |
ENSDART00000109070
ENSDART00000182002 ENSDART00000192852 |
gigyf1b
|
GRB10 interacting GYF protein 1b |
| chr6_-_16394528 | 0.21 |
ENSDART00000089445
|
agap1
|
ArfGAP with GTPase domain, ankyrin repeat and PH domain 1 |
| chr15_+_36941490 | 0.21 |
ENSDART00000172664
|
kirrel3l
|
kirre like nephrin family adhesion molecule 3, like |
| chr8_-_38506339 | 0.21 |
ENSDART00000054558
ENSDART00000141623 |
foxb2
|
forkhead box B2 |
| chr9_+_33158191 | 0.21 |
ENSDART00000180786
|
dopey2
|
dopey family member 2 |
| chr3_+_60721342 | 0.21 |
ENSDART00000157772
|
foxj1a
|
forkhead box J1a |
| chr14_-_47391084 | 0.21 |
ENSDART00000159608
|
fstl5
|
follistatin-like 5 |
| chr14_+_25986895 | 0.21 |
ENSDART00000149087
|
slc36a1
|
solute carrier family 36 (proton/amino acid symporter), member 1 |
| chr7_+_11197940 | 0.20 |
ENSDART00000081346
|
cemip
|
cell migration inducing protein, hyaluronan binding |
| chr2_+_25657958 | 0.20 |
ENSDART00000161407
|
tnika
|
TRAF2 and NCK interacting kinase a |
| chr23_+_21544227 | 0.20 |
ENSDART00000140253
|
arhgef10lb
|
Rho guanine nucleotide exchange factor (GEF) 10-like b |
| chr9_+_3429662 | 0.20 |
ENSDART00000160977
ENSDART00000114168 ENSDART00000082153 |
CU469503.1
itga6a
|
integrin, alpha 6a |
| chr6_+_49551614 | 0.20 |
ENSDART00000022581
|
rab22a
|
RAB22A, member RAS oncogene family |
| chr13_+_6092135 | 0.20 |
ENSDART00000162738
|
fam120b
|
family with sequence similarity 120B |
| chr4_-_5019113 | 0.20 |
ENSDART00000189321
ENSDART00000081990 |
strip2
|
striatin interacting protein 2 |
| chr4_-_75157223 | 0.19 |
ENSDART00000174127
|
CABZ01066312.1
|
|
| chr10_+_15967643 | 0.19 |
ENSDART00000136709
|
apbb1
|
amyloid beta (A4) precursor protein-binding, family B, member 1 (Fe65) |
| chr2_-_22530969 | 0.19 |
ENSDART00000159641
|
znf644a
|
zinc finger protein 644a |
| chr11_-_889845 | 0.19 |
ENSDART00000162152
|
atg7
|
ATG7 autophagy related 7 homolog (S. cerevisiae) |
| chr1_-_1627487 | 0.19 |
ENSDART00000166094
|
clic6
|
chloride intracellular channel 6 |
| chr11_+_14199802 | 0.19 |
ENSDART00000102520
ENSDART00000133172 |
palm1a
|
paralemmin 1a |
| chr6_+_13726844 | 0.18 |
ENSDART00000055833
|
wnt6a
|
wingless-type MMTV integration site family, member 6a |
| chr18_+_17827149 | 0.18 |
ENSDART00000190237
ENSDART00000189345 |
ZNF423
|
si:ch211-216l23.1 |
| chr4_+_25912308 | 0.18 |
ENSDART00000167845
ENSDART00000136927 |
vezt
|
vezatin, adherens junctions transmembrane protein |
| chr21_-_38619305 | 0.18 |
ENSDART00000139178
|
slc25a14
|
solute carrier family 25 (mitochondrial carrier, brain), member 14 |
| chr2_-_52365251 | 0.18 |
ENSDART00000097716
|
tle2c
|
transducin like enhancer of split 2c |
| chr13_+_4883377 | 0.18 |
ENSDART00000181530
ENSDART00000092521 |
micu1
|
mitochondrial calcium uptake 1 |
| chr4_-_74892355 | 0.18 |
ENSDART00000188725
|
PHF21B
|
PHD finger protein 21B |
| chr3_+_62339264 | 0.18 |
ENSDART00000174569
|
BX470259.3
|
|
| chr22_+_39054012 | 0.18 |
ENSDART00000191326
|
ip6k1
|
inositol hexakisphosphate kinase 1 |
| chr15_-_33807758 | 0.18 |
ENSDART00000158445
|
pds5b
|
PDS5 cohesin associated factor B |
| chr10_-_20706270 | 0.17 |
ENSDART00000166540
|
kcnip3b
|
Kv channel interacting protein 3b, calsenilin |
| chr8_+_28358161 | 0.17 |
ENSDART00000062682
|
adipor1b
|
adiponectin receptor 1b |
| chr8_-_29930821 | 0.17 |
ENSDART00000125173
|
ercc6l2
|
excision repair cross-complementation group 6-like 2 |
| chr13_-_17943135 | 0.17 |
ENSDART00000176027
|
march8
|
membrane-associated ring finger (C3HC4) 8 |
| chr15_+_33843596 | 0.17 |
ENSDART00000169037
|
inpp5ka
|
inositol polyphosphate-5-phosphatase Ka |
| chr16_-_36979592 | 0.17 |
ENSDART00000168443
|
snrnp48
|
small nuclear ribonucleoprotein 48 (U11/U12) |
| chr8_-_2100808 | 0.16 |
ENSDART00000123804
|
cltcl1
|
clathrin, heavy chain-like 1 |
| chr18_-_16885362 | 0.16 |
ENSDART00000132778
|
swap70b
|
switching B cell complex subunit SWAP70b |
| chr4_-_75158035 | 0.16 |
ENSDART00000174353
|
CABZ01066312.1
|
|
| chr4_-_2440175 | 0.16 |
ENSDART00000058059
|
OSBPL8
|
oxysterol binding protein like 8 |
| chr3_+_60828813 | 0.16 |
ENSDART00000128260
|
CABZ01087514.1
|
|
| chr5_+_5398966 | 0.16 |
ENSDART00000139553
|
mapkap1
|
mitogen-activated protein kinase associated protein 1 |
| chr11_-_26832685 | 0.16 |
ENSDART00000153519
|
iqsec1b
|
IQ motif and Sec7 domain 1b |
| chr21_-_32487061 | 0.16 |
ENSDART00000114359
ENSDART00000131591 ENSDART00000131477 |
si:dkeyp-72g9.4
|
si:dkeyp-72g9.4 |
| chr5_+_50371951 | 0.16 |
ENSDART00000184488
ENSDART00000171295 |
slf1
|
SMC5-SMC6 complex localization factor 1 |
| chr4_-_25911609 | 0.16 |
ENSDART00000186958
ENSDART00000175487 |
fgd6
|
FYVE, RhoGEF and PH domain containing 6 |
| chr11_+_2687395 | 0.15 |
ENSDART00000082510
|
b3galt6
|
UDP-Gal:betaGal beta 1,3-galactosyltransferase polypeptide 6 |
| chr18_-_22414565 | 0.15 |
ENSDART00000079158
|
tmem170a
|
transmembrane protein 170A |
| chr19_-_46018152 | 0.15 |
ENSDART00000159206
|
krit1
|
KRIT1, ankyrin repeat containing |
| chr12_+_14556092 | 0.15 |
ENSDART00000115237
|
becn1
|
beclin 1, autophagy related |
| chr5_+_19320554 | 0.15 |
ENSDART00000165119
|
rusc2
|
RUN and SH3 domain containing 2 |
| chr21_-_3606539 | 0.15 |
ENSDART00000062418
|
dym
|
dymeclin |
| chr24_-_3426620 | 0.15 |
ENSDART00000184346
|
nck1b
|
NCK adaptor protein 1b |
| chr25_+_34407740 | 0.15 |
ENSDART00000012677
|
OTUD7A
|
si:dkey-37f18.2 |
| chr8_+_9715010 | 0.15 |
ENSDART00000139414
|
gripap1
|
GRIP1 associated protein 1 |
| chr12_-_18414536 | 0.15 |
ENSDART00000132956
ENSDART00000078724 |
atpaf2
|
ATP synthase mitochondrial F1 complex assembly factor 2 |
| chr19_+_15521997 | 0.15 |
ENSDART00000003164
|
ppp1r8a
|
protein phosphatase 1, regulatory subunit 8a |
| chr4_-_72296520 | 0.15 |
ENSDART00000182638
|
si:cabz01071911.3
|
si:cabz01071911.3 |
| chr9_+_33009284 | 0.15 |
ENSDART00000036926
|
vangl1
|
VANGL planar cell polarity protein 1 |
| chr23_-_20051369 | 0.14 |
ENSDART00000049836
|
bgnb
|
biglycan b |
| chr25_+_10762016 | 0.14 |
ENSDART00000131231
|
ap3s2
|
adaptor-related protein complex 3, sigma 2 subunit |
| chr3_+_24511959 | 0.14 |
ENSDART00000133898
|
dnal4a
|
dynein, axonemal, light chain 4a |
| chr11_+_24819324 | 0.14 |
ENSDART00000184549
|
kdm5ba
|
lysine (K)-specific demethylase 5Ba |
| chr13_-_5978433 | 0.14 |
ENSDART00000102555
|
actr2b
|
ARP2 actin related protein 2b homolog |
| chr12_-_13650344 | 0.14 |
ENSDART00000124364
ENSDART00000124638 ENSDART00000171929 |
stat5b
|
signal transducer and activator of transcription 5b |
| chr12_-_34258384 | 0.13 |
ENSDART00000109196
|
pgs1
|
phosphatidylglycerophosphate synthase 1 |
| chr17_-_7060477 | 0.13 |
ENSDART00000081797
|
sash1b
|
SAM and SH3 domain containing 1b |
| chr18_-_17000842 | 0.13 |
ENSDART00000079832
|
tph2
|
tryptophan hydroxylase 2 (tryptophan 5-monooxygenase) |
| chr4_-_55178568 | 0.13 |
ENSDART00000165288
|
znf1039
|
zinc finger protein 1039 |
| chr21_+_45758498 | 0.13 |
ENSDART00000162548
|
h2afy
|
H2A histone family, member Y |
| chr12_-_19346678 | 0.13 |
ENSDART00000044860
|
maff
|
v-maf avian musculoaponeurotic fibrosarcoma oncogene homolog F |
| chr15_-_1733235 | 0.13 |
ENSDART00000023153
ENSDART00000154668 |
rabgef1l
|
RAB guanine nucleotide exchange factor (GEF) 1, like |
| chr17_-_18797245 | 0.13 |
ENSDART00000045991
|
vrk1
|
vaccinia related kinase 1 |
| chr21_+_34976600 | 0.13 |
ENSDART00000191672
ENSDART00000139635 |
rbm11
|
RNA binding motif protein 11 |
| chr4_+_23127284 | 0.13 |
ENSDART00000122675
|
mdm2
|
MDM2 oncogene, E3 ubiquitin protein ligase |
Network of associatons between targets according to the STRING database.
First level regulatory network of arid3a+arid3b+arid3c
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.4 | 1.9 | GO:0008591 | regulation of Wnt signaling pathway, calcium modulating pathway(GO:0008591) negative regulation of non-canonical Wnt signaling pathway(GO:2000051) |
| 0.3 | 2.4 | GO:0045299 | otolith mineralization(GO:0045299) |
| 0.2 | 1.1 | GO:0046952 | ketone body catabolic process(GO:0046952) |
| 0.2 | 0.5 | GO:0042040 | establishment of synaptic specificity at neuromuscular junction(GO:0007529) molybdenum incorporation into molybdenum-molybdopterin complex(GO:0018315) metal incorporation into metallo-molybdopterin complex(GO:0042040) glycine receptor clustering(GO:0072579) |
| 0.1 | 1.8 | GO:0001574 | ganglioside biosynthetic process(GO:0001574) |
| 0.1 | 1.1 | GO:0050975 | sensory perception of touch(GO:0050975) |
| 0.1 | 0.4 | GO:0021730 | trigeminal sensory nucleus development(GO:0021730) |
| 0.1 | 0.3 | GO:2000378 | negative regulation of reactive oxygen species metabolic process(GO:2000378) |
| 0.1 | 0.7 | GO:0060612 | adipose tissue development(GO:0060612) |
| 0.1 | 0.4 | GO:0098529 | neuromuscular junction development, skeletal muscle fiber(GO:0098529) |
| 0.1 | 0.2 | GO:0043416 | regulation of skeletal muscle tissue regeneration(GO:0043416) |
| 0.1 | 0.3 | GO:0010874 | regulation of cholesterol efflux(GO:0010874) |
| 0.1 | 0.2 | GO:0015808 | L-alanine transport(GO:0015808) proline transport(GO:0015824) |
| 0.1 | 0.3 | GO:0036149 | phosphatidylinositol acyl-chain remodeling(GO:0036149) |
| 0.1 | 0.4 | GO:0006501 | C-terminal protein lipidation(GO:0006501) |
| 0.1 | 0.3 | GO:0010752 | regulation of cGMP-mediated signaling(GO:0010752) |
| 0.1 | 0.4 | GO:0051013 | microtubule severing(GO:0051013) |
| 0.1 | 0.2 | GO:0030857 | negative regulation of epithelial cell differentiation(GO:0030857) regulation of endothelial cell differentiation(GO:0045601) |
| 0.0 | 0.2 | GO:0032615 | interleukin-12 production(GO:0032615) regulation of interleukin-12 production(GO:0032655) |
| 0.0 | 0.1 | GO:0032049 | phosphatidylglycerol biosynthetic process(GO:0006655) cardiolipin biosynthetic process(GO:0032049) |
| 0.0 | 0.3 | GO:0097477 | spinal cord motor neuron migration(GO:0097476) lateral motor column neuron migration(GO:0097477) |
| 0.0 | 0.2 | GO:0002495 | antigen processing and presentation of peptide antigen via MHC class II(GO:0002495) |
| 0.0 | 0.2 | GO:1902019 | regulation of cilium movement involved in cell motility(GO:0060295) regulation of cilium beat frequency involved in ciliary motility(GO:0060296) regulation of cilium-dependent cell motility(GO:1902019) |
| 0.0 | 0.3 | GO:1901339 | regulation of store-operated calcium channel activity(GO:1901339) |
| 0.0 | 0.1 | GO:0034214 | protein hexamerization(GO:0034214) mitotic spindle disassembly(GO:0051228) spindle disassembly(GO:0051230) |
| 0.0 | 0.2 | GO:0099563 | modification of synaptic structure(GO:0099563) |
| 0.0 | 0.2 | GO:0070272 | proton-transporting ATP synthase complex assembly(GO:0043461) proton-transporting ATP synthase complex biogenesis(GO:0070272) |
| 0.0 | 2.3 | GO:0018107 | peptidyl-threonine phosphorylation(GO:0018107) |
| 0.0 | 0.2 | GO:0015744 | thiosulfate transport(GO:0015709) oxaloacetate transport(GO:0015729) malate transport(GO:0015743) succinate transport(GO:0015744) succinate transmembrane transport(GO:0071422) malate transmembrane transport(GO:0071423) |
| 0.0 | 0.2 | GO:0097355 | protein localization to heterochromatin(GO:0097355) |
| 0.0 | 0.1 | GO:0021611 | facial nerve formation(GO:0021611) |
| 0.0 | 0.2 | GO:0006703 | estrogen biosynthetic process(GO:0006703) |
| 0.0 | 0.1 | GO:0048308 | organelle inheritance(GO:0048308) Golgi inheritance(GO:0048313) |
| 0.0 | 0.3 | GO:0006798 | polyphosphate metabolic process(GO:0006797) polyphosphate catabolic process(GO:0006798) |
| 0.0 | 1.2 | GO:1901385 | regulation of voltage-gated calcium channel activity(GO:1901385) |
| 0.0 | 0.2 | GO:0051561 | positive regulation of mitochondrial calcium ion concentration(GO:0051561) |
| 0.0 | 0.6 | GO:0043153 | entrainment of circadian clock by photoperiod(GO:0043153) |
| 0.0 | 0.3 | GO:0050732 | negative regulation of peptidyl-tyrosine phosphorylation(GO:0050732) negative regulation of protein tyrosine kinase activity(GO:0061099) |
| 0.0 | 0.1 | GO:1903358 | regulation of Golgi organization(GO:1903358) |
| 0.0 | 0.2 | GO:0048714 | positive regulation of glial cell differentiation(GO:0045687) positive regulation of oligodendrocyte differentiation(GO:0048714) |
| 0.0 | 0.2 | GO:0090133 | establishment or maintenance of cytoskeleton polarity(GO:0030952) mesendoderm migration(GO:0090133) cell migration involved in mesendoderm migration(GO:0090134) |
| 0.0 | 0.5 | GO:0032467 | positive regulation of cytokinesis(GO:0032467) |
| 0.0 | 0.5 | GO:0098703 | calcium ion import across plasma membrane(GO:0098703) calcium ion import into cell(GO:1990035) |
| 0.0 | 0.1 | GO:1904036 | negative regulation of epithelial cell apoptotic process(GO:1904036) |
| 0.0 | 1.1 | GO:0006904 | vesicle docking involved in exocytosis(GO:0006904) |
| 0.0 | 0.1 | GO:0044108 | calcitriol biosynthetic process from calciol(GO:0036378) vitamin D biosynthetic process(GO:0042368) cellular alcohol metabolic process(GO:0044107) cellular alcohol biosynthetic process(GO:0044108) |
| 0.0 | 0.7 | GO:0072348 | sulfur compound transport(GO:0072348) |
| 0.0 | 0.1 | GO:2000582 | positive regulation of microtubule motor activity(GO:2000576) regulation of ATP-dependent microtubule motor activity, plus-end-directed(GO:2000580) positive regulation of ATP-dependent microtubule motor activity, plus-end-directed(GO:2000582) |
| 0.0 | 0.1 | GO:0060049 | regulation of protein glycosylation(GO:0060049) |
| 0.0 | 0.1 | GO:0060397 | JAK-STAT cascade involved in growth hormone signaling pathway(GO:0060397) |
| 0.0 | 0.4 | GO:0006208 | pyrimidine nucleobase catabolic process(GO:0006208) |
| 0.0 | 0.1 | GO:1901162 | serotonin biosynthetic process(GO:0042427) primary amino compound biosynthetic process(GO:1901162) |
| 0.0 | 0.1 | GO:0061469 | regulation of type B pancreatic cell proliferation(GO:0061469) |
| 0.0 | 0.5 | GO:0021654 | rhombomere boundary formation(GO:0021654) |
| 0.0 | 0.3 | GO:0070816 | phosphorylation of RNA polymerase II C-terminal domain(GO:0070816) |
| 0.0 | 0.1 | GO:0034421 | post-translational protein acetylation(GO:0034421) |
| 0.0 | 0.3 | GO:0016486 | peptide hormone processing(GO:0016486) |
| 0.0 | 0.1 | GO:0097428 | protein maturation by iron-sulfur cluster transfer(GO:0097428) |
| 0.0 | 0.1 | GO:0031087 | nuclear-transcribed mRNA catabolic process, deadenylation-independent decay(GO:0031086) deadenylation-independent decapping of nuclear-transcribed mRNA(GO:0031087) |
| 0.0 | 0.4 | GO:0060088 | auditory receptor cell stereocilium organization(GO:0060088) |
| 0.0 | 0.1 | GO:1905168 | positive regulation of double-strand break repair via homologous recombination(GO:1905168) |
| 0.0 | 0.1 | GO:0000448 | cleavage in ITS2 between 5.8S rRNA and LSU-rRNA of tricistronic rRNA transcript (SSU-rRNA, 5.8S rRNA, LSU-rRNA)(GO:0000448) |
| 0.0 | 0.4 | GO:0050650 | chondroitin sulfate proteoglycan biosynthetic process(GO:0050650) |
| 0.0 | 1.2 | GO:0006414 | translational elongation(GO:0006414) |
| 0.0 | 0.1 | GO:0045604 | regulation of epidermal cell differentiation(GO:0045604) |
| 0.0 | 0.3 | GO:0032483 | regulation of Rab protein signal transduction(GO:0032483) |
| 0.0 | 0.1 | GO:0010847 | regulation of chromatin assembly(GO:0010847) |
| 0.0 | 0.2 | GO:2000781 | positive regulation of double-strand break repair(GO:2000781) |
| 0.0 | 0.2 | GO:0033211 | adiponectin-activated signaling pathway(GO:0033211) |
| 0.0 | 0.6 | GO:0035118 | embryonic pectoral fin morphogenesis(GO:0035118) |
| 0.0 | 0.3 | GO:0098962 | regulation of postsynaptic neurotransmitter receptor activity(GO:0098962) |
| 0.0 | 0.2 | GO:0007158 | neuron cell-cell adhesion(GO:0007158) |
| 0.0 | 0.2 | GO:0071501 | response to sterol depletion(GO:0006991) SREBP signaling pathway(GO:0032933) cellular response to sterol depletion(GO:0071501) |
| 0.0 | 0.1 | GO:0060282 | positive regulation of oocyte development(GO:0060282) |
| 0.0 | 1.2 | GO:0009142 | nucleoside triphosphate biosynthetic process(GO:0009142) |
| 0.0 | 0.1 | GO:0045721 | negative regulation of gluconeogenesis(GO:0045721) |
| 0.0 | 0.2 | GO:0071786 | endoplasmic reticulum tubular network organization(GO:0071786) |
| 0.0 | 0.1 | GO:0003261 | cardiac muscle progenitor cell migration to the midline involved in heart field formation(GO:0003261) |
| 0.0 | 0.1 | GO:1900145 | nodal signaling pathway involved in determination of left/right asymmetry(GO:0038107) regulation of nodal signaling pathway involved in determination of left/right asymmetry(GO:1900145) |
| 0.0 | 0.2 | GO:0055117 | regulation of cardiac muscle contraction(GO:0055117) |
| 0.0 | 0.3 | GO:0043249 | erythrocyte maturation(GO:0043249) |
| 0.0 | 0.3 | GO:0010507 | negative regulation of autophagy(GO:0010507) |
| 0.0 | 0.2 | GO:0072176 | pronephric duct development(GO:0039022) nephric duct development(GO:0072176) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.7 | GO:0005790 | smooth endoplasmic reticulum(GO:0005790) |
| 0.1 | 0.3 | GO:0044233 | ER-mitochondrion membrane contact site(GO:0044233) |
| 0.0 | 0.2 | GO:0008091 | spectrin(GO:0008091) |
| 0.0 | 0.3 | GO:0000439 | core TFIIH complex(GO:0000439) |
| 0.0 | 0.3 | GO:0032045 | guanyl-nucleotide exchange factor complex(GO:0032045) |
| 0.0 | 0.3 | GO:0005614 | interstitial matrix(GO:0005614) |
| 0.0 | 0.2 | GO:0071439 | clathrin complex(GO:0071439) |
| 0.0 | 0.2 | GO:0034272 | phosphatidylinositol 3-kinase complex, class III, type I(GO:0034271) phosphatidylinositol 3-kinase complex, class III, type II(GO:0034272) |
| 0.0 | 0.1 | GO:0090443 | FAR/SIN/STRIPAK complex(GO:0090443) |
| 0.0 | 0.2 | GO:1990726 | Lsm1-7-Pat1 complex(GO:1990726) |
| 0.0 | 0.2 | GO:0098837 | postsynaptic recycling endosome(GO:0098837) |
| 0.0 | 0.1 | GO:0042584 | chromaffin granule membrane(GO:0042584) |
| 0.0 | 0.1 | GO:0031501 | mannosyltransferase complex(GO:0031501) |
| 0.0 | 0.4 | GO:0000407 | pre-autophagosomal structure(GO:0000407) |
| 0.0 | 0.3 | GO:0030992 | intraciliary transport particle B(GO:0030992) |
| 0.0 | 0.2 | GO:1990246 | uniplex complex(GO:1990246) |
| 0.0 | 0.4 | GO:0031594 | neuromuscular junction(GO:0031594) |
| 0.0 | 0.1 | GO:0000126 | transcription factor TFIIIB complex(GO:0000126) |
| 0.0 | 0.3 | GO:0032420 | stereocilium(GO:0032420) |
| 0.0 | 0.1 | GO:0005851 | eukaryotic translation initiation factor 2B complex(GO:0005851) |
| 0.0 | 0.1 | GO:0035145 | exon-exon junction complex(GO:0035145) |
| 0.0 | 0.5 | GO:0030496 | midbody(GO:0030496) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 1.7 | GO:0043295 | glutathione binding(GO:0043295) |
| 0.2 | 1.8 | GO:0001665 | alpha-N-acetylgalactosaminide alpha-2,6-sialyltransferase activity(GO:0001665) |
| 0.2 | 1.1 | GO:0008260 | 3-oxoacid CoA-transferase activity(GO:0008260) |
| 0.2 | 0.5 | GO:0061599 | molybdopterin adenylyltransferase activity(GO:0061598) molybdopterin molybdotransferase activity(GO:0061599) |
| 0.1 | 0.7 | GO:0008281 | sulfonylurea receptor activity(GO:0008281) |
| 0.1 | 0.5 | GO:0015018 | galactosylgalactosylxylosylprotein 3-beta-glucuronosyltransferase activity(GO:0015018) |
| 0.1 | 0.9 | GO:0072320 | volume-sensitive chloride channel activity(GO:0072320) |
| 0.1 | 0.4 | GO:0019778 | Atg12 activating enzyme activity(GO:0019778) Atg8 activating enzyme activity(GO:0019779) |
| 0.1 | 0.5 | GO:0004865 | protein serine/threonine phosphatase inhibitor activity(GO:0004865) |
| 0.1 | 0.6 | GO:0016805 | dipeptidase activity(GO:0016805) |
| 0.1 | 0.2 | GO:0047035 | testosterone dehydrogenase (NAD+) activity(GO:0047035) |
| 0.1 | 0.7 | GO:0004687 | myosin light chain kinase activity(GO:0004687) |
| 0.0 | 1.3 | GO:0004550 | nucleoside diphosphate kinase activity(GO:0004550) |
| 0.0 | 0.2 | GO:0015193 | L-alanine transmembrane transporter activity(GO:0015180) L-proline transmembrane transporter activity(GO:0015193) alanine transmembrane transporter activity(GO:0022858) |
| 0.0 | 0.5 | GO:0005432 | calcium:sodium antiporter activity(GO:0005432) |
| 0.0 | 1.7 | GO:0003746 | translation elongation factor activity(GO:0003746) |
| 0.0 | 0.2 | GO:0015140 | thiosulfate transmembrane transporter activity(GO:0015117) oxaloacetate transmembrane transporter activity(GO:0015131) malate transmembrane transporter activity(GO:0015140) |
| 0.0 | 0.4 | GO:0004157 | dihydropyrimidinase activity(GO:0004157) |
| 0.0 | 0.3 | GO:0004309 | exopolyphosphatase activity(GO:0004309) |
| 0.0 | 0.9 | GO:0008331 | high voltage-gated calcium channel activity(GO:0008331) |
| 0.0 | 0.1 | GO:0048487 | beta-tubulin binding(GO:0048487) |
| 0.0 | 0.2 | GO:0016312 | inositol bisphosphate phosphatase activity(GO:0016312) |
| 0.0 | 0.1 | GO:0004510 | tryptophan 5-monooxygenase activity(GO:0004510) |
| 0.0 | 0.1 | GO:0004689 | phosphorylase kinase activity(GO:0004689) |
| 0.0 | 0.1 | GO:0030343 | vitamin D3 25-hydroxylase activity(GO:0030343) cholesterol 26-hydroxylase activity(GO:0031073) |
| 0.0 | 0.7 | GO:1901682 | sulfur compound transmembrane transporter activity(GO:1901682) |
| 0.0 | 0.2 | GO:0034647 | histone demethylase activity (H3-trimethyl-K4 specific)(GO:0034647) |
| 0.0 | 0.3 | GO:0001591 | dopamine neurotransmitter receptor activity, coupled via Gi/Go(GO:0001591) |
| 0.0 | 0.8 | GO:0004683 | calmodulin-dependent protein kinase activity(GO:0004683) |
| 0.0 | 0.4 | GO:0042056 | chemoattractant activity(GO:0042056) |
| 0.0 | 0.1 | GO:0005017 | platelet-derived growth factor-activated receptor activity(GO:0005017) platelet-derived growth factor binding(GO:0048407) |
| 0.0 | 0.2 | GO:0004169 | dolichyl-phosphate-mannose-protein mannosyltransferase activity(GO:0004169) |
| 0.0 | 0.3 | GO:0031682 | G-protein gamma-subunit binding(GO:0031682) |
| 0.0 | 0.1 | GO:0017169 | CDP-alcohol phosphatidyltransferase activity(GO:0017169) |
| 0.0 | 0.6 | GO:0017075 | syntaxin-1 binding(GO:0017075) |
| 0.0 | 0.4 | GO:0005154 | epidermal growth factor receptor binding(GO:0005154) |
| 0.0 | 0.1 | GO:0000995 | transcription factor activity, core RNA polymerase III binding(GO:0000995) |
| 0.0 | 0.0 | GO:1990174 | phosphodiesterase decapping endonuclease activity(GO:1990174) |
| 0.0 | 0.3 | GO:0071617 | lysophospholipid acyltransferase activity(GO:0071617) |
| 0.0 | 0.3 | GO:0019210 | protein kinase inhibitor activity(GO:0004860) kinase inhibitor activity(GO:0019210) |
| 0.0 | 0.1 | GO:0001609 | G-protein coupled adenosine receptor activity(GO:0001609) |
| 0.0 | 0.3 | GO:0016854 | racemase and epimerase activity(GO:0016854) |
| 0.0 | 0.2 | GO:0032051 | clathrin light chain binding(GO:0032051) |
| 0.0 | 0.1 | GO:0051731 | polynucleotide 5'-hydroxyl-kinase activity(GO:0051731) |
| 0.0 | 0.2 | GO:0000828 | inositol hexakisphosphate kinase activity(GO:0000828) |
| 0.0 | 0.6 | GO:0071949 | FAD binding(GO:0071949) |
| 0.0 | 0.1 | GO:0001042 | RNA polymerase I core binding(GO:0001042) |
| 0.0 | 0.1 | GO:0004376 | glycolipid mannosyltransferase activity(GO:0004376) |
| 0.0 | 0.6 | GO:0005200 | structural constituent of cytoskeleton(GO:0005200) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.4 | PID INTEGRIN A4B1 PATHWAY | Alpha4 beta1 integrin signaling events |
| 0.0 | 0.5 | PID ALPHA SYNUCLEIN PATHWAY | Alpha-synuclein signaling |
| 0.0 | 0.1 | PID AR NONGENOMIC PATHWAY | Nongenotropic Androgen signaling |
| 0.0 | 0.3 | PID IL8 CXCR2 PATHWAY | IL8- and CXCR2-mediated signaling events |
| 0.0 | 0.1 | PID IL5 PATHWAY | IL5-mediated signaling events |
| 0.0 | 0.1 | PID S1P S1P4 PATHWAY | S1P4 pathway |
| 0.0 | 0.2 | PID CXCR3 PATHWAY | CXCR3-mediated signaling events |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 1.8 | REACTOME TERMINATION OF O GLYCAN BIOSYNTHESIS | Genes involved in Termination of O-glycan biosynthesis |
| 0.1 | 0.3 | REACTOME ACYL CHAIN REMODELLING OF PI | Genes involved in Acyl chain remodelling of PI |
| 0.1 | 1.3 | REACTOME SYNTHESIS AND INTERCONVERSION OF NUCLEOTIDE DI AND TRIPHOSPHATES | Genes involved in Synthesis and interconversion of nucleotide di- and triphosphates |
| 0.0 | 0.6 | REACTOME SIGNAL ATTENUATION | Genes involved in Signal attenuation |
| 0.0 | 0.4 | REACTOME SIGNALING BY NODAL | Genes involved in Signaling by NODAL |
| 0.0 | 0.7 | REACTOME A TETRASACCHARIDE LINKER SEQUENCE IS REQUIRED FOR GAG SYNTHESIS | Genes involved in A tetrasaccharide linker sequence is required for GAG synthesis |
| 0.0 | 1.1 | REACTOME SIGNALING BY ROBO RECEPTOR | Genes involved in Signaling by Robo receptor |
| 0.0 | 0.2 | REACTOME GAP JUNCTION DEGRADATION | Genes involved in Gap junction degradation |
| 0.0 | 0.5 | REACTOME ABC FAMILY PROTEINS MEDIATED TRANSPORT | Genes involved in ABC-family proteins mediated transport |
| 0.0 | 0.2 | REACTOME INTERACTION BETWEEN L1 AND ANKYRINS | Genes involved in Interaction between L1 and Ankyrins |
| 0.0 | 0.1 | REACTOME PROLACTIN RECEPTOR SIGNALING | Genes involved in Prolactin receptor signaling |
| 0.0 | 0.1 | REACTOME AMINE DERIVED HORMONES | Genes involved in Amine-derived hormones |
| 0.0 | 0.4 | REACTOME FORMATION OF THE HIV1 EARLY ELONGATION COMPLEX | Genes involved in Formation of the HIV-1 Early Elongation Complex |
| 0.0 | 0.2 | REACTOME CD28 DEPENDENT PI3K AKT SIGNALING | Genes involved in CD28 dependent PI3K/Akt signaling |
| 0.0 | 0.3 | REACTOME THE ROLE OF NEF IN HIV1 REPLICATION AND DISEASE PATHOGENESIS | Genes involved in The role of Nef in HIV-1 replication and disease pathogenesis |