Project
PRJNA207719: Tissue specific transcriptome profiling
Navigation
Downloads
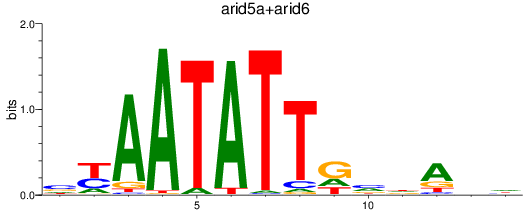
Results for arid5a+arid6
Z-value: 0.70
Transcription factors associated with arid5a+arid6
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
arid6
|
ENSDARG00000069988 | AT-rich interaction domain 6 |
|
arid5a
|
ENSDARG00000077120 | AT rich interactive domain 5A (MRF1-like) |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| arid6 | dr11_v1_chr21_+_11248448_11248448 | 0.97 | 5.1e-03 | Click! |
| arid5a | dr11_v1_chr8_+_52377516_52377516 | 0.71 | 1.8e-01 | Click! |
Activity profile of arid5a+arid6 motif
Sorted Z-values of arid5a+arid6 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr4_-_8043839 | 0.60 |
ENSDART00000190047
ENSDART00000057567 |
si:ch211-240l19.5
|
si:ch211-240l19.5 |
| chr9_+_33207574 | 0.50 |
ENSDART00000055897
ENSDART00000166030 |
si:ch211-125e6.11
|
si:ch211-125e6.11 |
| chr3_+_57820913 | 0.44 |
ENSDART00000168101
|
CU571328.1
|
|
| chr15_-_452347 | 0.43 |
ENSDART00000115233
|
VSTM5
|
V-set and transmembrane domain containing 5 |
| chr12_+_46462090 | 0.41 |
ENSDART00000130748
|
BX005305.2
|
|
| chr12_+_46404307 | 0.41 |
ENSDART00000185011
|
BX005305.4
|
|
| chr12_+_46512881 | 0.38 |
ENSDART00000105454
|
CU855711.1
|
|
| chr24_-_26328721 | 0.36 |
ENSDART00000125468
|
apodb
|
apolipoprotein Db |
| chr16_-_46660680 | 0.34 |
ENSDART00000159209
ENSDART00000191929 |
tmem176l.4
|
transmembrane protein 176l.4 |
| chr12_+_46483618 | 0.34 |
ENSDART00000186970
|
BX005305.5
|
|
| chr22_+_7439186 | 0.33 |
ENSDART00000190667
|
zgc:92041
|
zgc:92041 |
| chr12_+_46425800 | 0.33 |
ENSDART00000191965
|
BX005305.1
|
|
| chr12_+_46443477 | 0.33 |
ENSDART00000191873
|
BX005305.1
|
|
| chr12_+_46386983 | 0.33 |
ENSDART00000183982
|
BX005305.3
|
Danio rerio legumain (LOC100005356), mRNA. |
| chr2_-_32262287 | 0.32 |
ENSDART00000056621
ENSDART00000039717 |
fam49ba
|
family with sequence similarity 49, member Ba |
| chr21_-_5077715 | 0.31 |
ENSDART00000081954
|
haus1
|
HAUS augmin-like complex, subunit 1 |
| chr7_-_12968689 | 0.29 |
ENSDART00000173115
ENSDART00000013690 |
rplp2l
|
ribosomal protein, large P2, like |
| chr19_-_48336535 | 0.29 |
ENSDART00000162752
|
si:ch73-359m17.6
|
si:ch73-359m17.6 |
| chr14_+_11458044 | 0.28 |
ENSDART00000186425
|
si:ch211-153b23.5
|
si:ch211-153b23.5 |
| chr8_+_53344726 | 0.28 |
ENSDART00000184395
ENSDART00000170212 |
CU914536.1
|
|
| chr16_+_28754403 | 0.27 |
ENSDART00000103340
|
s100v1
|
S100 calcium binding protein V1 |
| chr14_+_11457500 | 0.27 |
ENSDART00000169202
|
si:ch211-153b23.5
|
si:ch211-153b23.5 |
| chr21_-_42876565 | 0.26 |
ENSDART00000126480
|
zmp:0000001268
|
zmp:0000001268 |
| chr11_+_44135351 | 0.25 |
ENSDART00000182914
|
FO704721.1
|
|
| chr22_+_1170294 | 0.25 |
ENSDART00000159761
ENSDART00000169809 |
irf6
|
interferon regulatory factor 6 |
| chr21_+_11560153 | 0.24 |
ENSDART00000065842
|
cd8a
|
CD8a molecule |
| chr23_+_5631381 | 0.24 |
ENSDART00000149143
|
pkp1a
|
plakophilin 1a |
| chr12_-_30558694 | 0.24 |
ENSDART00000153417
|
si:ch211-28p3.3
|
si:ch211-28p3.3 |
| chr7_+_6879534 | 0.24 |
ENSDART00000157731
|
zgc:175248
|
zgc:175248 |
| chr2_+_49580450 | 0.23 |
ENSDART00000056252
|
sema4e
|
semaphorin 4e |
| chr17_+_6585333 | 0.23 |
ENSDART00000124293
|
adgrf3a
|
adhesion G protein-coupled receptor F3a |
| chr3_-_50139860 | 0.23 |
ENSDART00000101563
|
btr02
|
bloodthirsty-related gene family, member 2 |
| chr19_-_5769553 | 0.22 |
ENSDART00000175003
|
si:ch211-264f5.6
|
si:ch211-264f5.6 |
| chr3_-_55104310 | 0.22 |
ENSDART00000101713
|
hbba1
|
hemoglobin, beta adult 1 |
| chr17_-_10838434 | 0.22 |
ENSDART00000064597
|
lgals3b
|
lectin, galactoside binding soluble 3b |
| chr4_-_13518381 | 0.22 |
ENSDART00000067153
|
ifng1-1
|
interferon, gamma 1-1 |
| chr4_+_42450386 | 0.22 |
ENSDART00000168211
|
si:dkey-11d20.1
|
si:dkey-11d20.1 |
| chr21_-_7940043 | 0.22 |
ENSDART00000099733
ENSDART00000136671 |
f2rl1.1
|
coagulation factor II (thrombin) receptor-like 1, tandem duplicate 1 |
| chr21_-_32436679 | 0.21 |
ENSDART00000076974
|
gfpt2
|
glutamine-fructose-6-phosphate transaminase 2 |
| chr12_+_22588923 | 0.21 |
ENSDART00000184862
|
capgb
|
capping protein (actin filament), gelsolin-like b |
| chr3_-_33970190 | 0.21 |
ENSDART00000151238
|
ighj2-5
|
ighj2-5 |
| chr2_+_40181633 | 0.21 |
ENSDART00000185494
|
FP236331.1
|
|
| chr21_-_22689805 | 0.21 |
ENSDART00000157560
ENSDART00000110792 |
gig2e
|
grass carp reovirus (GCRV)-induced gene 2e |
| chr24_+_39695688 | 0.21 |
ENSDART00000109747
|
zgc:153659
|
zgc:153659 |
| chr18_+_30567945 | 0.21 |
ENSDART00000078894
|
irf8
|
interferon regulatory factor 8 |
| chr19_+_43119014 | 0.20 |
ENSDART00000023156
|
eef1a1l1
|
eukaryotic translation elongation factor 1 alpha 1, like 1 |
| chr12_-_17863467 | 0.20 |
ENSDART00000042006
|
baiap2l1a
|
BAI1-associated protein 2-like 1a |
| chr13_+_46941930 | 0.20 |
ENSDART00000056962
|
fbxo5
|
F-box protein 5 |
| chr10_+_17088261 | 0.20 |
ENSDART00000132103
|
si:dkey-106l3.7
|
si:dkey-106l3.7 |
| chr13_+_22675802 | 0.20 |
ENSDART00000145538
ENSDART00000143312 |
zgc:193505
|
zgc:193505 |
| chr9_-_56272465 | 0.19 |
ENSDART00000039235
|
lcp1
|
lymphocyte cytosolic protein 1 (L-plastin) |
| chr20_-_39367895 | 0.19 |
ENSDART00000136476
ENSDART00000021788 ENSDART00000180784 |
pbk
|
PDZ binding kinase |
| chr17_-_15546862 | 0.19 |
ENSDART00000091021
|
col10a1a
|
collagen, type X, alpha 1a |
| chr7_+_8361083 | 0.18 |
ENSDART00000102091
|
jac7
|
jacalin 7 |
| chr8_-_14209852 | 0.18 |
ENSDART00000005359
|
def6a
|
differentially expressed in FDCP 6a homolog (mouse) |
| chr13_-_35892243 | 0.18 |
ENSDART00000002750
ENSDART00000122810 ENSDART00000162399 |
tacc3
|
transforming, acidic coiled-coil containing protein 3 |
| chr20_-_26822522 | 0.18 |
ENSDART00000146326
ENSDART00000046764 ENSDART00000103234 ENSDART00000143267 |
gmds
|
GDP-mannose 4,6-dehydratase |
| chr18_+_19456648 | 0.18 |
ENSDART00000079695
|
zwilch
|
zwilch kinetochore protein |
| chr1_+_26667872 | 0.18 |
ENSDART00000152803
ENSDART00000152144 ENSDART00000152785 ENSDART00000152393 |
hemgn
|
hemogen |
| chr16_-_42004544 | 0.17 |
ENSDART00000034544
|
caspa
|
caspase a |
| chr3_-_62403550 | 0.17 |
ENSDART00000055055
|
sox8b
|
SRY (sex determining region Y)-box 8b |
| chr19_-_17210760 | 0.17 |
ENSDART00000007906
|
stmn1a
|
stathmin 1a |
| chr23_+_642395 | 0.17 |
ENSDART00000186995
|
irf10
|
interferon regulatory factor 10 |
| chr2_+_36015049 | 0.17 |
ENSDART00000158276
|
lamc2
|
laminin, gamma 2 |
| chr12_-_611367 | 0.17 |
ENSDART00000152286
|
wu:fj29h11
|
wu:fj29h11 |
| chr4_+_15944245 | 0.17 |
ENSDART00000134594
|
si:dkey-117n7.3
|
si:dkey-117n7.3 |
| chr15_-_12360409 | 0.17 |
ENSDART00000164596
|
tmprss13a
|
transmembrane protease, serine 13a |
| chr24_-_31306724 | 0.17 |
ENSDART00000165399
|
acp5b
|
acid phosphatase 5b, tartrate resistant |
| chr6_-_11091449 | 0.17 |
ENSDART00000127209
|
pttg1ipa
|
PTTG1 interacting protein a |
| chr2_+_36109002 | 0.16 |
ENSDART00000158978
|
traj28
|
T-cell receptor alpha joining 28 |
| chr16_+_29492749 | 0.16 |
ENSDART00000179680
|
ctsk
|
cathepsin K |
| chr14_-_22015232 | 0.16 |
ENSDART00000137795
|
ssrp1a
|
structure specific recognition protein 1a |
| chr22_-_24880824 | 0.16 |
ENSDART00000061165
|
vtg2
|
vitellogenin 2 |
| chr25_-_35095129 | 0.16 |
ENSDART00000099866
ENSDART00000099868 |
kif15
|
kinesin family member 15 |
| chr22_+_25715925 | 0.16 |
ENSDART00000150650
|
si:dkeyp-98a7.7
|
si:dkeyp-98a7.7 |
| chr24_+_26328787 | 0.16 |
ENSDART00000003884
|
mynn
|
myoneurin |
| chr8_-_19977088 | 0.16 |
ENSDART00000139647
|
lpxn
|
leupaxin |
| chr24_-_32582378 | 0.16 |
ENSDART00000066590
|
rdh12l
|
retinol dehydrogenase 12, like |
| chr22_-_17489040 | 0.15 |
ENSDART00000141286
|
si:ch211-197g15.6
|
si:ch211-197g15.6 |
| chr13_-_35892051 | 0.15 |
ENSDART00000145884
|
tacc3
|
transforming, acidic coiled-coil containing protein 3 |
| chr1_+_23563691 | 0.15 |
ENSDART00000142879
|
ncapg
|
non-SMC condensin I complex, subunit G |
| chr20_-_29499363 | 0.15 |
ENSDART00000152889
ENSDART00000153252 ENSDART00000170972 ENSDART00000166420 ENSDART00000163079 |
rrm2
|
ribonucleotide reductase M2 polypeptide |
| chr7_+_30626378 | 0.15 |
ENSDART00000173533
ENSDART00000052541 |
ccnb2
|
cyclin B2 |
| chr5_-_14521500 | 0.15 |
ENSDART00000176565
|
si:ch211-244o22.2
|
si:ch211-244o22.2 |
| chr24_-_35561672 | 0.15 |
ENSDART00000058564
|
mcm4
|
minichromosome maintenance complex component 4 |
| chr22_+_19247255 | 0.15 |
ENSDART00000144053
|
si:dkey-21e2.10
|
si:dkey-21e2.10 |
| chr19_+_4076259 | 0.15 |
ENSDART00000160633
|
zgc:173581
|
zgc:173581 |
| chr11_-_438294 | 0.14 |
ENSDART00000040812
|
nuf2
|
NUF2, NDC80 kinetochore complex component, homolog |
| chr17_-_38291065 | 0.14 |
ENSDART00000152056
|
pax9
|
paired box 9 |
| chr8_-_46455874 | 0.14 |
ENSDART00000146985
|
sult1st7
|
sulfotransferase family 1, cytosolic sulfotransferase 7 |
| chr17_-_4395373 | 0.14 |
ENSDART00000015923
|
klhl10a
|
kelch-like family member 10a |
| chr3_-_29506960 | 0.14 |
ENSDART00000141720
|
cyth4a
|
cytohesin 4a |
| chr19_-_2318391 | 0.14 |
ENSDART00000012791
|
sp8a
|
sp8 transcription factor a |
| chr7_-_50367642 | 0.14 |
ENSDART00000134941
|
prc1b
|
protein regulator of cytokinesis 1b |
| chr13_+_17694845 | 0.14 |
ENSDART00000079778
|
ifit8
|
interferon-induced protein with tetratricopeptide repeats 8 |
| chr22_+_25693295 | 0.14 |
ENSDART00000123888
ENSDART00000150783 |
si:dkeyp-98a7.4
si:dkeyp-98a7.3
|
si:dkeyp-98a7.4 si:dkeyp-98a7.3 |
| chr9_-_5263947 | 0.13 |
ENSDART00000088342
|
cytip
|
cytohesin 1 interacting protein |
| chr25_-_29074064 | 0.13 |
ENSDART00000165603
|
arid3b
|
AT rich interactive domain 3B (BRIGHT-like) |
| chr14_-_1635296 | 0.13 |
ENSDART00000186658
|
LO018564.1
|
|
| chr18_+_44532370 | 0.13 |
ENSDART00000086952
|
st14a
|
suppression of tumorigenicity 14 (colon carcinoma) a |
| chr12_-_10567188 | 0.13 |
ENSDART00000144283
|
myof
|
myoferlin |
| chr22_+_19552987 | 0.13 |
ENSDART00000105315
|
hsd11b1la
|
hydroxysteroid (11-beta) dehydrogenase 1-like a |
| chr15_+_36457888 | 0.13 |
ENSDART00000155100
|
si:dkey-262k9.2
|
si:dkey-262k9.2 |
| chr18_+_29898955 | 0.13 |
ENSDART00000064080
|
cenpn
|
centromere protein N |
| chr2_+_19195841 | 0.13 |
ENSDART00000163137
ENSDART00000161095 |
elovl1a
|
ELOVL fatty acid elongase 1a |
| chr10_+_25204626 | 0.13 |
ENSDART00000024514
|
chordc1b
|
cysteine and histidine-rich domain (CHORD) containing 1b |
| chr12_+_2665081 | 0.13 |
ENSDART00000147532
|
rbp3
|
retinol binding protein 3 |
| chr15_-_25556074 | 0.12 |
ENSDART00000124677
|
mmp20a
|
matrix metallopeptidase 20a (enamelysin) |
| chr7_+_10610791 | 0.12 |
ENSDART00000166064
|
fah
|
fumarylacetoacetate hydrolase (fumarylacetoacetase) |
| chr4_-_12718367 | 0.12 |
ENSDART00000035259
|
mgst1.1
|
microsomal glutathione S-transferase 1.1 |
| chr8_+_22359881 | 0.12 |
ENSDART00000187867
|
zgc:153631
|
zgc:153631 |
| chr14_-_4120636 | 0.12 |
ENSDART00000059230
|
irf2
|
interferon regulatory factor 2 |
| chr4_+_42604252 | 0.12 |
ENSDART00000184850
|
CR925768.1
|
|
| chr9_-_11549379 | 0.12 |
ENSDART00000187074
|
fev
|
FEV (ETS oncogene family) |
| chr12_-_46145909 | 0.12 |
ENSDART00000139094
|
zgc:153932
|
zgc:153932 |
| chr7_-_8504355 | 0.12 |
ENSDART00000173067
|
loc564660
|
hypothetical protein LOC564660 |
| chr9_+_30478768 | 0.12 |
ENSDART00000101097
|
acp6
|
acid phosphatase 6, lysophosphatidic |
| chr3_-_36209936 | 0.12 |
ENSDART00000175208
|
csnk1da
|
casein kinase 1, delta a |
| chr1_+_53714734 | 0.12 |
ENSDART00000114689
|
pus10
|
pseudouridylate synthase 10 |
| chr16_-_30878521 | 0.12 |
ENSDART00000141403
|
dennd3b
|
DENN/MADD domain containing 3b |
| chr25_+_20694177 | 0.12 |
ENSDART00000073648
|
kxd1
|
KxDL motif containing 1 |
| chr4_+_30776883 | 0.12 |
ENSDART00000169519
|
si:dkey-11d20.1
|
si:dkey-11d20.1 |
| chr7_+_30178253 | 0.12 |
ENSDART00000002075
|
si:ch211-107o10.3
|
si:ch211-107o10.3 |
| chr9_-_14075868 | 0.11 |
ENSDART00000146449
|
fer1l6
|
fer-1-like family member 6 |
| chr15_-_21155641 | 0.11 |
ENSDART00000061098
ENSDART00000046443 |
A2ML1 (1 of many)
|
si:dkey-105h12.2 |
| chr19_+_31061718 | 0.11 |
ENSDART00000145971
|
sostdc1b
|
sclerostin domain containing 1b |
| chr25_+_3099073 | 0.11 |
ENSDART00000022506
|
rab3il1
|
RAB3A interacting protein (rabin3)-like 1 |
| chr22_-_24858042 | 0.11 |
ENSDART00000137998
ENSDART00000078216 ENSDART00000138378 |
vtg7
|
vitellogenin 7 |
| chr16_-_48400639 | 0.11 |
ENSDART00000159372
|
eif3ha
|
eukaryotic translation initiation factor 3, subunit H, a |
| chr22_+_6471584 | 0.11 |
ENSDART00000133974
|
si:ch211-105n15.1
|
si:ch211-105n15.1 |
| chr23_+_28381260 | 0.11 |
ENSDART00000162722
|
zgc:153867
|
zgc:153867 |
| chr13_-_23612324 | 0.11 |
ENSDART00000136406
ENSDART00000005004 |
prim2
|
DNA primase subunit 2 |
| chr11_-_44906703 | 0.11 |
ENSDART00000180591
|
zgc:171772
|
zgc:171772 |
| chr6_+_4160579 | 0.10 |
ENSDART00000105278
ENSDART00000187932 ENSDART00000111817 |
trim25l
|
tripartite motif containing 25, like |
| chr2_+_48074243 | 0.10 |
ENSDART00000056291
|
klf6b
|
Kruppel-like factor 6b |
| chr4_+_391297 | 0.10 |
ENSDART00000030215
|
rpl18a
|
ribosomal protein L18a |
| chr4_-_3811017 | 0.10 |
ENSDART00000056913
|
si:dkey-61f9.1
|
si:dkey-61f9.1 |
| chr14_+_164556 | 0.10 |
ENSDART00000185606
|
WDR1
|
WD repeat domain 1 |
| chr14_+_15620640 | 0.10 |
ENSDART00000188867
|
si:dkey-203a12.9
|
si:dkey-203a12.9 |
| chr9_-_11550711 | 0.10 |
ENSDART00000093343
|
fev
|
FEV (ETS oncogene family) |
| chr18_+_20468157 | 0.10 |
ENSDART00000100665
ENSDART00000147867 ENSDART00000060302 ENSDART00000180370 |
ddb2
|
damage-specific DNA binding protein 2 |
| chr9_+_13986427 | 0.10 |
ENSDART00000147200
|
cd28
|
CD28 molecule |
| chr14_-_1454045 | 0.10 |
ENSDART00000161460
|
pmt
|
phosphoethanolamine methyltransferase |
| chr18_+_44532883 | 0.10 |
ENSDART00000121994
|
st14a
|
suppression of tumorigenicity 14 (colon carcinoma) a |
| chr5_-_66429357 | 0.10 |
ENSDART00000160189
|
hip1rb
|
huntingtin interacting protein 1 related b |
| chr19_+_47405867 | 0.10 |
ENSDART00000041114
|
psmb2
|
proteasome subunit beta 2 |
| chr21_+_41468210 | 0.10 |
ENSDART00000192287
|
bod1
|
biorientation of chromosomes in cell division 1 |
| chr22_+_25687525 | 0.10 |
ENSDART00000135717
|
si:dkeyp-98a7.3
|
si:dkeyp-98a7.3 |
| chr4_+_74928675 | 0.10 |
ENSDART00000192755
|
nup50
|
nucleoporin 50 |
| chr19_+_14113886 | 0.10 |
ENSDART00000169343
|
kdf1b
|
keratinocyte differentiation factor 1b |
| chr11_+_1575435 | 0.10 |
ENSDART00000155713
ENSDART00000156562 |
si:dkey-40c23.1
|
si:dkey-40c23.1 |
| chr22_+_25681911 | 0.10 |
ENSDART00000113381
|
si:dkeyp-98a7.3
|
si:dkeyp-98a7.3 |
| chr25_+_7591293 | 0.10 |
ENSDART00000130416
|
FO704779.1
|
|
| chr7_+_38936132 | 0.10 |
ENSDART00000173945
|
creb3l1
|
cAMP responsive element binding protein 3-like 1 |
| chr2_+_36121373 | 0.10 |
ENSDART00000187002
|
CT867973.2
|
|
| chr19_+_30884706 | 0.09 |
ENSDART00000052126
|
yars
|
tyrosyl-tRNA synthetase |
| chr24_-_17039638 | 0.09 |
ENSDART00000048823
|
c8g
|
complement component 8, gamma polypeptide |
| chr7_-_30926030 | 0.09 |
ENSDART00000075421
|
sord
|
sorbitol dehydrogenase |
| chr18_+_30998472 | 0.09 |
ENSDART00000154993
ENSDART00000099333 |
cd151l
|
CD151 antigen, like |
| chr8_-_25002182 | 0.09 |
ENSDART00000078792
|
ahcyl1
|
adenosylhomocysteinase-like 1 |
| chr8_+_23861461 | 0.09 |
ENSDART00000037109
|
srpk1a
|
SRSF protein kinase 1a |
| chr23_-_36446307 | 0.09 |
ENSDART00000136623
|
zgc:174906
|
zgc:174906 |
| chr16_+_52847079 | 0.09 |
ENSDART00000163151
|
cep72
|
centrosomal protein 72 |
| chr14_-_4121052 | 0.09 |
ENSDART00000167074
|
irf2
|
interferon regulatory factor 2 |
| chr7_+_30178453 | 0.09 |
ENSDART00000174420
|
si:ch211-107o10.3
|
si:ch211-107o10.3 |
| chr23_-_27101600 | 0.09 |
ENSDART00000139231
|
stat6
|
signal transducer and activator of transcription 6, interleukin-4 induced |
| chr21_-_40173821 | 0.09 |
ENSDART00000180667
|
slco2b1
|
solute carrier organic anion transporter family, member 2B1 |
| chr20_+_18993452 | 0.09 |
ENSDART00000025509
|
tdh
|
L-threonine dehydrogenase |
| chr6_-_24053404 | 0.09 |
ENSDART00000168511
|
si:dkey-44g17.6
|
si:dkey-44g17.6 |
| chr19_+_816208 | 0.09 |
ENSDART00000093304
|
nrm
|
nurim |
| chr22_+_31039091 | 0.09 |
ENSDART00000060005
|
rpl32
|
ribosomal protein L32 |
| chr7_+_20917966 | 0.09 |
ENSDART00000129161
|
wrap53
|
WD repeat containing, antisense to TP53 |
| chr3_-_10677890 | 0.09 |
ENSDART00000155382
ENSDART00000171319 |
si:ch1073-144j5.2
|
si:ch1073-144j5.2 |
| chr5_+_12743640 | 0.09 |
ENSDART00000081411
|
pole
|
polymerase (DNA directed), epsilon |
| chr24_-_14209202 | 0.09 |
ENSDART00000180674
|
xkr9
|
XK, Kell blood group complex subunit-related family, member 9 |
| chr19_+_30884960 | 0.09 |
ENSDART00000140603
ENSDART00000183224 ENSDART00000135484 ENSDART00000139599 |
yars
|
tyrosyl-tRNA synthetase |
| chr4_+_71578099 | 0.09 |
ENSDART00000180472
ENSDART00000162184 |
si:dkey-27n6.4
|
si:dkey-27n6.4 |
| chr25_-_7670616 | 0.09 |
ENSDART00000131583
ENSDART00000142794 |
bet1l
|
Bet1 golgi vesicular membrane trafficking protein-like |
| chr6_+_51713076 | 0.09 |
ENSDART00000146281
|
ripor3
|
RIPOR family member 3 |
| chr17_-_5424175 | 0.09 |
ENSDART00000171063
ENSDART00000188102 |
supt3h
|
SPT3 homolog, SAGA and STAGA complex component |
| chr3_+_58656312 | 0.09 |
ENSDART00000148094
|
FP016018.1
|
|
| chr16_-_45917683 | 0.09 |
ENSDART00000184289
|
afp4
|
antifreeze protein type IV |
| chr24_+_20927135 | 0.09 |
ENSDART00000144883
ENSDART00000131829 |
fam162a
|
family with sequence similarity 162, member A |
| chr17_+_53418445 | 0.08 |
ENSDART00000097631
|
slc9a1b
|
solute carrier family 9 member A1b |
| chr11_-_24191928 | 0.08 |
ENSDART00000136827
|
sox12
|
SRY (sex determining region Y)-box 12 |
| chr22_+_25704430 | 0.08 |
ENSDART00000143776
|
si:dkeyp-98a7.3
|
si:dkeyp-98a7.3 |
| chr14_+_21159693 | 0.08 |
ENSDART00000192678
|
zgc:136929
|
zgc:136929 |
| chr7_-_2129076 | 0.08 |
ENSDART00000182385
|
si:cabz01007807.1
|
si:cabz01007807.1 |
| chr19_-_24988640 | 0.08 |
ENSDART00000104033
|
xkr8.1
|
XK, Kell blood group complex subunit-related family, member 8, tandem duplicate 1 |
| chr10_-_39130839 | 0.08 |
ENSDART00000061274
ENSDART00000148648 |
rps25
|
ribosomal protein S25 |
| chr8_+_41229233 | 0.08 |
ENSDART00000131135
|
zgc:152830
|
zgc:152830 |
| chr15_-_43284021 | 0.08 |
ENSDART00000041677
|
serpine2
|
serpin peptidase inhibitor, clade E (nexin, plasminogen activator inhibitor type 1), member 2 |
| chr18_+_44532199 | 0.08 |
ENSDART00000135386
|
st14a
|
suppression of tumorigenicity 14 (colon carcinoma) a |
| chr9_-_25989989 | 0.08 |
ENSDART00000090052
|
arhgap15
|
Rho GTPase activating protein 15 |
| chr13_+_29292011 | 0.08 |
ENSDART00000115023
|
parga
|
poly (ADP-ribose) glycohydrolase a |
| chr3_+_26322596 | 0.08 |
ENSDART00000172700
|
si:ch211-156b7.5
|
si:ch211-156b7.5 |
| chr19_-_10295173 | 0.08 |
ENSDART00000136697
|
cnot3b
|
CCR4-NOT transcription complex, subunit 3b |
| chr10_+_20128267 | 0.08 |
ENSDART00000064615
|
dmtn
|
dematin actin binding protein |
| chr6_+_29086594 | 0.08 |
ENSDART00000127695
|
rpl5b
|
ribosomal protein L5b |
| chr16_+_40576679 | 0.08 |
ENSDART00000169412
ENSDART00000193464 |
ccne2
|
cyclin E2 |
Network of associatons between targets according to the STRING database.
First level regulatory network of arid5a+arid6
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.2 | GO:0045649 | regulation of macrophage differentiation(GO:0045649) |
| 0.1 | 0.2 | GO:1904667 | negative regulation of ligase activity(GO:0051352) negative regulation of ubiquitin-protein transferase activity(GO:0051444) negative regulation of ubiquitin protein ligase activity(GO:1904667) |
| 0.1 | 0.2 | GO:0021564 | vagus nerve development(GO:0021564) |
| 0.0 | 0.2 | GO:0010693 | regulation of alkaline phosphatase activity(GO:0010692) negative regulation of alkaline phosphatase activity(GO:0010693) |
| 0.0 | 0.2 | GO:0072677 | eosinophil chemotaxis(GO:0048245) eosinophil migration(GO:0072677) |
| 0.0 | 0.2 | GO:0045110 | intermediate filament bundle assembly(GO:0045110) |
| 0.0 | 0.0 | GO:0045109 | intermediate filament organization(GO:0045109) |
| 0.0 | 0.2 | GO:0070493 | thrombin receptor signaling pathway(GO:0070493) |
| 0.0 | 0.2 | GO:0050764 | regulation of phagocytosis(GO:0050764) |
| 0.0 | 0.1 | GO:0006269 | DNA replication, synthesis of RNA primer(GO:0006269) |
| 0.0 | 0.2 | GO:2000251 | positive regulation of actin cytoskeleton reorganization(GO:2000251) |
| 0.0 | 0.1 | GO:1902975 | cell cycle DNA replication initiation(GO:1902292) nuclear cell cycle DNA replication initiation(GO:1902315) mitotic DNA replication initiation(GO:1902975) |
| 0.0 | 0.1 | GO:0006297 | leading strand elongation(GO:0006272) nucleotide-excision repair, DNA gap filling(GO:0006297) |
| 0.0 | 0.1 | GO:0031120 | snRNA pseudouridine synthesis(GO:0031120) |
| 0.0 | 0.1 | GO:0006572 | tyrosine catabolic process(GO:0006572) |
| 0.0 | 0.2 | GO:0019405 | alditol catabolic process(GO:0019405) |
| 0.0 | 0.1 | GO:0019262 | N-acetylneuraminate catabolic process(GO:0019262) |
| 0.0 | 0.0 | GO:0071267 | L-methionine biosynthetic process from methylthioadenosine(GO:0019509) amino acid salvage(GO:0043102) L-methionine biosynthetic process(GO:0071265) L-methionine salvage(GO:0071267) |
| 0.0 | 0.1 | GO:0031119 | tRNA pseudouridine synthesis(GO:0031119) |
| 0.0 | 0.2 | GO:0072395 | signal transduction involved in cell cycle checkpoint(GO:0072395) |
| 0.0 | 0.1 | GO:0010481 | epidermal cell division(GO:0010481) regulation of epidermal cell division(GO:0010482) |
| 0.0 | 0.1 | GO:0030576 | nuclear body organization(GO:0030575) Cajal body organization(GO:0030576) |
| 0.0 | 0.1 | GO:0090200 | regulation of release of cytochrome c from mitochondria(GO:0090199) positive regulation of release of cytochrome c from mitochondria(GO:0090200) |
| 0.0 | 0.0 | GO:0043654 | recognition of apoptotic cell(GO:0043654) |
| 0.0 | 0.2 | GO:0006048 | UDP-N-acetylglucosamine biosynthetic process(GO:0006048) |
| 0.0 | 0.1 | GO:0051315 | attachment of mitotic spindle microtubules to kinetochore(GO:0051315) |
| 0.0 | 0.1 | GO:0033353 | S-adenosylmethionine cycle(GO:0033353) |
| 0.0 | 0.2 | GO:0070782 | phosphatidylserine exposure on apoptotic cell surface(GO:0070782) |
| 0.0 | 0.1 | GO:0060397 | JAK-STAT cascade involved in growth hormone signaling pathway(GO:0060397) |
| 0.0 | 0.2 | GO:0045453 | bone resorption(GO:0045453) |
| 0.0 | 0.1 | GO:0015670 | carbon dioxide transport(GO:0015670) |
| 0.0 | 0.1 | GO:0000012 | single strand break repair(GO:0000012) |
| 0.0 | 0.1 | GO:0009263 | deoxyribonucleotide biosynthetic process(GO:0009263) |
| 0.0 | 0.1 | GO:0052651 | phosphatidylserine catabolic process(GO:0006660) monoacylglycerol metabolic process(GO:0046462) monoacylglycerol catabolic process(GO:0052651) |
| 0.0 | 0.1 | GO:2000036 | regulation of stem cell population maintenance(GO:2000036) |
| 0.0 | 0.1 | GO:0051231 | mitotic spindle elongation(GO:0000022) spindle elongation(GO:0051231) mitotic spindle midzone assembly(GO:0051256) |
| 0.0 | 0.1 | GO:0032515 | negative regulation of phosphoprotein phosphatase activity(GO:0032515) |
| 0.0 | 0.1 | GO:0035307 | positive regulation of dephosphorylation(GO:0035306) positive regulation of protein dephosphorylation(GO:0035307) |
| 0.0 | 0.1 | GO:0043137 | DNA replication, removal of RNA primer(GO:0043137) |
| 0.0 | 0.2 | GO:0007076 | mitotic chromosome condensation(GO:0007076) |
| 0.0 | 0.4 | GO:0007568 | aging(GO:0007568) |
| 0.0 | 0.1 | GO:0006566 | threonine metabolic process(GO:0006566) threonine catabolic process(GO:0006567) |
| 0.0 | 0.2 | GO:0015669 | gas transport(GO:0015669) |
| 0.0 | 0.1 | GO:0050907 | detection of chemical stimulus involved in sensory perception(GO:0050907) |
| 0.0 | 0.0 | GO:0045428 | regulation of nitric oxide biosynthetic process(GO:0045428) positive regulation of nitric oxide biosynthetic process(GO:0045429) positive regulation of reactive oxygen species biosynthetic process(GO:1903428) positive regulation of nitric oxide metabolic process(GO:1904407) positive regulation of reactive oxygen species metabolic process(GO:2000379) |
| 0.0 | 0.0 | GO:1901232 | regulation of convergent extension involved in axis elongation(GO:1901232) positive regulation of epiboly involved in gastrulation with mouth forming second(GO:1904088) |
| 0.0 | 0.0 | GO:0060829 | negative regulation of canonical Wnt signaling pathway involved in neural plate anterior/posterior pattern formation(GO:0060829) |
| 0.0 | 0.1 | GO:2000406 | positive regulation of lymphocyte migration(GO:2000403) positive regulation of T cell migration(GO:2000406) |
| 0.0 | 0.1 | GO:0034498 | early endosome to Golgi transport(GO:0034498) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.2 | GO:0061702 | inflammasome complex(GO:0061702) |
| 0.0 | 0.1 | GO:0032299 | ribonuclease H2 complex(GO:0032299) |
| 0.0 | 0.1 | GO:1990077 | primosome complex(GO:1990077) |
| 0.0 | 0.3 | GO:0070652 | HAUS complex(GO:0070652) |
| 0.0 | 0.1 | GO:0070319 | Golgi to plasma membrane transport vesicle(GO:0070319) |
| 0.0 | 0.2 | GO:0001772 | immunological synapse(GO:0001772) |
| 0.0 | 0.1 | GO:0031262 | Ndc80 complex(GO:0031262) |
| 0.0 | 0.1 | GO:0008622 | epsilon DNA polymerase complex(GO:0008622) |
| 0.0 | 0.1 | GO:0097134 | cyclin E1-CDK2 complex(GO:0097134) |
| 0.0 | 0.2 | GO:0000796 | condensin complex(GO:0000796) |
| 0.0 | 0.1 | GO:0005577 | fibrinogen complex(GO:0005577) |
| 0.0 | 0.2 | GO:0005833 | hemoglobin complex(GO:0005833) |
| 0.0 | 0.1 | GO:0035327 | transcriptionally active chromatin(GO:0035327) |
| 0.0 | 0.1 | GO:0072588 | box H/ACA snoRNP complex(GO:0031429) box H/ACA RNP complex(GO:0072588) |
| 0.0 | 0.2 | GO:0035101 | FACT complex(GO:0035101) |
| 0.0 | 0.0 | GO:0034663 | endoplasmic reticulum chaperone complex(GO:0034663) |
| 0.0 | 0.2 | GO:0030057 | desmosome(GO:0030057) |
| 0.0 | 0.1 | GO:0000138 | Golgi trans cisterna(GO:0000138) |
| 0.0 | 0.1 | GO:0030914 | STAGA complex(GO:0030914) |
| 0.0 | 0.1 | GO:0042788 | polysomal ribosome(GO:0042788) |
| 0.0 | 0.1 | GO:1990023 | mitotic spindle midzone(GO:1990023) |
| 0.0 | 0.1 | GO:0042555 | MCM complex(GO:0042555) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.2 | GO:0070548 | L-glutamine aminotransferase activity(GO:0070548) |
| 0.0 | 0.2 | GO:0019863 | IgE binding(GO:0019863) immunoglobulin binding(GO:0019865) |
| 0.0 | 0.2 | GO:0070891 | lipoteichoic acid binding(GO:0070891) |
| 0.0 | 0.1 | GO:0098809 | nitrite reductase activity(GO:0098809) |
| 0.0 | 0.1 | GO:0016802 | adenosylhomocysteinase activity(GO:0004013) trialkylsulfonium hydrolase activity(GO:0016802) |
| 0.0 | 0.2 | GO:0015057 | thrombin receptor activity(GO:0015057) |
| 0.0 | 0.1 | GO:0015562 | efflux transmembrane transporter activity(GO:0015562) |
| 0.0 | 0.1 | GO:0016822 | hydrolase activity, acting on acid carbon-carbon bonds(GO:0016822) hydrolase activity, acting on acid carbon-carbon bonds, in ketonic substances(GO:0016823) |
| 0.0 | 0.1 | GO:0008310 | single-stranded DNA 3'-5' exodeoxyribonuclease activity(GO:0008310) |
| 0.0 | 0.1 | GO:0061731 | ribonucleoside-diphosphate reductase activity, thioredoxin disulfide as acceptor(GO:0004748) oxidoreductase activity, acting on CH or CH2 groups, disulfide as acceptor(GO:0016728) ribonucleoside-diphosphate reductase activity(GO:0061731) |
| 0.0 | 0.1 | GO:0004649 | poly(ADP-ribose) glycohydrolase activity(GO:0004649) |
| 0.0 | 0.2 | GO:0008199 | ferric iron binding(GO:0008199) |
| 0.0 | 0.1 | GO:0008454 | alpha-1,3-mannosylglycoprotein 4-beta-N-acetylglucosaminyltransferase activity(GO:0008454) |
| 0.0 | 0.1 | GO:0045735 | nutrient reservoir activity(GO:0045735) |
| 0.0 | 0.1 | GO:0008743 | L-threonine 3-dehydrogenase activity(GO:0008743) |
| 0.0 | 0.0 | GO:1990715 | mRNA CDS binding(GO:1990715) |
| 0.0 | 0.1 | GO:0008097 | 5S rRNA binding(GO:0008097) |
| 0.0 | 0.0 | GO:0061629 | RNA polymerase II sequence-specific DNA binding transcription factor binding(GO:0061629) |
| 0.0 | 0.1 | GO:0004185 | serine-type carboxypeptidase activity(GO:0004185) |
| 0.0 | 0.1 | GO:0070004 | cysteine-type exopeptidase activity(GO:0070004) |
| 0.0 | 0.1 | GO:0004062 | aryl sulfotransferase activity(GO:0004062) |
| 0.0 | 0.1 | GO:0047192 | 1-acylglycerophosphocholine O-acyltransferase activity(GO:0047184) 1-alkylglycerophosphocholine O-acetyltransferase activity(GO:0047192) |
| 0.0 | 0.1 | GO:0019841 | retinol binding(GO:0019841) |
| 0.0 | 0.2 | GO:0097200 | cysteine-type endopeptidase activity involved in execution phase of apoptosis(GO:0097200) |
| 0.0 | 0.1 | GO:0004385 | guanylate kinase activity(GO:0004385) |
| 0.0 | 0.1 | GO:0038132 | neuregulin receptor activity(GO:0038131) neuregulin binding(GO:0038132) |
| 0.0 | 0.0 | GO:0042134 | rRNA primary transcript binding(GO:0042134) |
| 0.0 | 0.2 | GO:0019825 | oxygen transporter activity(GO:0005344) oxygen binding(GO:0019825) |
| 0.0 | 0.2 | GO:0008417 | fucosyltransferase activity(GO:0008417) |
| 0.0 | 0.1 | GO:0004523 | RNA-DNA hybrid ribonuclease activity(GO:0004523) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.2 | PID INTEGRIN4 PATHWAY | Alpha6 beta4 integrin-ligand interactions |
| 0.0 | 0.1 | PID IL12 STAT4 PATHWAY | IL12 signaling mediated by STAT4 |
| 0.0 | 0.1 | SIG IL4RECEPTOR IN B LYPHOCYTES | Genes related to IL4 rceptor signaling in B lymphocytes |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.7 | REACTOME INTERFERON GAMMA SIGNALING | Genes involved in Interferon gamma signaling |
| 0.0 | 0.2 | REACTOME IMMUNOREGULATORY INTERACTIONS BETWEEN A LYMPHOID AND A NON LYMPHOID CELL | Genes involved in Immunoregulatory interactions between a Lymphoid and a non-Lymphoid cell |
| 0.0 | 0.3 | REACTOME G1 S SPECIFIC TRANSCRIPTION | Genes involved in G1/S-Specific Transcription |
| 0.0 | 0.2 | REACTOME TRAFFICKING AND PROCESSING OF ENDOSOMAL TLR | Genes involved in Trafficking and processing of endosomal TLR |
| 0.0 | 0.1 | REACTOME ACYL CHAIN REMODELLING OF PG | Genes involved in Acyl chain remodelling of PG |
| 0.0 | 0.1 | REACTOME UNWINDING OF DNA | Genes involved in Unwinding of DNA |
| 0.0 | 0.1 | REACTOME TRANSPORT OF ORGANIC ANIONS | Genes involved in Transport of organic anions |
| 0.0 | 0.0 | REACTOME ACYL CHAIN REMODELLING OF PI | Genes involved in Acyl chain remodelling of PI |
| 0.0 | 0.1 | REACTOME CYCLIN A B1 ASSOCIATED EVENTS DURING G2 M TRANSITION | Genes involved in Cyclin A/B1 associated events during G2/M transition |
| 0.0 | 0.3 | REACTOME EXTENSION OF TELOMERES | Genes involved in Extension of Telomeres |