Project
PRJNA207719: Tissue specific transcriptome profiling
Navigation
Downloads
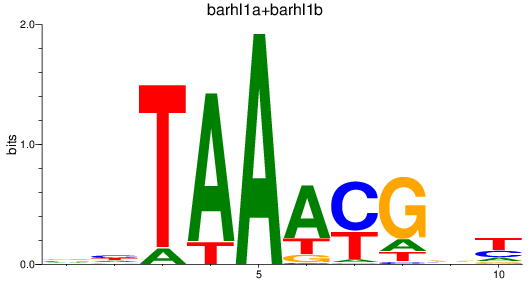
Results for barhl1a+barhl1b
Z-value: 1.70
Transcription factors associated with barhl1a+barhl1b
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
barhl1b
|
ENSDARG00000019013 | BarH-like homeobox 1b |
|
barhl1a
|
ENSDARG00000035508 | BarH-like homeobox 1a |
|
barhl1a
|
ENSDARG00000110061 | BarH-like homeobox 1a |
|
barhl1a
|
ENSDARG00000112355 | BarH-like homeobox 1a |
|
barhl1b
|
ENSDARG00000113145 | BarH-like homeobox 1b |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| barhl1a | dr11_v1_chr5_-_29750377_29750377 | 0.19 | 7.6e-01 | Click! |
| barhl1b | dr11_v1_chr21_-_17482465_17482465 | 0.09 | 8.8e-01 | Click! |
Activity profile of barhl1a+barhl1b motif
Sorted Z-values of barhl1a+barhl1b motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr24_-_4765740 | 0.78 |
ENSDART00000121576
|
cpb1
|
carboxypeptidase B1 (tissue) |
| chr23_+_35713557 | 0.75 |
ENSDART00000123518
|
tuba1c
|
tubulin, alpha 1c |
| chr19_-_5380770 | 0.68 |
ENSDART00000000221
|
krt91
|
keratin 91 |
| chr22_+_11756040 | 0.65 |
ENSDART00000105808
|
krt97
|
keratin 97 |
| chr2_-_32387441 | 0.63 |
ENSDART00000148202
|
ubtfl
|
upstream binding transcription factor, like |
| chr24_-_41797681 | 0.60 |
ENSDART00000169643
|
arhgap28
|
Rho GTPase activating protein 28 |
| chr25_-_20666328 | 0.53 |
ENSDART00000098076
|
csk
|
C-terminal Src kinase |
| chr16_+_49601838 | 0.47 |
ENSDART00000168570
ENSDART00000159236 |
si:dkey-82o10.4
|
si:dkey-82o10.4 |
| chr23_+_19813677 | 0.47 |
ENSDART00000139192
ENSDART00000142308 |
emd
|
emerin (Emery-Dreifuss muscular dystrophy) |
| chr11_+_6136220 | 0.46 |
ENSDART00000082223
|
tax1bp3
|
Tax1 (human T-cell leukemia virus type I) binding protein 3 |
| chr5_+_13373593 | 0.45 |
ENSDART00000051668
ENSDART00000183883 |
ccl19a.2
|
chemokine (C-C motif) ligand 19a, tandem duplicate 2 |
| chr13_+_22717939 | 0.44 |
ENSDART00000188288
|
nfkb2
|
nuclear factor of kappa light polypeptide gene enhancer in B-cells 2 (p49/p100) |
| chr20_+_33294428 | 0.43 |
ENSDART00000024104
|
mycn
|
MYCN proto-oncogene, bHLH transcription factor |
| chr19_+_29798064 | 0.41 |
ENSDART00000167803
ENSDART00000051804 |
marcksl1b
|
MARCKS-like 1b |
| chr17_+_24446353 | 0.40 |
ENSDART00000140467
|
ugp2b
|
UDP-glucose pyrophosphorylase 2b |
| chr5_-_22082918 | 0.39 |
ENSDART00000020908
|
zc4h2
|
zinc finger, C4H2 domain containing |
| chr3_+_35005730 | 0.39 |
ENSDART00000029451
|
prkcbb
|
protein kinase C, beta b |
| chr11_+_37612214 | 0.38 |
ENSDART00000172899
ENSDART00000077496 |
hp1bp3
|
heterochromatin protein 1, binding protein 3 |
| chr16_-_31824525 | 0.37 |
ENSDART00000058737
|
cdc42l
|
cell division cycle 42, like |
| chr17_+_24445818 | 0.36 |
ENSDART00000139963
|
ugp2b
|
UDP-glucose pyrophosphorylase 2b |
| chr5_-_42180205 | 0.36 |
ENSDART00000145247
|
fam222ba
|
family with sequence similarity 222, member Ba |
| chr18_+_31410652 | 0.35 |
ENSDART00000098504
|
def8
|
differentially expressed in FDCP 8 homolog (mouse) |
| chr6_+_168962 | 0.35 |
ENSDART00000182631
|
CABZ01066694.1
|
|
| chr9_+_14010823 | 0.35 |
ENSDART00000143837
|
si:ch211-67e16.3
|
si:ch211-67e16.3 |
| chr2_-_37140423 | 0.35 |
ENSDART00000144220
|
tspan37
|
tetraspanin 37 |
| chr2_-_26469065 | 0.35 |
ENSDART00000099247
ENSDART00000099248 |
rabggtb
|
Rab geranylgeranyltransferase, beta subunit |
| chr20_+_2460864 | 0.35 |
ENSDART00000131642
|
akap7
|
A kinase (PRKA) anchor protein 7 |
| chr17_+_24446533 | 0.34 |
ENSDART00000131417
|
ugp2b
|
UDP-glucose pyrophosphorylase 2b |
| chr16_+_43347966 | 0.34 |
ENSDART00000171308
|
zmp:0000000930
|
zmp:0000000930 |
| chr19_+_342094 | 0.33 |
ENSDART00000151013
ENSDART00000187622 |
ensaa
|
endosulfine alpha a |
| chr12_-_13318944 | 0.33 |
ENSDART00000152201
ENSDART00000041394 |
emc9
|
ER membrane protein complex subunit 9 |
| chr19_+_7124337 | 0.33 |
ENSDART00000031380
|
tap2a
|
transporter associated with antigen processing, subunit type a |
| chr16_-_35415004 | 0.33 |
ENSDART00000170401
ENSDART00000157832 ENSDART00000170048 |
taf12
|
TAF12 RNA polymerase II, TATA box binding protein (TBP)-associated factor |
| chr8_+_25761654 | 0.32 |
ENSDART00000137899
ENSDART00000062403 |
tmem9
|
transmembrane protein 9 |
| chr6_-_6976096 | 0.32 |
ENSDART00000151822
ENSDART00000039443 ENSDART00000177960 |
tuba8l4
|
tubulin, alpha 8 like 4 |
| chr3_-_52683241 | 0.32 |
ENSDART00000155248
|
si:dkey-210j14.5
|
si:dkey-210j14.5 |
| chr5_+_32141790 | 0.32 |
ENSDART00000041504
|
tescb
|
tescalcin b |
| chr21_-_33478164 | 0.32 |
ENSDART00000191542
|
si:ch73-42p12.2
|
si:ch73-42p12.2 |
| chr25_+_1732838 | 0.31 |
ENSDART00000159555
ENSDART00000168161 |
FBLN1
|
fibulin 1 |
| chr3_-_36210344 | 0.31 |
ENSDART00000025326
|
csnk1da
|
casein kinase 1, delta a |
| chr3_-_20106233 | 0.30 |
ENSDART00000145285
|
selenow2b
|
selenoprotein W, 2b |
| chr7_+_24645186 | 0.30 |
ENSDART00000150118
ENSDART00000150233 ENSDART00000087691 |
gba2
|
glucosidase, beta (bile acid) 2 |
| chr19_+_29808471 | 0.30 |
ENSDART00000186428
|
hdac1
|
histone deacetylase 1 |
| chr19_-_13950231 | 0.30 |
ENSDART00000168665
|
sh3bgrl3
|
SH3 domain binding glutamate-rich protein like 3 |
| chr1_-_55166511 | 0.30 |
ENSDART00000150430
ENSDART00000035725 |
pane1
|
proliferation associated nuclear element |
| chr3_+_24050043 | 0.30 |
ENSDART00000151788
|
cbx1a
|
chromobox homolog 1a (HP1 beta homolog Drosophila) |
| chr25_-_26753196 | 0.30 |
ENSDART00000155698
|
usp3
|
ubiquitin specific peptidase 3 |
| chr5_+_36599107 | 0.30 |
ENSDART00000097677
ENSDART00000138147 |
micu2
|
mitochondrial calcium uptake 2 |
| chr6_+_48348415 | 0.30 |
ENSDART00000064826
|
mov10a
|
Mov10 RISC complex RNA helicase a |
| chr4_+_15006217 | 0.29 |
ENSDART00000090837
|
zc3hc1
|
zinc finger, C3HC-type containing 1 |
| chr19_+_18739085 | 0.29 |
ENSDART00000188868
|
spaca4l
|
sperm acrosome associated 4 like |
| chr7_+_25000060 | 0.29 |
ENSDART00000039265
ENSDART00000141814 |
naa40
|
N(alpha)-acetyltransferase 40, NatD catalytic subunit, homolog (S. cerevisiae) |
| chr19_-_33274978 | 0.29 |
ENSDART00000020301
ENSDART00000114714 |
fam92a1
|
family with sequence similarity 92, member A1 |
| chr3_+_45365098 | 0.29 |
ENSDART00000052746
ENSDART00000156555 |
ube2ia
|
ubiquitin-conjugating enzyme E2Ia |
| chr12_-_6063328 | 0.28 |
ENSDART00000002583
|
aarsd1
|
alanyl-tRNA synthetase domain containing 1 |
| chr3_-_31783737 | 0.28 |
ENSDART00000090809
|
kcnh6a
|
potassium voltage-gated channel, subfamily H (eag-related), member 6a |
| chr12_-_27031060 | 0.28 |
ENSDART00000076103
|
chmp2a
|
charged multivesicular body protein 2A |
| chr3_+_27798094 | 0.28 |
ENSDART00000075100
ENSDART00000151437 |
carhsp1
|
calcium regulated heat stable protein 1 |
| chr3_-_32859335 | 0.28 |
ENSDART00000158916
|
si:dkey-16l2.20
|
si:dkey-16l2.20 |
| chr18_+_6536598 | 0.28 |
ENSDART00000149350
|
fkbp4
|
FK506 binding protein 4 |
| chr20_-_46554440 | 0.28 |
ENSDART00000043298
ENSDART00000060680 |
fosab
|
v-fos FBJ murine osteosarcoma viral oncogene homolog Ab |
| chr25_-_31118923 | 0.28 |
ENSDART00000009126
ENSDART00000188286 |
kras
|
v-Ki-ras2 Kirsten rat sarcoma viral oncogene homolog |
| chr13_-_28263856 | 0.27 |
ENSDART00000041036
ENSDART00000079806 |
smap1
|
small ArfGAP 1 |
| chr4_-_20051141 | 0.26 |
ENSDART00000066963
|
atp6v1f
|
ATPase H+ transporting V1 subunit F |
| chr6_-_21873266 | 0.26 |
ENSDART00000151658
ENSDART00000151152 ENSDART00000151179 |
si:dkey-19e4.5
|
si:dkey-19e4.5 |
| chr3_-_37082618 | 0.26 |
ENSDART00000026701
ENSDART00000110716 |
tubg1
|
tubulin, gamma 1 |
| chr21_-_38730557 | 0.26 |
ENSDART00000150984
ENSDART00000111885 |
taf9
|
TAF9 RNA polymerase II, TATA box binding protein (TBP)-associated factor |
| chr14_+_21222287 | 0.26 |
ENSDART00000159905
|
si:ch211-175m2.4
|
si:ch211-175m2.4 |
| chr5_+_36900157 | 0.25 |
ENSDART00000183533
ENSDART00000051184 |
hnrnpl
|
heterogeneous nuclear ribonucleoprotein L |
| chr24_-_22756508 | 0.25 |
ENSDART00000035409
ENSDART00000146247 |
zc2hc1a
|
zinc finger, C2HC-type containing 1A |
| chr16_-_11779508 | 0.25 |
ENSDART00000136329
ENSDART00000060145 ENSDART00000141101 |
pafah1b3
|
platelet-activating factor acetylhydrolase, isoform Ib, gamma subunit |
| chr23_+_42272588 | 0.24 |
ENSDART00000164907
|
CABZ01065131.1
|
|
| chr19_+_41509956 | 0.24 |
ENSDART00000171318
ENSDART00000172311 |
si:ch211-57n23.1
|
si:ch211-57n23.1 |
| chr24_-_20599781 | 0.24 |
ENSDART00000179664
ENSDART00000141823 |
zbtb47b
|
zinc finger and BTB domain containing 47b |
| chr8_-_7847921 | 0.24 |
ENSDART00000188094
|
zgc:113363
|
zgc:113363 |
| chr23_-_24682244 | 0.24 |
ENSDART00000104035
|
ctdsp2
|
CTD (carboxy-terminal domain, RNA polymerase II, polypeptide A) small phosphatase 2 |
| chr22_-_33916620 | 0.24 |
ENSDART00000191276
|
CR974456.1
|
|
| chr3_-_19368435 | 0.24 |
ENSDART00000132987
|
s1pr5a
|
sphingosine-1-phosphate receptor 5a |
| chr2_-_10098191 | 0.24 |
ENSDART00000138081
|
bcl6ab
|
B-cell CLL/lymphoma 6a, genome duplicate b |
| chr3_-_20091964 | 0.24 |
ENSDART00000029386
ENSDART00000020253 ENSDART00000124326 |
slc4a1a
|
solute carrier family 4 (anion exchanger), member 1a (Diego blood group) |
| chr1_-_33380011 | 0.23 |
ENSDART00000141347
ENSDART00000136383 |
cd99
|
CD99 molecule |
| chr3_+_45364849 | 0.23 |
ENSDART00000153974
|
ube2ia
|
ubiquitin-conjugating enzyme E2Ia |
| chr2_-_32386791 | 0.23 |
ENSDART00000056634
|
ubtfl
|
upstream binding transcription factor, like |
| chr23_-_29824146 | 0.23 |
ENSDART00000020616
|
zgc:194189
|
zgc:194189 |
| chr17_+_50657509 | 0.23 |
ENSDART00000179957
|
ddhd1a
|
DDHD domain containing 1a |
| chr3_-_16110100 | 0.23 |
ENSDART00000051807
|
lasp1
|
LIM and SH3 protein 1 |
| chr4_-_75172216 | 0.23 |
ENSDART00000127522
|
naa40
|
N(alpha)-acetyltransferase 40, NatD catalytic subunit, homolog (S. cerevisiae) |
| chr23_-_10722664 | 0.22 |
ENSDART00000146526
ENSDART00000129022 ENSDART00000104985 |
foxp1a
|
forkhead box P1a |
| chr23_-_35790235 | 0.22 |
ENSDART00000142369
ENSDART00000141141 ENSDART00000011004 |
mfsd5
|
major facilitator superfamily domain containing 5 |
| chr19_-_3488860 | 0.22 |
ENSDART00000172520
|
hivep1
|
human immunodeficiency virus type I enhancer binding protein 1 |
| chr20_-_30938184 | 0.22 |
ENSDART00000147359
ENSDART00000062552 |
wtap
|
WT1 associated protein |
| chr13_+_9612395 | 0.22 |
ENSDART00000136689
ENSDART00000138362 |
slf2
|
SMC5-SMC6 complex localization factor 2 |
| chr11_+_11719575 | 0.22 |
ENSDART00000003891
|
jupa
|
junction plakoglobin a |
| chr25_+_753364 | 0.22 |
ENSDART00000183804
|
TWF1 (1 of many)
|
twinfilin actin binding protein 1 |
| chr23_-_17509656 | 0.22 |
ENSDART00000148423
|
dnajc5ab
|
DnaJ (Hsp40) homolog, subfamily C, member 5ab |
| chr18_+_808911 | 0.21 |
ENSDART00000172518
|
cox5ab
|
cytochrome c oxidase subunit Vab |
| chr10_-_11397590 | 0.21 |
ENSDART00000064212
|
srek1ip1
|
SREK1-interacting protein 1 |
| chr10_-_8032885 | 0.21 |
ENSDART00000188619
|
atp6v0a2a
|
ATPase H+ transporting V0 subunit a2a |
| chr2_-_30460293 | 0.21 |
ENSDART00000113193
|
cbln2a
|
cerebellin 2a precursor |
| chr8_+_39674707 | 0.21 |
ENSDART00000126301
ENSDART00000040330 |
prkab1b
|
protein kinase, AMP-activated, beta 1 non-catalytic subunit, b |
| chr19_-_47571456 | 0.21 |
ENSDART00000158071
ENSDART00000165841 |
rrm2
|
ribonucleotide reductase M2 polypeptide |
| chr21_+_35215810 | 0.21 |
ENSDART00000135256
|
ubtd2
|
ubiquitin domain containing 2 |
| chr5_-_28915130 | 0.20 |
ENSDART00000078592
|
npdc1b
|
neural proliferation, differentiation and control, 1b |
| chr2_+_49457449 | 0.20 |
ENSDART00000185470
|
sh3gl1a
|
SH3-domain GRB2-like 1a |
| chr14_+_21755469 | 0.20 |
ENSDART00000186326
|
kdm2ab
|
lysine (K)-specific demethylase 2Ab |
| chr3_+_32791786 | 0.20 |
ENSDART00000180174
|
tbc1d10b
|
TBC1 domain family, member 10b |
| chr21_-_22715297 | 0.20 |
ENSDART00000065548
|
c1qb
|
complement component 1, q subcomponent, B chain |
| chr15_-_14098522 | 0.19 |
ENSDART00000156947
|
si:ch211-116o3.5
|
si:ch211-116o3.5 |
| chr6_-_55309190 | 0.19 |
ENSDART00000162117
|
ube2c
|
ubiquitin-conjugating enzyme E2C |
| chr20_+_26556174 | 0.19 |
ENSDART00000138492
|
irf4b
|
interferon regulatory factor 4b |
| chr2_-_38080075 | 0.19 |
ENSDART00000056544
|
tox4a
|
TOX high mobility group box family member 4 a |
| chr6_-_42336987 | 0.19 |
ENSDART00000128777
ENSDART00000075601 |
fancd2
|
Fanconi anemia, complementation group D2 |
| chr9_+_500052 | 0.19 |
ENSDART00000166707
|
CU984600.1
|
|
| chr19_-_20777351 | 0.19 |
ENSDART00000019206
|
ngly1
|
N-glycanase 1 |
| chr22_+_10215558 | 0.19 |
ENSDART00000063274
|
kctd6a
|
potassium channel tetramerization domain containing 6a |
| chr12_+_28910762 | 0.19 |
ENSDART00000076342
ENSDART00000160939 ENSDART00000076572 |
rnf40
|
ring finger protein 40 |
| chr7_-_31921687 | 0.19 |
ENSDART00000146720
|
lin7c
|
lin-7 homolog C (C. elegans) |
| chr15_-_47338769 | 0.19 |
ENSDART00000164983
ENSDART00000176289 |
anapc15
|
anaphase promoting complex subunit 15 |
| chr7_+_26466826 | 0.19 |
ENSDART00000058908
|
mpdu1b
|
mannose-P-dolichol utilization defect 1b |
| chr6_-_8362419 | 0.19 |
ENSDART00000142752
ENSDART00000135810 |
acp5a
|
acid phosphatase 5a, tartrate resistant |
| chr7_+_21887307 | 0.18 |
ENSDART00000052871
|
pop7
|
POP7 homolog, ribonuclease P/MRP subunit |
| chr11_-_33618612 | 0.18 |
ENSDART00000033980
|
lims1
|
LIM and senescent cell antigen-like domains 1 |
| chr5_-_32890807 | 0.18 |
ENSDART00000007512
|
pole3
|
polymerase (DNA directed), epsilon 3 (p17 subunit) |
| chr4_+_20051478 | 0.18 |
ENSDART00000143642
|
lamtor4
|
late endosomal/lysosomal adaptor, MAPK and MTOR activator 4 |
| chr1_-_37087966 | 0.17 |
ENSDART00000172111
ENSDART00000160056 |
nr3c2
|
nuclear receptor subfamily 3, group C, member 2 |
| chr5_+_24087035 | 0.17 |
ENSDART00000183644
|
tp53
|
tumor protein p53 |
| chr7_-_41812015 | 0.17 |
ENSDART00000174058
|
vps35
|
vacuolar protein sorting 35 homolog (S. cerevisiae) |
| chr1_+_29267841 | 0.17 |
ENSDART00000085648
|
lig4
|
ligase IV, DNA, ATP-dependent |
| chr19_+_3213914 | 0.17 |
ENSDART00000193144
|
zgc:86598
|
zgc:86598 |
| chr4_+_73606482 | 0.17 |
ENSDART00000150765
|
si:ch211-165i18.2
|
si:ch211-165i18.2 |
| chr21_-_30181732 | 0.17 |
ENSDART00000015636
|
hnrnph1l
|
heterogeneous nuclear ribonucleoprotein H1, like |
| chr17_+_43926523 | 0.17 |
ENSDART00000121550
ENSDART00000041447 |
ktn1
|
kinectin 1 |
| chr5_+_57714902 | 0.17 |
ENSDART00000182860
|
ufc1
|
ubiquitin-fold modifier conjugating enzyme 1 |
| chr23_+_24955394 | 0.17 |
ENSDART00000142124
|
nol9
|
nucleolar protein 9 |
| chr21_-_32082130 | 0.16 |
ENSDART00000003978
ENSDART00000182050 |
mat2b
|
methionine adenosyltransferase II, beta |
| chr12_+_14149686 | 0.16 |
ENSDART00000123741
|
kbtbd2
|
kelch repeat and BTB (POZ) domain containing 2 |
| chr1_+_8601935 | 0.16 |
ENSDART00000152367
|
si:ch211-160d14.6
|
si:ch211-160d14.6 |
| chr5_+_53580846 | 0.16 |
ENSDART00000184967
ENSDART00000161751 |
taf6
|
TAF6 RNA polymerase II, TATA box binding protein (TBP)-associated factor |
| chr23_+_7518294 | 0.16 |
ENSDART00000081536
|
hck
|
HCK proto-oncogene, Src family tyrosine kinase |
| chr19_+_46237665 | 0.16 |
ENSDART00000159391
|
vps28
|
vacuolar protein sorting 28 (yeast) |
| chr16_+_45746549 | 0.16 |
ENSDART00000190403
|
paqr6
|
progestin and adipoQ receptor family member VI |
| chr6_+_9107063 | 0.16 |
ENSDART00000083820
|
vps16
|
vacuolar protein sorting protein 16 |
| chr15_+_32249062 | 0.16 |
ENSDART00000133867
ENSDART00000152545 ENSDART00000082315 ENSDART00000152513 ENSDART00000152139 |
arfip2a
|
ADP-ribosylation factor interacting protein 2a |
| chr8_+_25190023 | 0.16 |
ENSDART00000047528
|
gnai3
|
guanine nucleotide binding protein (G protein), alpha inhibiting activity polypeptide 3 |
| chr2_+_32826235 | 0.16 |
ENSDART00000143127
|
si:dkey-154p10.3
|
si:dkey-154p10.3 |
| chr15_+_34699585 | 0.16 |
ENSDART00000017569
|
tnfb
|
tumor necrosis factor b (TNF superfamily, member 2) |
| chr13_+_9368621 | 0.16 |
ENSDART00000109126
|
alms1
|
Alstrom syndrome protein 1 |
| chr2_+_16449627 | 0.16 |
ENSDART00000005381
|
zgc:110269
|
zgc:110269 |
| chr3_-_52644737 | 0.16 |
ENSDART00000126180
|
si:dkey-210j14.3
|
si:dkey-210j14.3 |
| chr14_+_50937757 | 0.16 |
ENSDART00000163865
|
rnf44
|
ring finger protein 44 |
| chr7_+_65398161 | 0.15 |
ENSDART00000166109
ENSDART00000157399 |
usp47
|
ubiquitin specific peptidase 47 |
| chr6_+_19948043 | 0.15 |
ENSDART00000182636
|
pik3r5
|
phosphoinositide-3-kinase, regulatory subunit 5 |
| chr2_-_31614277 | 0.15 |
ENSDART00000137054
ENSDART00000131507 ENSDART00000137674 ENSDART00000147041 |
si:ch211-106h4.5
|
si:ch211-106h4.5 |
| chr4_-_9350585 | 0.15 |
ENSDART00000148155
|
si:ch211-125a15.1
|
si:ch211-125a15.1 |
| chr18_+_7639401 | 0.15 |
ENSDART00000092416
|
rabl2
|
RAB, member of RAS oncogene family-like 2 |
| chr25_+_5035343 | 0.15 |
ENSDART00000011751
|
parvb
|
parvin, beta |
| chr15_-_951118 | 0.15 |
ENSDART00000171427
|
alox5b.1
|
arachidonate 5-lipoxygenase b, tandem duplicate 1 |
| chr10_-_8033468 | 0.15 |
ENSDART00000140476
|
atp6v0a2a
|
ATPase H+ transporting V0 subunit a2a |
| chr4_-_18840487 | 0.15 |
ENSDART00000066978
|
cbll1
|
Cbl proto-oncogene-like 1, E3 ubiquitin protein ligase |
| chr12_+_22404108 | 0.15 |
ENSDART00000153055
|
hdlbpb
|
high density lipoprotein binding protein b |
| chr19_+_31904836 | 0.15 |
ENSDART00000162297
ENSDART00000088340 ENSDART00000151280 ENSDART00000151218 |
tpd52
|
tumor protein D52 |
| chr19_-_33850201 | 0.15 |
ENSDART00000168583
|
otud6b
|
OTU domain containing 6B |
| chr11_-_25461336 | 0.14 |
ENSDART00000014945
|
hcfc1a
|
host cell factor C1a |
| chr2_+_27386617 | 0.14 |
ENSDART00000134976
|
si:ch73-382f3.1
|
si:ch73-382f3.1 |
| chr14_+_1170968 | 0.14 |
ENSDART00000125203
ENSDART00000193575 |
hopx
|
HOP homeobox |
| chr3_+_13842554 | 0.14 |
ENSDART00000162317
ENSDART00000158068 |
ilf3b
|
interleukin enhancer binding factor 3b |
| chr9_-_33477588 | 0.14 |
ENSDART00000144150
|
caska
|
calcium/calmodulin-dependent serine protein kinase a |
| chr2_-_13268556 | 0.14 |
ENSDART00000127693
ENSDART00000079757 |
vps4b
|
vacuolar protein sorting 4b homolog B (S. cerevisiae) |
| chr1_+_19433004 | 0.14 |
ENSDART00000133959
|
clockb
|
clock circadian regulator b |
| chr25_-_3745393 | 0.14 |
ENSDART00000037973
|
morc2
|
MORC family CW-type zinc finger 2 |
| chr20_-_10120442 | 0.14 |
ENSDART00000144970
|
meis2b
|
Meis homeobox 2b |
| chr3_+_40170216 | 0.14 |
ENSDART00000011568
|
syngr3a
|
synaptogyrin 3a |
| chr2_-_13216269 | 0.14 |
ENSDART00000149947
|
bcl2b
|
BCL2, apoptosis regulator b |
| chr8_-_44223473 | 0.14 |
ENSDART00000098525
|
stx2b
|
syntaxin 2b |
| chr7_+_30178453 | 0.14 |
ENSDART00000174420
|
si:ch211-107o10.3
|
si:ch211-107o10.3 |
| chr2_-_16449504 | 0.14 |
ENSDART00000144801
|
atr
|
ATR serine/threonine kinase |
| chr8_-_30316436 | 0.14 |
ENSDART00000111358
ENSDART00000167065 |
zgc:162939
|
zgc:162939 |
| chr23_+_31913292 | 0.14 |
ENSDART00000136910
|
armc1l
|
armadillo repeat containing 1, like |
| chr19_+_7045033 | 0.13 |
ENSDART00000146579
|
mhc1uka
|
major histocompatibility complex class I UKA |
| chr7_-_40630698 | 0.13 |
ENSDART00000134547
|
ube3c
|
ubiquitin protein ligase E3C |
| chr11_-_7261717 | 0.13 |
ENSDART00000128959
|
zgc:113223
|
zgc:113223 |
| chr22_+_34701848 | 0.13 |
ENSDART00000082066
|
atpv0e2
|
ATPase H+ transporting V0 subunit e2 |
| chr23_-_31555696 | 0.13 |
ENSDART00000053539
|
tcf21
|
transcription factor 21 |
| chr8_+_47099033 | 0.13 |
ENSDART00000142979
|
arhgef16
|
Rho guanine nucleotide exchange factor (GEF) 16 |
| chr2_+_307665 | 0.13 |
ENSDART00000082083
|
zgc:113452
|
zgc:113452 |
| chr11_-_40519886 | 0.13 |
ENSDART00000172819
|
miip
|
migration and invasion inhibitory protein |
| chr1_-_52128425 | 0.13 |
ENSDART00000149939
|
rad23aa
|
RAD23 homolog A, nucleotide excision repair protein a |
| chr18_+_13837746 | 0.12 |
ENSDART00000169552
|
cdh13
|
cadherin 13, H-cadherin (heart) |
| chr14_+_7892383 | 0.12 |
ENSDART00000063837
|
ube2d2
|
ubiquitin-conjugating enzyme E2D 2 (UBC4/5 homolog, yeast) |
| chr11_+_6080528 | 0.12 |
ENSDART00000183099
|
ushbp1
|
Usher syndrome 1C binding protein 1 |
| chr22_+_28236737 | 0.12 |
ENSDART00000086868
|
senp7b
|
SUMO1/sentrin specific peptidase 7b |
| chr16_+_40576679 | 0.12 |
ENSDART00000169412
ENSDART00000193464 |
ccne2
|
cyclin E2 |
| chr11_+_8565323 | 0.12 |
ENSDART00000081607
ENSDART00000141612 |
tbl1xr1a
|
transducin (beta)-like 1 X-linked receptor 1a |
| chr16_-_11962959 | 0.12 |
ENSDART00000135379
|
cd4-2.2
|
CD4-2 molecule, tandem duplicate 2 |
| chr14_+_26247319 | 0.12 |
ENSDART00000192793
|
CCDC69
|
coiled-coil domain containing 69 |
| chr13_+_45980163 | 0.12 |
ENSDART00000074547
ENSDART00000005195 |
bsdc1
|
BSD domain containing 1 |
| chr20_-_8443425 | 0.12 |
ENSDART00000083908
|
dab1a
|
Dab, reelin signal transducer, homolog 1a (Drosophila) |
| chr6_+_296130 | 0.12 |
ENSDART00000073985
|
rbfox2
|
RNA binding fox-1 homolog 2 |
| chr21_-_22724980 | 0.12 |
ENSDART00000035469
|
c1qa
|
complement component 1, q subcomponent, A chain |
| chr6_+_3473657 | 0.12 |
ENSDART00000011785
|
rad54l
|
RAD54 like |
Network of associatons between targets according to the STRING database.
First level regulatory network of barhl1a+barhl1b
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 1.1 | GO:0006011 | UDP-glucose metabolic process(GO:0006011) |
| 0.2 | 0.5 | GO:2000008 | regulation of protein localization to cell surface(GO:2000008) negative regulation of protein localization to cell surface(GO:2000009) |
| 0.1 | 0.4 | GO:0030521 | androgen receptor signaling pathway(GO:0030521) regulation of androgen receptor signaling pathway(GO:0060765) |
| 0.1 | 0.5 | GO:0036306 | embryonic heart tube elongation(GO:0036306) |
| 0.1 | 0.5 | GO:0060368 | regulation of Fc receptor mediated stimulatory signaling pathway(GO:0060368) |
| 0.1 | 0.3 | GO:0006678 | glucosylceramide metabolic process(GO:0006678) |
| 0.1 | 0.3 | GO:0060306 | regulation of membrane repolarization(GO:0060306) regulation of ventricular cardiac muscle cell membrane repolarization(GO:0060307) regulation of cardiac muscle cell membrane repolarization(GO:0099623) ventricular cardiac muscle cell membrane repolarization(GO:0099625) |
| 0.1 | 0.5 | GO:0051204 | protein insertion into membrane from inner side(GO:0032978) protein insertion into mitochondrial membrane from inner side(GO:0032979) protein insertion into mitochondrial membrane(GO:0051204) |
| 0.1 | 0.3 | GO:0000212 | meiotic spindle organization(GO:0000212) |
| 0.1 | 0.3 | GO:0035994 | response to muscle stretch(GO:0035994) |
| 0.1 | 0.4 | GO:1901207 | regulation of heart looping(GO:1901207) |
| 0.1 | 0.2 | GO:1903504 | regulation of spindle checkpoint(GO:0090231) regulation of mitotic cell cycle spindle assembly checkpoint(GO:0090266) regulation of mitotic spindle checkpoint(GO:1903504) |
| 0.1 | 0.2 | GO:1901216 | regulation of transposition, RNA-mediated(GO:0010525) negative regulation of transposition, RNA-mediated(GO:0010526) transposition, RNA-mediated(GO:0032197) positive regulation of neuron apoptotic process(GO:0043525) positive regulation of neuron death(GO:1901216) |
| 0.1 | 0.3 | GO:0031507 | heterochromatin assembly(GO:0031507) |
| 0.1 | 0.3 | GO:1990592 | protein polyufmylation(GO:1990564) protein K69-linked ufmylation(GO:1990592) |
| 0.1 | 0.3 | GO:0010269 | response to selenium ion(GO:0010269) |
| 0.0 | 0.3 | GO:0018344 | protein geranylgeranylation(GO:0018344) |
| 0.0 | 0.4 | GO:0032488 | Cdc42 protein signal transduction(GO:0032488) |
| 0.0 | 0.2 | GO:0000448 | cleavage in ITS2 between 5.8S rRNA and LSU-rRNA of tricistronic rRNA transcript (SSU-rRNA, 5.8S rRNA, LSU-rRNA)(GO:0000448) |
| 0.0 | 0.2 | GO:2000391 | neutrophil extravasation(GO:0072672) regulation of neutrophil extravasation(GO:2000389) positive regulation of neutrophil extravasation(GO:2000391) |
| 0.0 | 0.2 | GO:0071387 | cellular response to cortisol stimulus(GO:0071387) |
| 0.0 | 0.2 | GO:0002159 | desmosome assembly(GO:0002159) |
| 0.0 | 0.3 | GO:0033687 | osteoblast proliferation(GO:0033687) |
| 0.0 | 0.2 | GO:0006266 | DNA ligation(GO:0006266) DNA ligation involved in DNA repair(GO:0051103) |
| 0.0 | 0.2 | GO:1990918 | double-strand break repair involved in meiotic recombination(GO:1990918) |
| 0.0 | 0.2 | GO:1903361 | protein localization to basolateral plasma membrane(GO:1903361) |
| 0.0 | 0.5 | GO:0006474 | N-terminal protein amino acid acetylation(GO:0006474) |
| 0.0 | 0.2 | GO:0080009 | mRNA methylation(GO:0080009) |
| 0.0 | 0.6 | GO:0051098 | regulation of binding(GO:0051098) |
| 0.0 | 0.2 | GO:0044342 | type B pancreatic cell proliferation(GO:0044342) |
| 0.0 | 0.4 | GO:0070828 | heterochromatin organization(GO:0070828) |
| 0.0 | 0.1 | GO:0000390 | spliceosomal complex disassembly(GO:0000390) |
| 0.0 | 0.1 | GO:0003096 | renal sodium ion transport(GO:0003096) renal sodium ion absorption(GO:0070294) |
| 0.0 | 0.3 | GO:0006450 | regulation of translational fidelity(GO:0006450) |
| 0.0 | 0.2 | GO:0009312 | oligosaccharide biosynthetic process(GO:0009312) |
| 0.0 | 0.2 | GO:0045003 | double-strand break repair via synthesis-dependent strand annealing(GO:0045003) |
| 0.0 | 0.4 | GO:0051452 | vacuolar acidification(GO:0007035) pH reduction(GO:0045851) intracellular pH reduction(GO:0051452) |
| 0.0 | 0.3 | GO:0051123 | RNA polymerase II transcriptional preinitiation complex assembly(GO:0051123) |
| 0.0 | 0.1 | GO:1902262 | apoptotic process involved in patterning of blood vessels(GO:1902262) |
| 0.0 | 0.1 | GO:0048739 | cardiac muscle fiber development(GO:0048739) |
| 0.0 | 0.2 | GO:0006556 | S-adenosylmethionine biosynthetic process(GO:0006556) |
| 0.0 | 0.1 | GO:0018342 | protein prenylation(GO:0018342) prenylation(GO:0097354) |
| 0.0 | 0.2 | GO:1900026 | positive regulation of substrate adhesion-dependent cell spreading(GO:1900026) |
| 0.0 | 0.1 | GO:0034551 | respiratory chain complex III assembly(GO:0017062) mitochondrial respiratory chain complex III assembly(GO:0034551) mitochondrial respiratory chain complex III biogenesis(GO:0097033) |
| 0.0 | 0.1 | GO:0060571 | invagination involved in gastrulation with mouth forming second(GO:0055109) morphogenesis of an epithelial fold(GO:0060571) |
| 0.0 | 0.3 | GO:0061026 | cardiac muscle tissue regeneration(GO:0061026) |
| 0.0 | 0.2 | GO:0019883 | antigen processing and presentation of endogenous peptide antigen(GO:0002483) antigen processing and presentation of endogenous antigen(GO:0019883) antigen processing and presentation of endogenous peptide antigen via MHC class I(GO:0019885) |
| 0.0 | 0.3 | GO:0046686 | response to cadmium ion(GO:0046686) |
| 0.0 | 0.2 | GO:0006517 | protein deglycosylation(GO:0006517) |
| 0.0 | 0.1 | GO:0031591 | wybutosine metabolic process(GO:0031590) wybutosine biosynthetic process(GO:0031591) |
| 0.0 | 0.2 | GO:0009263 | deoxyribonucleotide biosynthetic process(GO:0009263) |
| 0.0 | 0.1 | GO:0006404 | RNA import into nucleus(GO:0006404) snRNA import into nucleus(GO:0061015) |
| 0.0 | 0.2 | GO:0006123 | mitochondrial electron transport, cytochrome c to oxygen(GO:0006123) |
| 0.0 | 0.1 | GO:0032196 | transposition(GO:0032196) |
| 0.0 | 0.2 | GO:0043328 | protein targeting to vacuole involved in ubiquitin-dependent protein catabolic process via the multivesicular body sorting pathway(GO:0043328) |
| 0.0 | 0.2 | GO:0042144 | vacuole fusion, non-autophagic(GO:0042144) |
| 0.0 | 0.2 | GO:0045453 | bone resorption(GO:0045453) |
| 0.0 | 0.3 | GO:0031468 | nuclear envelope reassembly(GO:0031468) |
| 0.0 | 0.3 | GO:0036444 | calcium ion transmembrane import into mitochondrion(GO:0036444) |
| 0.0 | 0.3 | GO:2000369 | regulation of clathrin-mediated endocytosis(GO:2000369) |
| 0.0 | 0.1 | GO:0006369 | termination of RNA polymerase II transcription(GO:0006369) |
| 0.0 | 0.1 | GO:0097477 | spinal cord motor neuron migration(GO:0097476) lateral motor column neuron migration(GO:0097477) |
| 0.0 | 0.1 | GO:0021825 | substrate-dependent cell migration(GO:0006929) pallium development(GO:0021543) cerebral cortex cell migration(GO:0021795) cerebral cortex tangential migration(GO:0021800) cerebral cortex tangential migration using cell-axon interactions(GO:0021824) substrate-dependent cerebral cortex tangential migration(GO:0021825) gonadotrophin-releasing hormone neuronal migration to the hypothalamus(GO:0021828) hypothalamic tangential migration using cell-axon interactions(GO:0021856) cerebral cortex development(GO:0021987) |
| 0.0 | 0.0 | GO:0042772 | DNA damage response, signal transduction by p53 class mediator resulting in transcription of p21 class mediator(GO:0006978) DNA damage response, signal transduction resulting in transcription(GO:0042772) |
| 0.0 | 0.4 | GO:0042632 | cholesterol homeostasis(GO:0042632) sterol homeostasis(GO:0055092) |
| 0.0 | 0.1 | GO:0045039 | protein import into mitochondrial inner membrane(GO:0045039) |
| 0.0 | 0.1 | GO:0051121 | lipoxygenase pathway(GO:0019372) linoleic acid metabolic process(GO:0043651) hepoxilin metabolic process(GO:0051121) hepoxilin biosynthetic process(GO:0051122) |
| 0.0 | 0.1 | GO:1902101 | positive regulation of mitotic metaphase/anaphase transition(GO:0045842) positive regulation of mitotic sister chromatid separation(GO:1901970) positive regulation of metaphase/anaphase transition of cell cycle(GO:1902101) |
| 0.0 | 0.1 | GO:0000395 | mRNA 5'-splice site recognition(GO:0000395) |
| 0.0 | 0.3 | GO:0035305 | negative regulation of dephosphorylation(GO:0035305) negative regulation of protein dephosphorylation(GO:0035308) |
| 0.0 | 0.2 | GO:0015701 | bicarbonate transport(GO:0015701) |
| 0.0 | 0.1 | GO:0046606 | negative regulation of centrosome duplication(GO:0010826) negative regulation of centrosome cycle(GO:0046606) |
| 0.0 | 0.1 | GO:0045292 | mRNA cis splicing, via spliceosome(GO:0045292) |
| 0.0 | 0.4 | GO:0048247 | lymphocyte chemotaxis(GO:0048247) |
| 0.0 | 0.2 | GO:1901642 | nucleoside transport(GO:0015858) nucleoside transmembrane transport(GO:1901642) |
| 0.0 | 0.4 | GO:0021522 | spinal cord motor neuron differentiation(GO:0021522) |
| 0.0 | 0.1 | GO:0046548 | retinal rod cell development(GO:0046548) |
| 0.0 | 0.3 | GO:0007286 | spermatid development(GO:0007286) |
| 0.0 | 0.2 | GO:0090502 | RNA phosphodiester bond hydrolysis, endonucleolytic(GO:0090502) |
| 0.0 | 0.2 | GO:1904262 | negative regulation of TORC1 signaling(GO:1904262) |
| 0.0 | 0.2 | GO:0016926 | protein desumoylation(GO:0016926) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.5 | GO:0046695 | SLIK (SAGA-like) complex(GO:0046695) |
| 0.1 | 0.3 | GO:0005968 | Rab-protein geranylgeranyltransferase complex(GO:0005968) |
| 0.1 | 0.2 | GO:0030906 | retromer, cargo-selective complex(GO:0030906) |
| 0.1 | 0.2 | GO:0005958 | DNA-dependent protein kinase-DNA ligase 4 complex(GO:0005958) |
| 0.1 | 0.2 | GO:0045293 | MIS complex(GO:0036396) mRNA editing complex(GO:0045293) |
| 0.0 | 0.2 | GO:0072357 | PTW/PP1 phosphatase complex(GO:0072357) |
| 0.0 | 0.2 | GO:1990513 | CLOCK-BMAL transcription complex(GO:1990513) |
| 0.0 | 0.2 | GO:0097025 | MPP7-DLG1-LIN7 complex(GO:0097025) |
| 0.0 | 0.2 | GO:0008622 | epsilon DNA polymerase complex(GO:0008622) |
| 0.0 | 0.2 | GO:0048269 | methionine adenosyltransferase complex(GO:0048269) |
| 0.0 | 0.1 | GO:0033065 | Rad51C-XRCC3 complex(GO:0033065) |
| 0.0 | 0.4 | GO:0000220 | vacuolar proton-transporting V-type ATPase, V0 domain(GO:0000220) |
| 0.0 | 0.2 | GO:0033263 | CORVET complex(GO:0033263) |
| 0.0 | 0.1 | GO:0097134 | cyclin E1-CDK2 complex(GO:0097134) |
| 0.0 | 0.3 | GO:0072546 | ER membrane protein complex(GO:0072546) |
| 0.0 | 0.2 | GO:0071986 | Ragulator complex(GO:0071986) |
| 0.0 | 0.1 | GO:0033179 | proton-transporting V-type ATPase, V0 domain(GO:0033179) |
| 0.0 | 0.3 | GO:0000930 | gamma-tubulin complex(GO:0000930) |
| 0.0 | 0.3 | GO:0000124 | SAGA complex(GO:0000124) |
| 0.0 | 0.3 | GO:1990246 | uniplex complex(GO:1990246) |
| 0.0 | 0.3 | GO:0000815 | ESCRT III complex(GO:0000815) |
| 0.0 | 0.1 | GO:0042719 | mitochondrial intermembrane space protein transporter complex(GO:0042719) |
| 0.0 | 0.1 | GO:0000941 | condensed nuclear chromosome inner kinetochore(GO:0000941) |
| 0.0 | 0.2 | GO:0031588 | nucleotide-activated protein kinase complex(GO:0031588) |
| 0.0 | 0.3 | GO:0033180 | proton-transporting V-type ATPase, V1 domain(GO:0033180) |
| 0.0 | 0.2 | GO:0005751 | mitochondrial respiratory chain complex IV(GO:0005751) |
| 0.0 | 0.2 | GO:0014704 | intercalated disc(GO:0014704) |
| 0.0 | 0.1 | GO:0030670 | phagocytic vesicle membrane(GO:0030670) |
| 0.0 | 0.4 | GO:0005680 | anaphase-promoting complex(GO:0005680) |
| 0.0 | 0.2 | GO:0005956 | protein kinase CK2 complex(GO:0005956) |
| 0.0 | 1.3 | GO:0005882 | intermediate filament(GO:0005882) |
| 0.0 | 0.2 | GO:0030057 | desmosome(GO:0030057) |
| 0.0 | 0.2 | GO:0048188 | Set1C/COMPASS complex(GO:0048188) |
| 0.0 | 0.1 | GO:0032045 | guanyl-nucleotide exchange factor complex(GO:0032045) |
| 0.0 | 0.1 | GO:0019773 | proteasome core complex, alpha-subunit complex(GO:0019773) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 1.1 | GO:0003983 | UTP:glucose-1-phosphate uridylyltransferase activity(GO:0003983) UTP-monosaccharide-1-phosphate uridylyltransferase activity(GO:0051748) |
| 0.1 | 0.5 | GO:0061656 | SUMO conjugating enzyme activity(GO:0061656) |
| 0.1 | 0.5 | GO:0043998 | H2A histone acetyltransferase activity(GO:0043998) |
| 0.1 | 0.3 | GO:0032574 | 5'-3' RNA helicase activity(GO:0032574) |
| 0.1 | 0.3 | GO:0004663 | Rab geranylgeranyltransferase activity(GO:0004663) |
| 0.1 | 0.2 | GO:0003909 | DNA ligase activity(GO:0003909) DNA ligase (ATP) activity(GO:0003910) |
| 0.1 | 0.3 | GO:0005528 | macrolide binding(GO:0005527) FK506 binding(GO:0005528) |
| 0.1 | 0.4 | GO:0035173 | histone kinase activity(GO:0035173) |
| 0.1 | 0.2 | GO:0071568 | UFM1 transferase activity(GO:0071568) |
| 0.1 | 0.4 | GO:0070888 | E-box binding(GO:0070888) |
| 0.0 | 0.2 | GO:0070182 | DNA polymerase binding(GO:0070182) |
| 0.0 | 0.2 | GO:0097016 | L27 domain binding(GO:0097016) |
| 0.0 | 0.3 | GO:0004045 | aminoacyl-tRNA hydrolase activity(GO:0004045) |
| 0.0 | 0.1 | GO:0001160 | transcription termination site sequence-specific DNA binding(GO:0001147) transcription termination site DNA binding(GO:0001160) |
| 0.0 | 0.2 | GO:0051731 | polynucleotide 5'-hydroxyl-kinase activity(GO:0051731) |
| 0.0 | 0.2 | GO:0016728 | ribonucleoside-diphosphate reductase activity, thioredoxin disulfide as acceptor(GO:0004748) oxidoreductase activity, acting on CH or CH2 groups, disulfide as acceptor(GO:0016728) ribonucleoside-diphosphate reductase activity(GO:0061731) |
| 0.0 | 0.2 | GO:0038036 | sphingosine-1-phosphate receptor activity(GO:0038036) |
| 0.0 | 0.3 | GO:0002161 | aminoacyl-tRNA editing activity(GO:0002161) |
| 0.0 | 0.7 | GO:0004181 | metallocarboxypeptidase activity(GO:0004181) |
| 0.0 | 0.2 | GO:0008420 | CTD phosphatase activity(GO:0008420) |
| 0.0 | 0.3 | GO:0017025 | TBP-class protein binding(GO:0017025) |
| 0.0 | 0.1 | GO:0000702 | oxidized base lesion DNA N-glycosylase activity(GO:0000702) |
| 0.0 | 0.3 | GO:0008307 | structural constituent of muscle(GO:0008307) |
| 0.0 | 0.2 | GO:0045294 | alpha-catenin binding(GO:0045294) |
| 0.0 | 0.4 | GO:0051117 | ATPase binding(GO:0051117) |
| 0.0 | 0.2 | GO:0003993 | acid phosphatase activity(GO:0003993) |
| 0.0 | 1.3 | GO:0005200 | structural constituent of cytoskeleton(GO:0005200) |
| 0.0 | 0.1 | GO:0008821 | crossover junction endodeoxyribonuclease activity(GO:0008821) |
| 0.0 | 0.2 | GO:0004523 | RNA-DNA hybrid ribonuclease activity(GO:0004523) |
| 0.0 | 0.3 | GO:0051721 | protein phosphatase 2A binding(GO:0051721) |
| 0.0 | 0.3 | GO:0032041 | histone deacetylase activity (H3-K14 specific)(GO:0031078) NAD-dependent histone deacetylase activity (H3-K14 specific)(GO:0032041) |
| 0.0 | 0.1 | GO:0004051 | arachidonate 5-lipoxygenase activity(GO:0004051) |
| 0.0 | 0.4 | GO:0016251 | obsolete general RNA polymerase II transcription factor activity(GO:0016251) |
| 0.0 | 0.2 | GO:0008504 | monoamine transmembrane transporter activity(GO:0008504) |
| 0.0 | 0.1 | GO:0016635 | oxidoreductase activity, acting on the CH-CH group of donors, quinone or related compound as acceptor(GO:0016635) |
| 0.0 | 0.1 | GO:0042030 | ATPase inhibitor activity(GO:0042030) |
| 0.0 | 0.1 | GO:0080019 | fatty-acyl-CoA reductase (alcohol-forming) activity(GO:0080019) |
| 0.0 | 0.2 | GO:1990239 | steroid hormone binding(GO:1990239) |
| 0.0 | 0.2 | GO:0004526 | ribonuclease P activity(GO:0004526) |
| 0.0 | 0.1 | GO:0016262 | protein N-acetylglucosaminyltransferase activity(GO:0016262) protein O-GlcNAc transferase activity(GO:0097363) |
| 0.0 | 0.1 | GO:0070139 | ubiquitin-like protein-specific endopeptidase activity(GO:0070137) SUMO-specific endopeptidase activity(GO:0070139) |
| 0.0 | 0.1 | GO:0003980 | UDP-glucose:glycoprotein glucosyltransferase activity(GO:0003980) |
| 0.0 | 0.3 | GO:0046961 | proton-transporting ATPase activity, rotational mechanism(GO:0046961) |
| 0.0 | 0.1 | GO:0035925 | AU-rich element binding(GO:0017091) mRNA 3'-UTR AU-rich region binding(GO:0035925) |
| 0.0 | 0.1 | GO:0008568 | microtubule-severing ATPase activity(GO:0008568) |
| 0.0 | 0.1 | GO:0039706 | co-receptor binding(GO:0039706) |
| 0.0 | 0.1 | GO:0008318 | protein prenyltransferase activity(GO:0008318) |
| 0.0 | 0.6 | GO:0031491 | nucleosome binding(GO:0031491) |
| 0.0 | 0.2 | GO:0005452 | inorganic anion exchanger activity(GO:0005452) |
| 0.0 | 0.4 | GO:0048020 | CCR chemokine receptor binding(GO:0048020) |
| 0.0 | 0.8 | GO:0004715 | non-membrane spanning protein tyrosine kinase activity(GO:0004715) |
| 0.0 | 0.3 | GO:0015035 | protein disulfide oxidoreductase activity(GO:0015035) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.3 | PID TCR RAS PATHWAY | Ras signaling in the CD4+ TCR pathway |
| 0.0 | 0.4 | PID RANBP2 PATHWAY | Sumoylation by RanBP2 regulates transcriptional repression |
| 0.0 | 1.1 | ST B CELL ANTIGEN RECEPTOR | B Cell Antigen Receptor |
| 0.0 | 0.6 | PID MYC PATHWAY | C-MYC pathway |
| 0.0 | 0.2 | PID S1P S1P4 PATHWAY | S1P4 pathway |
| 0.0 | 0.2 | PID SMAD2 3PATHWAY | Regulation of cytoplasmic and nuclear SMAD2/3 signaling |
| 0.0 | 0.5 | PID BARD1 PATHWAY | BARD1 signaling events |
| 0.0 | 0.3 | PID PRL SIGNALING EVENTS PATHWAY | Signaling events mediated by PRL |
| 0.0 | 0.3 | PID EPHA FWDPATHWAY | EPHA forward signaling |
| 0.0 | 0.2 | SA REG CASCADE OF CYCLIN EXPR | Expression of cyclins regulates progression through the cell cycle by activating cyclin-dependent kinases. |
| 0.0 | 0.2 | PID DNA PK PATHWAY | DNA-PK pathway in nonhomologous end joining |
| 0.0 | 0.4 | PID AJDISS 2PATHWAY | Posttranslational regulation of adherens junction stability and dissassembly |
| 0.0 | 0.2 | PID LIS1 PATHWAY | Lissencephaly gene (LIS1) in neuronal migration and development |
| 0.0 | 0.3 | PID ILK PATHWAY | Integrin-linked kinase signaling |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.5 | REACTOME PD1 SIGNALING | Genes involved in PD-1 signaling |
| 0.0 | 0.8 | REACTOME POST CHAPERONIN TUBULIN FOLDING PATHWAY | Genes involved in Post-chaperonin tubulin folding pathway |
| 0.0 | 0.3 | REACTOME RAF MAP KINASE CASCADE | Genes involved in RAF/MAP kinase cascade |
| 0.0 | 0.3 | REACTOME REGULATION OF THE FANCONI ANEMIA PATHWAY | Genes involved in Regulation of the Fanconi anemia pathway |
| 0.0 | 0.2 | REACTOME G BETA GAMMA SIGNALLING THROUGH PI3KGAMMA | Genes involved in G beta:gamma signalling through PI3Kgamma |
| 0.0 | 0.3 | REACTOME CREATION OF C4 AND C2 ACTIVATORS | Genes involved in Creation of C4 and C2 activators |
| 0.0 | 0.2 | REACTOME INTEGRATION OF PROVIRUS | Genes involved in Integration of provirus |
| 0.0 | 0.3 | REACTOME RECRUITMENT OF NUMA TO MITOTIC CENTROSOMES | Genes involved in Recruitment of NuMA to mitotic centrosomes |
| 0.0 | 0.3 | REACTOME TRANSFERRIN ENDOCYTOSIS AND RECYCLING | Genes involved in Transferrin endocytosis and recycling |
| 0.0 | 0.3 | REACTOME CELL EXTRACELLULAR MATRIX INTERACTIONS | Genes involved in Cell-extracellular matrix interactions |
| 0.0 | 0.4 | REACTOME RIP MEDIATED NFKB ACTIVATION VIA DAI | Genes involved in RIP-mediated NFkB activation via DAI |
| 0.0 | 0.6 | REACTOME ENDOSOMAL SORTING COMPLEX REQUIRED FOR TRANSPORT ESCRT | Genes involved in Endosomal Sorting Complex Required For Transport (ESCRT) |
| 0.0 | 0.2 | REACTOME ACTIVATION OF BH3 ONLY PROTEINS | Genes involved in Activation of BH3-only proteins |
| 0.0 | 0.3 | REACTOME PHOSPHORYLATION OF THE APC C | Genes involved in Phosphorylation of the APC/C |
| 0.0 | 0.3 | REACTOME DOWNREGULATION OF SMAD2 3 SMAD4 TRANSCRIPTIONAL ACTIVITY | Genes involved in Downregulation of SMAD2/3:SMAD4 transcriptional activity |
| 0.0 | 0.7 | REACTOME RNA POL II TRANSCRIPTION PRE INITIATION AND PROMOTER OPENING | Genes involved in RNA Polymerase II Transcription Pre-Initiation And Promoter Opening |
| 0.0 | 0.1 | REACTOME HORMONE SENSITIVE LIPASE HSL MEDIATED TRIACYLGLYCEROL HYDROLYSIS | Genes involved in Hormone-sensitive lipase (HSL)-mediated triacylglycerol hydrolysis |
| 0.0 | 0.2 | REACTOME ADP SIGNALLING THROUGH P2RY12 | Genes involved in ADP signalling through P2Y purinoceptor 12 |
| 0.0 | 0.1 | REACTOME OXYGEN DEPENDENT PROLINE HYDROXYLATION OF HYPOXIA INDUCIBLE FACTOR ALPHA | Genes involved in Oxygen-dependent Proline Hydroxylation of Hypoxia-inducible Factor Alpha |
| 0.0 | 0.1 | REACTOME MICRORNA MIRNA BIOGENESIS | Genes involved in MicroRNA (miRNA) Biogenesis |
| 0.0 | 0.1 | REACTOME RESOLUTION OF AP SITES VIA THE SINGLE NUCLEOTIDE REPLACEMENT PATHWAY | Genes involved in Resolution of AP sites via the single-nucleotide replacement pathway |