Project
PRJNA207719: Tissue specific transcriptome profiling
Navigation
Downloads
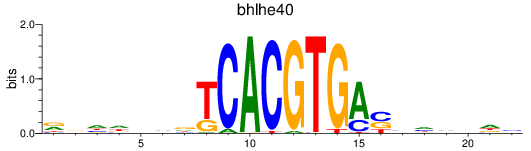
Results for bhlhe40
Z-value: 0.44
Transcription factors associated with bhlhe40
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
bhlhe40
|
ENSDARG00000004060 | basic helix-loop-helix family, member e40 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| bhlhe40 | dr11_v1_chr11_-_35763323_35763323 | 0.36 | 5.5e-01 | Click! |
Activity profile of bhlhe40 motif
Sorted Z-values of bhlhe40 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr10_+_5268054 | 0.22 |
ENSDART00000114491
|
ror2
|
receptor tyrosine kinase-like orphan receptor 2 |
| chr7_-_67214972 | 0.21 |
ENSDART00000156861
|
swap70a
|
switching B cell complex subunit SWAP70a |
| chr24_+_7631797 | 0.19 |
ENSDART00000187464
|
cavin1b
|
caveolae associated protein 1b |
| chr9_+_6587364 | 0.19 |
ENSDART00000122279
|
fhl2a
|
four and a half LIM domains 2a |
| chr5_-_4532516 | 0.16 |
ENSDART00000192398
|
cst14b.1
|
cystatin 14b, tandem duplicate 1 |
| chr10_-_15644904 | 0.14 |
ENSDART00000138389
ENSDART00000101191 ENSDART00000186559 ENSDART00000122170 |
smc5
|
structural maintenance of chromosomes 5 |
| chr13_+_7164345 | 0.12 |
ENSDART00000022051
|
gins1
|
GINS complex subunit 1 (Psf1 homolog) |
| chr21_-_5393125 | 0.12 |
ENSDART00000146061
|
psmd5
|
proteasome 26S subunit, non-ATPase 5 |
| chr15_-_28596507 | 0.12 |
ENSDART00000156800
|
si:ch211-225b7.5
|
si:ch211-225b7.5 |
| chr17_+_25331576 | 0.11 |
ENSDART00000157309
|
tmem54a
|
transmembrane protein 54a |
| chr5_-_66798729 | 0.11 |
ENSDART00000113077
|
FERMT3 (1 of many)
|
im:7154036 |
| chr10_+_5422575 | 0.11 |
ENSDART00000063093
|
auh
|
AU RNA binding protein/enoyl-CoA hydratase |
| chr16_+_26824691 | 0.11 |
ENSDART00000135053
|
zmp:0000001316
|
zmp:0000001316 |
| chr24_-_16917086 | 0.11 |
ENSDART00000110715
|
cmbl
|
carboxymethylenebutenolidase homolog (Pseudomonas) |
| chr8_+_41229233 | 0.11 |
ENSDART00000131135
|
zgc:152830
|
zgc:152830 |
| chr15_-_1534232 | 0.11 |
ENSDART00000056763
ENSDART00000133943 |
ift80
|
intraflagellar transport 80 homolog (Chlamydomonas) |
| chr7_+_22293894 | 0.10 |
ENSDART00000056790
|
tmem256
|
transmembrane protein 256 |
| chr25_+_10547228 | 0.10 |
ENSDART00000067678
|
zgc:110339
|
zgc:110339 |
| chr8_-_25817106 | 0.10 |
ENSDART00000099364
|
taf10
|
TAF10 RNA polymerase II, TATA box binding protein (TBP)-associated factor |
| chr11_-_44945636 | 0.10 |
ENSDART00000157658
|
orc2
|
origin recognition complex, subunit 2 |
| chr6_+_149405 | 0.10 |
ENSDART00000161154
|
fdx1l
|
ferredoxin 1-like |
| chr15_+_59417 | 0.10 |
ENSDART00000187260
|
AXL
|
AXL receptor tyrosine kinase |
| chr20_+_46213553 | 0.10 |
ENSDART00000100532
|
stx7l
|
syntaxin 7-like |
| chr9_+_36946340 | 0.10 |
ENSDART00000135281
|
si:dkey-3d4.3
|
si:dkey-3d4.3 |
| chr16_-_7228276 | 0.09 |
ENSDART00000149030
|
nt5c3a
|
5'-nucleotidase, cytosolic IIIA |
| chr16_+_33143503 | 0.09 |
ENSDART00000058471
ENSDART00000179385 |
rhbdl2
|
rhomboid, veinlet-like 2 (Drosophila) |
| chr9_-_27868267 | 0.09 |
ENSDART00000079502
|
dbr1
|
debranching RNA lariats 1 |
| chr14_-_36437249 | 0.09 |
ENSDART00000016728
|
aga
|
aspartylglucosaminidase |
| chr23_+_35847200 | 0.09 |
ENSDART00000129222
|
rarga
|
retinoic acid receptor gamma a |
| chr12_+_18529228 | 0.08 |
ENSDART00000193931
ENSDART00000146260 |
meiob
msrb1b
|
meiosis specific with OB domains methionine sulfoxide reductase B1b |
| chr16_+_20895904 | 0.08 |
ENSDART00000052662
|
hoxa13b
|
homeobox A13b |
| chr19_-_27334394 | 0.08 |
ENSDART00000052359
|
gtf2h4
|
general transcription factor IIH, polypeptide 4 |
| chr5_+_26204561 | 0.07 |
ENSDART00000137178
|
marveld2b
|
MARVEL domain containing 2b |
| chr9_-_10805231 | 0.07 |
ENSDART00000193913
ENSDART00000078348 |
si:ch1073-416j23.1
|
si:ch1073-416j23.1 |
| chr11_+_42556395 | 0.06 |
ENSDART00000039206
|
rps23
|
ribosomal protein S23 |
| chr2_-_32486080 | 0.06 |
ENSDART00000110821
|
ttc19
|
tetratricopeptide repeat domain 19 |
| chr8_+_23738122 | 0.06 |
ENSDART00000062983
|
rpl10a
|
ribosomal protein L10a |
| chr19_+_791538 | 0.06 |
ENSDART00000146554
ENSDART00000138406 |
tmem79a
|
transmembrane protein 79a |
| chr7_-_14446512 | 0.06 |
ENSDART00000041577
|
kif7
|
kinesin family member 7 |
| chr22_+_16759010 | 0.06 |
ENSDART00000079638
ENSDART00000113099 |
tm2d1
|
TM2 domain containing 1 |
| chr23_+_26079467 | 0.06 |
ENSDART00000129617
|
atp6ap1b
|
ATPase H+ transporting accessory protein 1b |
| chr4_-_9196291 | 0.05 |
ENSDART00000153963
|
hcfc2
|
host cell factor C2 |
| chr22_+_30184039 | 0.05 |
ENSDART00000049075
|
add3a
|
adducin 3 (gamma) a |
| chr15_-_41807371 | 0.05 |
ENSDART00000156819
|
slc25a36b
|
solute carrier family 25 (pyrimidine nucleotide carrier ), member 36b |
| chr7_-_41881177 | 0.05 |
ENSDART00000174258
ENSDART00000018972 |
zgc:92818
|
zgc:92818 |
| chr16_+_5251768 | 0.05 |
ENSDART00000144558
|
plecb
|
plectin b |
| chr21_-_14251306 | 0.05 |
ENSDART00000114715
ENSDART00000181380 |
man1b1a
|
mannosidase, alpha, class 1B, member 1a |
| chr13_-_12389748 | 0.05 |
ENSDART00000141606
|
commd8
|
COMM domain containing 8 |
| chr20_-_51100669 | 0.05 |
ENSDART00000023488
|
atp6v1d
|
ATPase H+ transporting V1 subunit D |
| chr1_+_54766943 | 0.05 |
ENSDART00000144759
|
nlrc6
|
NLR family CARD domain containing 6 |
| chr5_+_57210237 | 0.05 |
ENSDART00000167660
|
pja2
|
praja ring finger ubiquitin ligase 2 |
| chr9_-_38368138 | 0.05 |
ENSDART00000059574
|
ccdc93
|
coiled-coil domain containing 93 |
| chr3_+_16922226 | 0.04 |
ENSDART00000017646
|
atp6v0a1a
|
ATPase H+ transporting V0 subunit a1a |
| chr19_-_5103313 | 0.04 |
ENSDART00000037007
|
tpi1a
|
triosephosphate isomerase 1a |
| chr12_+_17933775 | 0.04 |
ENSDART00000186047
ENSDART00000160586 |
trrap
|
transformation/transcription domain-associated protein |
| chr11_-_236984 | 0.04 |
ENSDART00000170778
|
dusp7
|
dual specificity phosphatase 7 |
| chr17_-_2126517 | 0.04 |
ENSDART00000013093
|
zfyve19
|
zinc finger, FYVE domain containing 19 |
| chr19_-_22346582 | 0.04 |
ENSDART00000045675
ENSDART00000169065 |
slc52a2
zgc:109744
|
solute carrier family 52 (riboflavin transporter), member 2 zgc:109744 |
| chr25_+_3788074 | 0.04 |
ENSDART00000154008
|
chid1
|
chitinase domain containing 1 |
| chr13_+_40692804 | 0.04 |
ENSDART00000109822
|
hps1
|
Hermansky-Pudlak syndrome 1 |
| chr21_+_11916788 | 0.04 |
ENSDART00000136103
|
ubap2a
|
ubiquitin associated protein 2a |
| chr25_-_6223567 | 0.04 |
ENSDART00000067512
|
psma4
|
proteasome subunit alpha 4 |
| chr23_-_31913231 | 0.03 |
ENSDART00000146852
ENSDART00000085054 |
mtfr2
|
mitochondrial fission regulator 2 |
| chr19_+_15440841 | 0.03 |
ENSDART00000182329
|
lin28a
|
lin-28 homolog A (C. elegans) |
| chr18_+_31280984 | 0.03 |
ENSDART00000170285
ENSDART00000150608 ENSDART00000159720 |
def8
|
differentially expressed in FDCP 8 homolog (mouse) |
| chr20_-_29683754 | 0.03 |
ENSDART00000130599
ENSDART00000015928 ENSDART00000131219 |
si:ch211-195d17.2
|
si:ch211-195d17.2 |
| chr17_+_30843881 | 0.03 |
ENSDART00000149600
ENSDART00000148547 |
tpp1
|
tripeptidyl peptidase I |
| chr20_-_51386511 | 0.03 |
ENSDART00000151429
ENSDART00000066072 |
rnf8
|
ring finger protein 8, E3 ubiquitin protein ligase |
| chr23_-_2901167 | 0.03 |
ENSDART00000165955
ENSDART00000190616 |
zhx3
|
zinc fingers and homeoboxes 3 |
| chr4_+_17353714 | 0.03 |
ENSDART00000136299
|
nup37
|
nucleoporin 37 |
| chr17_-_14705039 | 0.03 |
ENSDART00000154281
ENSDART00000123550 |
ptp4a2a
|
protein tyrosine phosphatase type IVA, member 2a |
| chr25_+_3788443 | 0.03 |
ENSDART00000189747
|
chid1
|
chitinase domain containing 1 |
| chr14_+_94946 | 0.03 |
ENSDART00000165766
ENSDART00000163778 |
mcm7
|
minichromosome maintenance complex component 7 |
| chr23_+_9035578 | 0.03 |
ENSDART00000140455
|
ccm2l
|
cerebral cavernous malformation 2-like |
| chr12_+_34891529 | 0.02 |
ENSDART00000015643
|
tbcc
|
tubulin folding cofactor C |
| chr5_-_22130937 | 0.02 |
ENSDART00000138606
|
las1l
|
LAS1-like, ribosome biogenesis factor |
| chr24_-_21498802 | 0.02 |
ENSDART00000181235
ENSDART00000153695 |
atp8a2
|
ATPase phospholipid transporting 8A2 |
| chr19_+_15441022 | 0.02 |
ENSDART00000098970
ENSDART00000140276 |
lin28a
|
lin-28 homolog A (C. elegans) |
| chr8_-_41228530 | 0.02 |
ENSDART00000165949
ENSDART00000173055 |
fahd2a
|
fumarylacetoacetate hydrolase domain containing 2A |
| chr12_+_19958845 | 0.02 |
ENSDART00000193248
|
ercc4
|
excision repair cross-complementation group 4 |
| chr15_+_6652396 | 0.02 |
ENSDART00000192813
ENSDART00000157678 |
nop53
|
NOP53 ribosome biogenesis factor |
| chr24_-_6024466 | 0.02 |
ENSDART00000040865
|
pdss1
|
prenyl (decaprenyl) diphosphate synthase, subunit 1 |
| chr24_-_35534273 | 0.02 |
ENSDART00000026578
|
ube2v2
|
ubiquitin-conjugating enzyme E2 variant 2 |
| chr12_-_43438148 | 0.02 |
ENSDART00000004122
|
ptprea
|
protein tyrosine phosphatase, receptor type, E, a |
| chr5_+_61839707 | 0.02 |
ENSDART00000163051
|
CR759879.1
|
Danio rerio interferon-induced protein with tetratricopeptide repeats 5 (LOC572297), mRNA. |
| chr18_+_39074139 | 0.02 |
ENSDART00000142390
|
gnb5b
|
guanine nucleotide binding protein (G protein), beta 5b |
| chr24_-_42072886 | 0.02 |
ENSDART00000171389
|
CABZ01095370.1
|
|
| chr6_+_12968101 | 0.02 |
ENSDART00000013781
|
mcm6
|
minichromosome maintenance complex component 6 |
| chr5_+_32835219 | 0.02 |
ENSDART00000140832
ENSDART00000186055 |
si:ch211-208h16.4
|
si:ch211-208h16.4 |
| chr23_+_31913292 | 0.02 |
ENSDART00000136910
|
armc1l
|
armadillo repeat containing 1, like |
| chr10_+_32050906 | 0.02 |
ENSDART00000137373
|
si:ch211-266i6.3
|
si:ch211-266i6.3 |
| chr16_+_7985886 | 0.02 |
ENSDART00000126041
|
ano10a
|
anoctamin 10a |
| chr4_+_17655872 | 0.01 |
ENSDART00000066999
|
washc3
|
WASH complex subunit 3 |
| chr16_+_19029297 | 0.01 |
ENSDART00000115263
ENSDART00000114954 |
rapgef5b
|
Rap guanine nucleotide exchange factor (GEF) 5b |
| chr23_+_32028574 | 0.01 |
ENSDART00000145501
ENSDART00000143121 ENSDART00000111877 |
tpx2
|
TPX2, microtubule-associated, homolog (Xenopus laevis) |
| chr16_+_25296389 | 0.01 |
ENSDART00000114528
|
tbc1d31
|
TBC1 domain family, member 31 |
| chr17_+_32622933 | 0.01 |
ENSDART00000077418
|
ctsba
|
cathepsin Ba |
| chr13_+_12389849 | 0.01 |
ENSDART00000086525
|
atp10d
|
ATPase phospholipid transporting 10D |
| chr6_+_7123980 | 0.01 |
ENSDART00000179738
ENSDART00000151311 |
si:ch211-237c6.4
|
si:ch211-237c6.4 |
| chr17_-_37299394 | 0.01 |
ENSDART00000154414
|
ptgr2
|
prostaglandin reductase 2 |
| chr19_+_9111550 | 0.01 |
ENSDART00000088336
|
setdb1a
|
SET domain, bifurcated 1a |
| chr20_-_45040916 | 0.01 |
ENSDART00000190001
|
klhl29
|
kelch-like family member 29 |
| chr23_+_42131509 | 0.01 |
ENSDART00000184544
|
SHISAL2A
|
shisa like 2A |
| chr10_-_24724388 | 0.00 |
ENSDART00000148582
|
smpd1
|
sphingomyelin phosphodiesterase 1, acid lysosomal |
| chr5_+_41476443 | 0.00 |
ENSDART00000145228
ENSDART00000137981 ENSDART00000142538 |
pias2
|
protein inhibitor of activated STAT, 2 |
| chr1_-_54765262 | 0.00 |
ENSDART00000150362
|
si:ch211-197k17.3
|
si:ch211-197k17.3 |
| chr23_-_22130778 | 0.00 |
ENSDART00000079212
|
phc2a
|
polyhomeotic homolog 2a (Drosophila) |
| chr12_-_7280551 | 0.00 |
ENSDART00000061633
|
zgc:171971
|
zgc:171971 |
| chr24_+_12989727 | 0.00 |
ENSDART00000126842
ENSDART00000129309 |
flj11011l
|
hypothetical protein FLJ11011-like (H. sapiens) |
| chr23_-_10786400 | 0.00 |
ENSDART00000055038
|
rybpa
|
RING1 and YY1 binding protein a |
| chr20_-_40766387 | 0.00 |
ENSDART00000061173
|
hsdl1
|
hydroxysteroid dehydrogenase like 1 |
| chr15_-_26570948 | 0.00 |
ENSDART00000156621
|
wdr81
|
WD repeat domain 81 |
| chr2_+_50967947 | 0.00 |
ENSDART00000162288
|
si:ch211-249o11.5
|
si:ch211-249o11.5 |
| chr3_-_25420931 | 0.00 |
ENSDART00000109601
ENSDART00000182184 |
bptf
|
bromodomain PHD finger transcription factor |
| chr15_+_1148074 | 0.00 |
ENSDART00000152638
ENSDART00000152466 ENSDART00000188011 |
mlf1
|
myeloid leukemia factor 1 |
| chr17_-_25331439 | 0.00 |
ENSDART00000155422
ENSDART00000082324 |
zpcx
|
zona pellucida protein C |
Network of associatons between targets according to the STRING database.
First level regulatory network of bhlhe40
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.2 | GO:0006361 | transcription initiation from RNA polymerase I promoter(GO:0006361) |
| 0.0 | 0.1 | GO:1990519 | pyrimidine nucleotide transport(GO:0006864) mitochondrial pyrimidine nucleotide import(GO:1990519) |
| 0.0 | 0.0 | GO:0044878 | abscission(GO:0009838) mitotic cytokinesis checkpoint(GO:0044878) |
| 0.0 | 0.1 | GO:1901207 | regulation of heart looping(GO:1901207) |
| 0.0 | 0.0 | GO:0019242 | methylglyoxal biosynthetic process(GO:0019242) glyceraldehyde-3-phosphate biosynthetic process(GO:0046166) |
| 0.0 | 0.1 | GO:0097033 | respiratory chain complex III assembly(GO:0017062) mitochondrial respiratory chain complex III assembly(GO:0034551) mitochondrial respiratory chain complex III biogenesis(GO:0097033) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | GO:0000811 | GINS complex(GO:0000811) |
| 0.0 | 0.1 | GO:0030915 | Smc5-Smc6 complex(GO:0030915) |
| 0.0 | 0.1 | GO:0035301 | Hedgehog signaling complex(GO:0035301) |
| 0.0 | 0.1 | GO:0000439 | core TFIIH complex(GO:0000439) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.2 | GO:0042134 | rRNA primary transcript binding(GO:0042134) |
| 0.0 | 0.1 | GO:0008310 | single-stranded DNA 3'-5' exodeoxyribonuclease activity(GO:0008310) L-methionine-(R)-S-oxide reductase activity(GO:0033745) |
| 0.0 | 0.1 | GO:0015218 | pyrimidine nucleotide transmembrane transporter activity(GO:0015218) |
| 0.0 | 0.1 | GO:0004300 | enoyl-CoA hydratase activity(GO:0004300) |
| 0.0 | 0.0 | GO:0004807 | triose-phosphate isomerase activity(GO:0004807) methylglyoxal synthase activity(GO:0008929) |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.2 | REACTOME UNWINDING OF DNA | Genes involved in Unwinding of DNA |