Project
PRJNA207719: Tissue specific transcriptome profiling
Navigation
Downloads
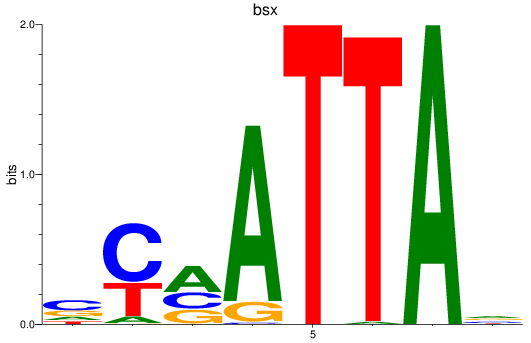
Results for bsx
Z-value: 0.48
Transcription factors associated with bsx
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
bsx
|
ENSDARG00000068976 | brain-specific homeobox |
|
bsx
|
ENSDARG00000110782 | brain-specific homeobox |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| bsx | dr11_v1_chr10_-_29892486_29892486 | 0.25 | 6.8e-01 | Click! |
Activity profile of bsx motif
Sorted Z-values of bsx motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr25_+_3326885 | 0.26 |
ENSDART00000104866
|
ldhbb
|
lactate dehydrogenase Bb |
| chr10_+_21867307 | 0.26 |
ENSDART00000126629
|
cbln17
|
cerebellin 17 |
| chr7_-_27685365 | 0.24 |
ENSDART00000188342
|
calca
|
calcitonin/calcitonin-related polypeptide, alpha |
| chr25_+_3327071 | 0.24 |
ENSDART00000136131
ENSDART00000133243 |
ldhbb
|
lactate dehydrogenase Bb |
| chr5_-_25723079 | 0.22 |
ENSDART00000014013
|
gda
|
guanine deaminase |
| chr11_-_37997419 | 0.21 |
ENSDART00000102870
|
slc41a1
|
solute carrier family 41 (magnesium transporter), member 1 |
| chr21_+_6751405 | 0.20 |
ENSDART00000037265
ENSDART00000146371 |
olfm1b
|
olfactomedin 1b |
| chr11_+_37178271 | 0.19 |
ENSDART00000161771
|
itih3b
|
inter-alpha-trypsin inhibitor heavy chain 3b |
| chr20_-_42702832 | 0.17 |
ENSDART00000134689
ENSDART00000045816 |
plg
|
plasminogen |
| chr13_-_20381485 | 0.17 |
ENSDART00000131351
|
si:ch211-270n8.1
|
si:ch211-270n8.1 |
| chr3_+_17456428 | 0.17 |
ENSDART00000090676
ENSDART00000182082 |
si:ch211-210g13.5
|
si:ch211-210g13.5 |
| chr5_-_8483457 | 0.17 |
ENSDART00000171886
ENSDART00000159477 ENSDART00000157423 |
lifra
|
leukemia inhibitory factor receptor alpha a |
| chr21_+_6751760 | 0.16 |
ENSDART00000135914
|
olfm1b
|
olfactomedin 1b |
| chr24_-_31846366 | 0.13 |
ENSDART00000155295
|
steap2
|
STEAP family member 2, metalloreductase |
| chr11_-_6452444 | 0.13 |
ENSDART00000137879
ENSDART00000134957 ENSDART00000004483 |
larp6b
|
La ribonucleoprotein domain family, member 6b |
| chr20_-_23426339 | 0.13 |
ENSDART00000004625
|
zar1
|
zygote arrest 1 |
| chr25_-_5119162 | 0.12 |
ENSDART00000153961
|
shisal1b
|
shisa like 1b |
| chr12_-_42368296 | 0.11 |
ENSDART00000171075
|
zgc:111868
|
zgc:111868 |
| chr10_+_35554219 | 0.11 |
ENSDART00000077373
|
zdhhc20a
|
zinc finger, DHHC-type containing 20a |
| chr1_-_47071979 | 0.11 |
ENSDART00000160817
|
itsn1
|
intersectin 1 (SH3 domain protein) |
| chr4_+_72797711 | 0.11 |
ENSDART00000190934
ENSDART00000163236 |
MYRFL
|
myelin regulatory factor-like |
| chr21_+_10794914 | 0.11 |
ENSDART00000084035
|
znf532
|
zinc finger protein 532 |
| chr3_+_46459540 | 0.10 |
ENSDART00000188150
|
si:ch211-66e2.5
|
si:ch211-66e2.5 |
| chr8_-_42238543 | 0.10 |
ENSDART00000062697
|
gfra2a
|
GDNF family receptor alpha 2a |
| chr9_-_20372977 | 0.10 |
ENSDART00000113418
|
igsf3
|
immunoglobulin superfamily, member 3 |
| chr15_-_34408777 | 0.10 |
ENSDART00000139934
|
agmo
|
alkylglycerol monooxygenase |
| chr21_-_43666420 | 0.10 |
ENSDART00000139008
ENSDART00000183996 ENSDART00000183395 |
si:dkey-229d11.3
si:dkey-229d11.5
|
si:dkey-229d11.3 si:dkey-229d11.5 |
| chr20_+_52442870 | 0.10 |
ENSDART00000163100
|
arhgap39
|
Rho GTPase activating protein 39 |
| chr5_-_9216758 | 0.10 |
ENSDART00000134896
ENSDART00000147000 |
lrp13
|
low-density lipoprotein receptor related-protein 13 |
| chr3_+_17537352 | 0.10 |
ENSDART00000104549
|
hcrt
|
hypocretin (orexin) neuropeptide precursor |
| chr14_-_1454045 | 0.10 |
ENSDART00000161460
|
pmt
|
phosphoethanolamine methyltransferase |
| chr16_+_5774977 | 0.10 |
ENSDART00000134202
|
ccka
|
cholecystokinin a |
| chr3_-_18711288 | 0.09 |
ENSDART00000183885
|
grid2ipa
|
glutamate receptor, ionotropic, delta 2 (Grid2) interacting protein, a |
| chr16_+_47207691 | 0.09 |
ENSDART00000062507
|
ica1
|
islet cell autoantigen 1 |
| chr24_-_41220538 | 0.09 |
ENSDART00000150207
|
acvr2ba
|
activin A receptor type 2Ba |
| chr19_+_14921000 | 0.09 |
ENSDART00000144052
|
oprd1a
|
opioid receptor, delta 1a |
| chr23_+_40460333 | 0.09 |
ENSDART00000184658
|
soga3b
|
SOGA family member 3b |
| chr16_-_29458806 | 0.09 |
ENSDART00000047931
|
lingo4b
|
leucine rich repeat and Ig domain containing 4b |
| chr19_-_5103313 | 0.09 |
ENSDART00000037007
|
tpi1a
|
triosephosphate isomerase 1a |
| chr8_+_36500061 | 0.09 |
ENSDART00000185840
|
slc7a4
|
solute carrier family 7, member 4 |
| chr18_-_8857137 | 0.09 |
ENSDART00000126331
|
prrt4
|
proline-rich transmembrane protein 4 |
| chr19_+_5543072 | 0.09 |
ENSDART00000082080
|
jupb
|
junction plakoglobin b |
| chr23_-_44677315 | 0.09 |
ENSDART00000143054
|
kif1c
|
kinesin family member 1C |
| chr11_-_12998400 | 0.09 |
ENSDART00000018614
|
chrna4b
|
cholinergic receptor, nicotinic, alpha 4b |
| chr14_-_1454413 | 0.09 |
ENSDART00000185403
ENSDART00000191357 |
pmt
|
phosphoethanolamine methyltransferase |
| chr9_+_17983463 | 0.09 |
ENSDART00000182150
|
akap11
|
A kinase (PRKA) anchor protein 11 |
| chr6_-_40352215 | 0.09 |
ENSDART00000103992
|
ttll3
|
tubulin tyrosine ligase-like family, member 3 |
| chr20_-_45722895 | 0.09 |
ENSDART00000153273
|
gpcpd1
|
glycerophosphocholine phosphodiesterase 1 |
| chr20_-_47188966 | 0.09 |
ENSDART00000152965
|
si:dkeyp-104f11.9
|
si:dkeyp-104f11.9 |
| chr5_+_37032038 | 0.08 |
ENSDART00000045036
|
nfkbib
|
nuclear factor of kappa light polypeptide gene enhancer in B-cells inhibitor, beta |
| chr19_-_5103141 | 0.08 |
ENSDART00000150952
|
tpi1a
|
triosephosphate isomerase 1a |
| chr24_-_41267184 | 0.08 |
ENSDART00000063504
|
xylb
|
xylulokinase homolog (H. influenzae) |
| chr5_+_50879545 | 0.08 |
ENSDART00000128402
|
nol6
|
nucleolar protein 6 (RNA-associated) |
| chr11_+_30057762 | 0.08 |
ENSDART00000164139
|
nhsb
|
Nance-Horan syndrome b (congenital cataracts and dental anomalies) |
| chr2_-_42843797 | 0.08 |
ENSDART00000137913
|
adcy8
|
adenylate cyclase 8 (brain) |
| chr6_-_50204262 | 0.08 |
ENSDART00000163648
|
raly
|
RALY heterogeneous nuclear ribonucleoprotein |
| chr12_-_42214 | 0.08 |
ENSDART00000045071
|
foxk2
|
forkhead box K2 |
| chr3_-_3448095 | 0.08 |
ENSDART00000078886
|
A2ML1 (1 of many)
|
si:dkey-46g23.5 |
| chr6_+_39232245 | 0.08 |
ENSDART00000187351
|
b4galnt1b
|
beta-1,4-N-acetyl-galactosaminyl transferase 1b |
| chr6_-_39649504 | 0.08 |
ENSDART00000179960
ENSDART00000190951 |
larp4ab
|
La ribonucleoprotein domain family, member 4Ab |
| chr18_-_46135927 | 0.08 |
ENSDART00000134537
ENSDART00000191481 |
SPTBN4
|
si:ch73-262h23.4 |
| chr8_-_1838315 | 0.08 |
ENSDART00000114476
ENSDART00000140077 |
pi4kab
|
phosphatidylinositol 4-kinase, catalytic, alpha b |
| chr9_+_21146862 | 0.08 |
ENSDART00000136365
|
hao2
|
hydroxyacid oxidase 2 (long chain) |
| chr2_-_52455525 | 0.08 |
ENSDART00000160768
ENSDART00000002241 |
chico
|
chico |
| chr20_-_45060241 | 0.08 |
ENSDART00000185227
|
klhl29
|
kelch-like family member 29 |
| chr5_-_69923241 | 0.08 |
ENSDART00000187389
|
fktn
|
fukutin |
| chr2_+_20332044 | 0.08 |
ENSDART00000112131
|
plppr4a
|
phospholipid phosphatase related 4a |
| chr17_+_37253706 | 0.08 |
ENSDART00000076004
|
tmem62
|
transmembrane protein 62 |
| chr6_-_50203682 | 0.08 |
ENSDART00000083999
ENSDART00000143050 |
raly
|
RALY heterogeneous nuclear ribonucleoprotein |
| chr15_-_16704417 | 0.08 |
ENSDART00000155163
|
caln1
|
calneuron 1 |
| chr16_+_39159752 | 0.07 |
ENSDART00000122081
|
sybu
|
syntabulin (syntaxin-interacting) |
| chr2_+_56891858 | 0.07 |
ENSDART00000159912
|
zgc:85843
|
zgc:85843 |
| chr13_+_25720969 | 0.07 |
ENSDART00000046050
|
pcbd1
|
pterin-4 alpha-carbinolamine dehydratase/dimerization cofactor of hepatocyte nuclear factor 1 alpha |
| chr23_+_45584223 | 0.07 |
ENSDART00000149367
|
si:ch73-290k24.5
|
si:ch73-290k24.5 |
| chr10_-_34772211 | 0.07 |
ENSDART00000145450
ENSDART00000134307 |
dclk1a
|
doublecortin-like kinase 1a |
| chr25_-_4482449 | 0.07 |
ENSDART00000056278
ENSDART00000149425 |
slc25a22a
|
solute carrier family 25 member 22a |
| chr12_+_5081759 | 0.07 |
ENSDART00000164178
|
prrt2
|
proline-rich transmembrane protein 2 |
| chr5_+_26805608 | 0.07 |
ENSDART00000111019
|
rabl6b
|
RAB, member RAS oncogene family-like 6b |
| chr17_-_200316 | 0.07 |
ENSDART00000190561
|
CABZ01083778.1
|
|
| chr21_-_42007213 | 0.07 |
ENSDART00000188804
ENSDART00000092821 ENSDART00000165743 |
gabrg2
|
gamma-aminobutyric acid (GABA) A receptor, gamma 2 |
| chr4_+_12615836 | 0.07 |
ENSDART00000003583
|
lmo3
|
LIM domain only 3 |
| chr15_+_29662401 | 0.07 |
ENSDART00000135540
|
nrip1a
|
nuclear receptor interacting protein 1a |
| chr19_-_25271155 | 0.07 |
ENSDART00000104027
|
rims3
|
regulating synaptic membrane exocytosis 3 |
| chr12_-_15620090 | 0.07 |
ENSDART00000038032
|
acbd4
|
acyl-CoA binding domain containing 4 |
| chr8_-_53166975 | 0.07 |
ENSDART00000114683
|
rbsn
|
rabenosyn, RAB effector |
| chr9_+_4250918 | 0.07 |
ENSDART00000164543
|
kalrna
|
kalirin RhoGEF kinase a |
| chr25_-_32869794 | 0.07 |
ENSDART00000162784
|
tmem266
|
transmembrane protein 266 |
| chr21_-_4764120 | 0.07 |
ENSDART00000129355
ENSDART00000102643 |
camsap1a
|
calmodulin regulated spectrin-associated protein 1a |
| chr24_-_10021341 | 0.07 |
ENSDART00000137250
|
zgc:173856
|
zgc:173856 |
| chr7_+_60551133 | 0.07 |
ENSDART00000148038
|
lrfn4b
|
leucine rich repeat and fibronectin type III domain containing 4b |
| chr2_+_34967210 | 0.07 |
ENSDART00000141796
|
astn1
|
astrotactin 1 |
| chr10_-_3258073 | 0.07 |
ENSDART00000113162
|
pi4kaa
|
phosphatidylinositol 4-kinase, catalytic, alpha a |
| chr11_+_1867613 | 0.07 |
ENSDART00000065470
|
rbms2a
|
RNA binding motif, single stranded interacting protein 2a |
| chr13_+_33282095 | 0.07 |
ENSDART00000135200
|
ccdc28b
|
coiled-coil domain containing 28B |
| chr4_-_14191434 | 0.07 |
ENSDART00000142374
ENSDART00000136730 |
pus7l
|
pseudouridylate synthase 7-like |
| chr5_+_32007627 | 0.07 |
ENSDART00000183061
|
scai
|
suppressor of cancer cell invasion |
| chr2_-_23541059 | 0.07 |
ENSDART00000133908
|
si:dkey-58b18.3
|
si:dkey-58b18.3 |
| chr17_-_12389259 | 0.07 |
ENSDART00000185724
|
snap25b
|
synaptosomal-associated protein, 25b |
| chr11_-_18323059 | 0.07 |
ENSDART00000182590
|
SFMBT1
|
Scm like with four mbt domains 1 |
| chr3_+_28860283 | 0.07 |
ENSDART00000077235
|
alg1
|
ALG1, chitobiosyldiphosphodolichol beta-mannosyltransferase |
| chr4_+_40265583 | 0.07 |
ENSDART00000151870
|
si:ch211-246n15.2
|
si:ch211-246n15.2 |
| chr2_-_38287987 | 0.06 |
ENSDART00000185329
ENSDART00000061677 |
si:ch211-14a17.6
|
si:ch211-14a17.6 |
| chr1_+_16397063 | 0.06 |
ENSDART00000159794
|
micu3a
|
mitochondrial calcium uptake family, member 3a |
| chr11_+_30000814 | 0.06 |
ENSDART00000191011
ENSDART00000189770 |
nhsb
|
Nance-Horan syndrome b (congenital cataracts and dental anomalies) |
| chr3_+_18807006 | 0.06 |
ENSDART00000180091
|
tnpo2
|
transportin 2 (importin 3, karyopherin beta 2b) |
| chr5_-_50640171 | 0.06 |
ENSDART00000183353
|
mctp1a
|
multiple C2 domains, transmembrane 1a |
| chr20_-_32110882 | 0.06 |
ENSDART00000030324
|
grm1a
|
glutamate receptor, metabotropic 1a |
| chr21_-_2322102 | 0.06 |
ENSDART00000162867
|
zgc:66483
|
zgc:66483 |
| chr14_-_2400668 | 0.06 |
ENSDART00000172717
ENSDART00000182882 |
si:ch73-233f7.1
|
si:ch73-233f7.1 |
| chr9_-_52962521 | 0.06 |
ENSDART00000170419
|
CU855885.1
|
|
| chr24_+_40860320 | 0.06 |
ENSDART00000161351
|
gorasp1b
|
golgi reassembly stacking protein 1b |
| chr23_-_14990865 | 0.06 |
ENSDART00000147799
|
ndrg3b
|
ndrg family member 3b |
| chr11_-_1024483 | 0.06 |
ENSDART00000017551
|
slc6a1b
|
solute carrier family 6 (neurotransmitter transporter), member 1b |
| chr11_+_25583950 | 0.06 |
ENSDART00000111961
|
ccdc120
|
coiled-coil domain containing 120 |
| chr11_+_34523132 | 0.06 |
ENSDART00000192257
|
zmat3
|
zinc finger, matrin-type 3 |
| chr8_+_53423408 | 0.06 |
ENSDART00000164792
|
cacna1db
|
calcium channel, voltage-dependent, L type, alpha 1D subunit, b |
| chr3_-_35800221 | 0.06 |
ENSDART00000031390
|
caskin1
|
CASK interacting protein 1 |
| chr14_-_24110251 | 0.06 |
ENSDART00000079226
|
cpeb4a
|
cytoplasmic polyadenylation element binding protein 4a |
| chr11_+_41089574 | 0.06 |
ENSDART00000170023
|
phf13
|
PHD finger protein 13 |
| chr25_-_27722614 | 0.06 |
ENSDART00000190154
|
zgc:153935
|
zgc:153935 |
| chr15_-_16177603 | 0.06 |
ENSDART00000156352
|
si:ch211-259g3.4
|
si:ch211-259g3.4 |
| chr21_-_30994577 | 0.06 |
ENSDART00000065503
|
pgap2
|
post-GPI attachment to proteins 2 |
| chr7_+_25059845 | 0.06 |
ENSDART00000077215
|
ppp2r5b
|
protein phosphatase 2, regulatory subunit B', beta |
| chr19_+_1831911 | 0.06 |
ENSDART00000166653
|
ptk2aa
|
protein tyrosine kinase 2aa |
| chr16_+_14707960 | 0.06 |
ENSDART00000137912
|
col14a1a
|
collagen, type XIV, alpha 1a |
| chr12_+_38563373 | 0.06 |
ENSDART00000134670
ENSDART00000193668 |
ttyh2
|
tweety family member 2 |
| chr1_-_12064715 | 0.06 |
ENSDART00000143628
ENSDART00000103406 |
pla2g12a
|
phospholipase A2, group XIIA |
| chr10_+_21730585 | 0.06 |
ENSDART00000188576
|
pcdh1g22
|
protocadherin 1 gamma 22 |
| chr1_-_39909985 | 0.06 |
ENSDART00000181673
|
stox2a
|
storkhead box 2a |
| chr5_-_10946232 | 0.06 |
ENSDART00000163139
ENSDART00000031265 |
rtn4r
|
reticulon 4 receptor |
| chr2_-_24444838 | 0.06 |
ENSDART00000147885
ENSDART00000164720 |
kcnn1a
|
potassium intermediate/small conductance calcium-activated channel, subfamily N, member 1a |
| chr10_+_42690374 | 0.06 |
ENSDART00000123496
|
rhobtb2b
|
Rho-related BTB domain containing 2b |
| chr13_-_49707885 | 0.06 |
ENSDART00000189805
ENSDART00000128415 |
tomm20a
|
translocase of outer mitochondrial membrane 20 |
| chr9_-_24031461 | 0.06 |
ENSDART00000021218
|
rpe
|
ribulose-5-phosphate-3-epimerase |
| chr7_-_72261721 | 0.06 |
ENSDART00000172229
|
RASGRP2
|
RAS guanyl releasing protein 2 |
| chr16_-_15988320 | 0.06 |
ENSDART00000160883
|
CABZ01060453.1
|
|
| chr13_-_37545487 | 0.06 |
ENSDART00000131269
|
syt16
|
synaptotagmin XVI |
| chr11_+_18130300 | 0.06 |
ENSDART00000169146
|
zgc:175135
|
zgc:175135 |
| chr4_-_193762 | 0.06 |
ENSDART00000169667
|
ptpro
|
protein tyrosine phosphatase, receptor type, O |
| chr8_+_7756893 | 0.06 |
ENSDART00000191894
|
fgd1
|
FYVE, RhoGEF and PH domain containing 1 |
| chr20_+_30445971 | 0.06 |
ENSDART00000153150
|
myt1la
|
myelin transcription factor 1-like, a |
| chr2_-_11662851 | 0.05 |
ENSDART00000145108
|
zgc:110130
|
zgc:110130 |
| chr19_+_3056450 | 0.05 |
ENSDART00000141324
ENSDART00000082353 |
hsf1
|
heat shock transcription factor 1 |
| chr3_-_39198113 | 0.05 |
ENSDART00000102690
|
RETSAT
|
zgc:154169 |
| chr17_-_37214196 | 0.05 |
ENSDART00000128715
|
kif3cb
|
kinesin family member 3Cb |
| chr14_+_34490445 | 0.05 |
ENSDART00000132193
ENSDART00000148044 |
wnt8a
|
wingless-type MMTV integration site family, member 8a |
| chr21_+_25643880 | 0.05 |
ENSDART00000192392
ENSDART00000145091 |
tmem151a
|
transmembrane protein 151A |
| chr24_-_10014512 | 0.05 |
ENSDART00000124341
ENSDART00000191630 |
zgc:171474
|
zgc:171474 |
| chr12_-_6818676 | 0.05 |
ENSDART00000106391
|
pcdh15b
|
protocadherin-related 15b |
| chr19_+_16015881 | 0.05 |
ENSDART00000187135
|
ctps1a
|
CTP synthase 1a |
| chr21_+_42226113 | 0.05 |
ENSDART00000170362
|
GABRB2 (1 of many)
|
gamma-aminobutyric acid type A receptor beta2 subunit |
| chr12_+_16087077 | 0.05 |
ENSDART00000141898
|
znf281b
|
zinc finger protein 281b |
| chr24_+_5208171 | 0.05 |
ENSDART00000155926
ENSDART00000154464 |
si:ch73-206p6.1
|
si:ch73-206p6.1 |
| chr19_-_32914227 | 0.05 |
ENSDART00000186115
ENSDART00000124246 |
mtdha
|
metadherin a |
| chr3_+_56366395 | 0.05 |
ENSDART00000154367
|
cacng5b
|
calcium channel, voltage-dependent, gamma subunit 5b |
| chr16_+_17389116 | 0.05 |
ENSDART00000103750
ENSDART00000173448 |
fam131bb
|
family with sequence similarity 131, member Bb |
| chr11_-_42980535 | 0.05 |
ENSDART00000181160
ENSDART00000192064 |
CABZ01069998.1
|
|
| chr13_-_50108337 | 0.05 |
ENSDART00000133308
|
nid1a
|
nidogen 1a |
| chr8_-_34051548 | 0.05 |
ENSDART00000105204
|
pbx3b
|
pre-B-cell leukemia homeobox 3b |
| chr13_+_29462249 | 0.05 |
ENSDART00000147903
|
lrit1a
|
leucine-rich repeat, immunoglobulin-like and transmembrane domains 1a |
| chr3_-_34296361 | 0.05 |
ENSDART00000181485
ENSDART00000113726 |
tnrc6c1
|
trinucleotide repeat containing 6C1 |
| chr25_+_35375848 | 0.05 |
ENSDART00000155721
|
ano3
|
anoctamin 3 |
| chr5_+_43965078 | 0.05 |
ENSDART00000113502
ENSDART00000187143 |
TMEM8B
|
si:dkey-84j12.1 |
| chr17_-_44247707 | 0.05 |
ENSDART00000126097
|
otx2b
|
orthodenticle homeobox 2b |
| chr14_-_46617228 | 0.05 |
ENSDART00000161010
ENSDART00000171054 |
prom1a
|
prominin 1a |
| chr2_+_50608099 | 0.05 |
ENSDART00000185805
ENSDART00000111135 |
neurod6b
|
neuronal differentiation 6b |
| chr20_-_19511016 | 0.05 |
ENSDART00000168521
|
snx17
|
sorting nexin 17 |
| chr1_+_40023640 | 0.05 |
ENSDART00000101623
|
lgi2b
|
leucine-rich repeat LGI family, member 2b |
| chr21_+_25777425 | 0.05 |
ENSDART00000021620
|
cldnd
|
claudin d |
| chr12_+_32073660 | 0.05 |
ENSDART00000153245
ENSDART00000153268 |
stxbp4
|
syntaxin binding protein 4 |
| chr6_+_21001264 | 0.05 |
ENSDART00000044519
ENSDART00000151278 |
cx44.2
|
connexin 44.2 |
| chr20_+_19238382 | 0.05 |
ENSDART00000136757
|
fndc4a
|
fibronectin type III domain containing 4a |
| chr13_+_2625150 | 0.05 |
ENSDART00000164177
|
plpp4
|
phospholipid phosphatase 4 |
| chr7_+_53498152 | 0.05 |
ENSDART00000184497
|
znf609b
|
zinc finger protein 609b |
| chr15_-_35031523 | 0.05 |
ENSDART00000154848
|
si:ch211-272b8.6
|
si:ch211-272b8.6 |
| chr2_+_26656283 | 0.05 |
ENSDART00000133202
ENSDART00000099208 |
asph
|
aspartate beta-hydroxylase |
| chr7_+_61050329 | 0.05 |
ENSDART00000115355
|
nwd2
|
NACHT and WD repeat domain containing 2 |
| chr24_+_12989727 | 0.05 |
ENSDART00000126842
ENSDART00000129309 |
flj11011l
|
hypothetical protein FLJ11011-like (H. sapiens) |
| chr15_-_22074315 | 0.05 |
ENSDART00000149830
|
drd2a
|
dopamine receptor D2a |
| chr8_+_30452945 | 0.05 |
ENSDART00000062303
|
foxd5
|
forkhead box D5 |
| chr24_+_34069675 | 0.05 |
ENSDART00000143995
|
si:ch211-190p8.2
|
si:ch211-190p8.2 |
| chr22_-_7129631 | 0.05 |
ENSDART00000171359
|
asic1b
|
acid-sensing (proton-gated) ion channel 1b |
| chr19_-_31402429 | 0.05 |
ENSDART00000137292
|
tmem106bb
|
transmembrane protein 106Bb |
| chr10_-_25217347 | 0.05 |
ENSDART00000036906
|
kpna7
|
karyopherin alpha 7 (importin alpha 8) |
| chr23_+_26946744 | 0.05 |
ENSDART00000115141
|
cacnb3b
|
calcium channel, voltage-dependent, beta 3b |
| chr3_-_21288202 | 0.05 |
ENSDART00000191766
ENSDART00000187319 |
fam171a2a
|
family with sequence similarity 171, member A2a |
| chr9_+_31282161 | 0.05 |
ENSDART00000010774
|
zic2a
|
zic family member 2 (odd-paired homolog, Drosophila), a |
| chr1_-_36152131 | 0.05 |
ENSDART00000182113
ENSDART00000182904 |
znf827
|
zinc finger protein 827 |
| chr3_+_34220194 | 0.05 |
ENSDART00000145859
|
slc25a23b
|
solute carrier family 25 (mitochondrial carrier; phosphate carrier), member 23b |
| chr24_+_24170914 | 0.05 |
ENSDART00000127842
|
si:dkey-226l10.6
|
si:dkey-226l10.6 |
| chr18_-_2433011 | 0.05 |
ENSDART00000181922
ENSDART00000193276 |
CR769778.1
|
|
| chr24_-_30843250 | 0.05 |
ENSDART00000162920
|
ptbp2a
|
polypyrimidine tract binding protein 2a |
| chr21_-_14174786 | 0.05 |
ENSDART00000145366
|
whrna
|
whirlin a |
| chr17_+_10501647 | 0.05 |
ENSDART00000140391
|
tyro3
|
TYRO3 protein tyrosine kinase |
| chr22_-_8725768 | 0.05 |
ENSDART00000189873
ENSDART00000181819 |
si:ch73-27e22.1
si:ch73-27e22.8
|
si:ch73-27e22.1 si:ch73-27e22.8 |
| chr13_-_44630111 | 0.05 |
ENSDART00000110092
|
mdga1
|
MAM domain containing glycosylphosphatidylinositol anchor 1 |
| chr13_-_36911118 | 0.04 |
ENSDART00000048739
|
trim9
|
tripartite motif containing 9 |
| chr2_-_38035235 | 0.04 |
ENSDART00000075904
|
cbln5
|
cerebellin 5 |
| chr7_+_32722227 | 0.04 |
ENSDART00000126565
|
si:ch211-150g13.3
|
si:ch211-150g13.3 |
| chr24_+_12835935 | 0.04 |
ENSDART00000114762
|
nanog
|
nanog homeobox |
Network of associatons between targets according to the STRING database.
First level regulatory network of bsx
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.2 | GO:0006145 | purine nucleobase catabolic process(GO:0006145) |
| 0.1 | 0.2 | GO:0042730 | fibrinolysis(GO:0042730) |
| 0.1 | 0.2 | GO:0033210 | leptin-mediated signaling pathway(GO:0033210) cellular response to leptin stimulus(GO:0044320) response to leptin(GO:0044321) |
| 0.0 | 0.2 | GO:0046166 | methylglyoxal biosynthetic process(GO:0019242) glyceraldehyde-3-phosphate biosynthetic process(GO:0046166) |
| 0.0 | 0.1 | GO:0015677 | copper ion import(GO:0015677) |
| 0.0 | 0.1 | GO:0005997 | xylulose metabolic process(GO:0005997) |
| 0.0 | 0.1 | GO:0006409 | tRNA export from nucleus(GO:0006409) tRNA-containing ribonucleoprotein complex export from nucleus(GO:0071431) |
| 0.0 | 0.1 | GO:0042755 | eating behavior(GO:0042755) |
| 0.0 | 0.1 | GO:0038003 | opioid receptor signaling pathway(GO:0038003) |
| 0.0 | 0.1 | GO:0046485 | ether lipid metabolic process(GO:0046485) |
| 0.0 | 0.1 | GO:0019323 | pentose catabolic process(GO:0019323) |
| 0.0 | 0.2 | GO:0010960 | magnesium ion homeostasis(GO:0010960) |
| 0.0 | 0.1 | GO:0060074 | synapse maturation(GO:0060074) |
| 0.0 | 0.1 | GO:0072111 | spinal cord anterior/posterior patterning(GO:0021512) cell proliferation involved in pronephros development(GO:0039015) cell proliferation involved in kidney development(GO:0072111) |
| 0.0 | 0.1 | GO:0002159 | desmosome assembly(GO:0002159) |
| 0.0 | 0.1 | GO:0060049 | regulation of protein glycosylation(GO:0060049) |
| 0.0 | 0.1 | GO:0016338 | calcium-independent cell-cell adhesion via plasma membrane cell-adhesion molecules(GO:0016338) |
| 0.0 | 0.0 | GO:1903441 | receptor localization to nonmotile primary cilium(GO:0097500) protein localization to ciliary membrane(GO:1903441) |
| 0.0 | 0.1 | GO:0070208 | protein heterotrimerization(GO:0070208) |
| 0.0 | 0.0 | GO:0021531 | spinal cord radial glial cell differentiation(GO:0021531) eye field cell fate commitment involved in camera-type eye formation(GO:0060898) |
| 0.0 | 0.1 | GO:0044210 | 'de novo' CTP biosynthetic process(GO:0044210) |
| 0.0 | 0.0 | GO:0006089 | lactate metabolic process(GO:0006089) |
| 0.0 | 0.0 | GO:0097065 | anterior head development(GO:0097065) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | GO:0034456 | CURI complex(GO:0032545) UTP-C complex(GO:0034456) |
| 0.0 | 0.1 | GO:0097189 | apoptotic body(GO:0097189) |
| 0.0 | 0.1 | GO:0097268 | cytoophidium(GO:0097268) |
| 0.0 | 0.1 | GO:0070032 | synaptobrevin 2-SNAP-25-syntaxin-1a-complexin I complex(GO:0070032) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.2 | GO:0031716 | calcitonin receptor binding(GO:0031716) |
| 0.1 | 0.5 | GO:0004459 | L-lactate dehydrogenase activity(GO:0004459) |
| 0.0 | 0.2 | GO:0004807 | triose-phosphate isomerase activity(GO:0004807) methylglyoxal synthase activity(GO:0008929) |
| 0.0 | 0.2 | GO:0004925 | prolactin receptor activity(GO:0004925) |
| 0.0 | 0.1 | GO:0052851 | cupric reductase activity(GO:0008823) ferric-chelate reductase (NADPH) activity(GO:0052851) |
| 0.0 | 0.1 | GO:0038046 | enkephalin receptor activity(GO:0038046) |
| 0.0 | 0.1 | GO:0070738 | protein-glycine ligase activity(GO:0070735) tubulin-glycine ligase activity(GO:0070738) |
| 0.0 | 0.1 | GO:0047389 | glycerophosphocholine phosphodiesterase activity(GO:0047389) |
| 0.0 | 0.1 | GO:0004430 | 1-phosphatidylinositol 4-kinase activity(GO:0004430) |
| 0.0 | 0.1 | GO:0009374 | biotin binding(GO:0009374) |
| 0.0 | 0.1 | GO:0051139 | calcium:proton antiporter activity(GO:0015369) metal ion:proton antiporter activity(GO:0051139) |
| 0.0 | 0.1 | GO:0008158 | hedgehog receptor activity(GO:0008158) |
| 0.0 | 0.1 | GO:0003883 | CTP synthase activity(GO:0003883) |
| 0.0 | 0.0 | GO:0004597 | peptide-aspartate beta-dioxygenase activity(GO:0004597) |
| 0.0 | 0.1 | GO:0016714 | oxidoreductase activity, acting on paired donors, with incorporation or reduction of molecular oxygen, reduced pteridine as one donor, and incorporation of one atom of oxygen(GO:0016714) |
| 0.0 | 0.0 | GO:0015228 | coenzyme A transmembrane transporter activity(GO:0015228) adenosine 3',5'-bisphosphate transmembrane transporter activity(GO:0071077) AMP transmembrane transporter activity(GO:0080122) |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.2 | REACTOME PURINE CATABOLISM | Genes involved in Purine catabolism |
| 0.0 | 0.2 | REACTOME DEGRADATION OF THE EXTRACELLULAR MATRIX | Genes involved in Degradation of the extracellular matrix |
| 0.0 | 0.1 | REACTOME ACYL CHAIN REMODELLING OF PI | Genes involved in Acyl chain remodelling of PI |