Project
PRJNA207719: Tissue specific transcriptome profiling
Navigation
Downloads
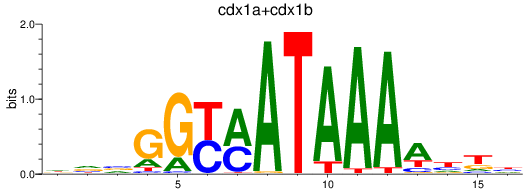
Results for cdx1a+cdx1b
Z-value: 0.75
Transcription factors associated with cdx1a+cdx1b
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
cdx1a
|
ENSDARG00000114554 | caudal type homeobox 1a |
|
cdx1b
|
ENSDARG00000116139 | caudal type homeobox 1 b |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| cdx1b | dr11_v1_chr21_-_43992027_43992027 | -0.34 | 5.8e-01 | Click! |
| cdx1a | dr11_v1_chr14_+_3287740_3287740 | -0.08 | 9.0e-01 | Click! |
Activity profile of cdx1a+cdx1b motif
Sorted Z-values of cdx1a+cdx1b motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr14_-_15171435 | 0.83 |
ENSDART00000159148
ENSDART00000166622 |
si:dkey-77g12.1
|
si:dkey-77g12.1 |
| chr24_+_39105051 | 0.64 |
ENSDART00000115297
|
mss51
|
MSS51 mitochondrial translational activator |
| chr13_-_36034582 | 0.60 |
ENSDART00000133565
|
si:dkey-157l19.2
|
si:dkey-157l19.2 |
| chr14_+_15495088 | 0.57 |
ENSDART00000165765
ENSDART00000188577 |
si:dkey-203a12.6
|
si:dkey-203a12.6 |
| chr11_-_25257595 | 0.53 |
ENSDART00000123567
|
snai1a
|
snail family zinc finger 1a |
| chr11_-_25257045 | 0.52 |
ENSDART00000130477
|
snai1a
|
snail family zinc finger 1a |
| chr23_+_36115541 | 0.50 |
ENSDART00000130090
|
hoxc6a
|
homeobox C6a |
| chr1_-_7603734 | 0.49 |
ENSDART00000009315
|
mxb
|
myxovirus (influenza) resistance B |
| chr23_+_7379728 | 0.49 |
ENSDART00000012194
|
gata5
|
GATA binding protein 5 |
| chr24_-_10014512 | 0.48 |
ENSDART00000124341
ENSDART00000191630 |
zgc:171474
|
zgc:171474 |
| chr20_+_52546186 | 0.48 |
ENSDART00000110777
ENSDART00000153377 ENSDART00000153013 ENSDART00000042704 |
eef1db
|
eukaryotic translation elongation factor 1 delta b (guanine nucleotide exchange protein) |
| chr14_+_15597049 | 0.48 |
ENSDART00000159732
|
si:dkey-203a12.8
|
si:dkey-203a12.8 |
| chr8_-_38159805 | 0.46 |
ENSDART00000112331
ENSDART00000180006 |
adgra2
|
adhesion G protein-coupled receptor A2 |
| chr25_-_29074064 | 0.46 |
ENSDART00000165603
|
arid3b
|
AT rich interactive domain 3B (BRIGHT-like) |
| chr10_+_27068251 | 0.45 |
ENSDART00000012717
|
ehd1b
|
EH-domain containing 1b |
| chr5_-_1963498 | 0.43 |
ENSDART00000073462
|
rplp0
|
ribosomal protein, large, P0 |
| chr15_-_28596507 | 0.43 |
ENSDART00000156800
|
si:ch211-225b7.5
|
si:ch211-225b7.5 |
| chr13_+_27329556 | 0.42 |
ENSDART00000140085
|
mb21d1
|
Mab-21 domain containing 1 |
| chr6_-_39727244 | 0.42 |
ENSDART00000065057
|
itgb7
|
integrin, beta 7 |
| chr24_-_9997948 | 0.41 |
ENSDART00000136274
|
si:ch211-146l10.7
|
si:ch211-146l10.7 |
| chr14_-_4120636 | 0.41 |
ENSDART00000059230
|
irf2
|
interferon regulatory factor 2 |
| chr14_-_4121052 | 0.40 |
ENSDART00000167074
|
irf2
|
interferon regulatory factor 2 |
| chr23_-_3758637 | 0.38 |
ENSDART00000131536
ENSDART00000139408 ENSDART00000137826 |
hmga1a
|
high mobility group AT-hook 1a |
| chr11_+_20896122 | 0.38 |
ENSDART00000162339
ENSDART00000181111 |
CABZ01008730.1
|
|
| chr9_+_17306162 | 0.37 |
ENSDART00000075926
|
scel
|
sciellin |
| chr7_-_48263516 | 0.37 |
ENSDART00000006619
ENSDART00000142370 ENSDART00000148273 ENSDART00000147968 |
rbpms2b
|
RNA binding protein with multiple splicing 2b |
| chr6_+_39114345 | 0.35 |
ENSDART00000136835
|
gpr84
|
G protein-coupled receptor 84 |
| chr3_-_53465223 | 0.34 |
ENSDART00000057123
ENSDART00000125515 ENSDART00000143096 |
nr5a5
|
nuclear receptor subfamily 5, group A, member 5 |
| chr11_+_12052791 | 0.34 |
ENSDART00000158479
|
si:ch211-156l18.8
|
si:ch211-156l18.8 |
| chr11_-_25538341 | 0.34 |
ENSDART00000171560
|
si:dkey-245f22.3
|
si:dkey-245f22.3 |
| chr22_+_34616151 | 0.34 |
ENSDART00000155399
ENSDART00000104705 |
si:ch1073-214b20.2
|
si:ch1073-214b20.2 |
| chr21_-_20840714 | 0.33 |
ENSDART00000144861
ENSDART00000139430 |
c6
|
complement component 6 |
| chr22_+_34615591 | 0.33 |
ENSDART00000189563
|
si:ch1073-214b20.2
|
si:ch1073-214b20.2 |
| chr16_+_52966812 | 0.33 |
ENSDART00000148435
ENSDART00000049099 ENSDART00000150117 |
trip13
|
thyroid hormone receptor interactor 13 |
| chr3_+_62161184 | 0.32 |
ENSDART00000090370
ENSDART00000192665 |
noxo1a
|
NADPH oxidase organizer 1a |
| chr10_+_40629616 | 0.32 |
ENSDART00000147476
|
CR396590.9
|
|
| chr15_+_22390076 | 0.32 |
ENSDART00000183764
|
oafa
|
OAF homolog a (Drosophila) |
| chr6_+_120181 | 0.32 |
ENSDART00000151209
ENSDART00000185930 |
cdkn2d
|
cyclin-dependent kinase inhibitor 2D (p19, inhibits CDK4) |
| chr5_+_38619813 | 0.31 |
ENSDART00000133314
|
si:ch211-271e10.3
|
si:ch211-271e10.3 |
| chr17_-_30652738 | 0.30 |
ENSDART00000154960
|
sh3yl1
|
SH3 and SYLF domain containing 1 |
| chr2_-_5840494 | 0.29 |
ENSDART00000154877
|
si:ch211-273k1.5
|
si:ch211-273k1.5 |
| chr24_-_10006158 | 0.29 |
ENSDART00000106244
|
zgc:171750
|
zgc:171750 |
| chr21_+_25236297 | 0.29 |
ENSDART00000112783
|
tmem45b
|
transmembrane protein 45B |
| chr25_+_20089986 | 0.28 |
ENSDART00000143441
ENSDART00000184073 |
tnni4b.2
|
troponin I4b, tandem duplicate 2 |
| chr11_-_36051004 | 0.28 |
ENSDART00000025033
|
gpx1a
|
glutathione peroxidase 1a |
| chr5_-_23783739 | 0.28 |
ENSDART00000139502
|
GBGT1 (1 of many)
|
si:ch211-287c22.1 |
| chr16_+_31542645 | 0.28 |
ENSDART00000163724
|
SLA (1 of many)
|
Src like adaptor |
| chr6_-_12588044 | 0.28 |
ENSDART00000047896
|
slc15a1b
|
solute carrier family 15 (oligopeptide transporter), member 1b |
| chr23_-_10722664 | 0.27 |
ENSDART00000146526
ENSDART00000129022 ENSDART00000104985 |
foxp1a
|
forkhead box P1a |
| chr14_-_2602445 | 0.27 |
ENSDART00000166910
|
etf1a
|
eukaryotic translation termination factor 1a |
| chr19_-_47571456 | 0.27 |
ENSDART00000158071
ENSDART00000165841 |
rrm2
|
ribonucleotide reductase M2 polypeptide |
| chr6_+_40629066 | 0.26 |
ENSDART00000103757
|
slc6a11a
|
solute carrier family 6 (neurotransmitter transporter), member 11a |
| chr12_-_16524727 | 0.26 |
ENSDART00000166645
|
FO704724.1
|
|
| chr22_+_10440991 | 0.26 |
ENSDART00000064805
|
cenpp
|
centromere protein P |
| chr10_-_15340362 | 0.26 |
ENSDART00000148119
ENSDART00000127277 ENSDART00000154037 ENSDART00000189109 |
pum3
|
pumilio RNA-binding family member 3 |
| chr7_-_60096318 | 0.26 |
ENSDART00000189125
|
BX511067.1
|
|
| chr23_+_36087219 | 0.26 |
ENSDART00000154825
|
hoxc3a
|
homeobox C3a |
| chr3_-_4557653 | 0.26 |
ENSDART00000111305
|
ftr50
|
finTRIM family, member 50 |
| chr12_-_33746111 | 0.26 |
ENSDART00000127203
|
llgl2
|
lethal giant larvae homolog 2 (Drosophila) |
| chr7_-_55454406 | 0.26 |
ENSDART00000108646
|
piezo1
|
piezo-type mechanosensitive ion channel component 1 |
| chr21_-_39024754 | 0.26 |
ENSDART00000056878
|
traf4b
|
tnf receptor-associated factor 4b |
| chr20_+_7398090 | 0.26 |
ENSDART00000108649
|
pcsk9
|
proprotein convertase subtilisin/kexin type 9 |
| chr10_-_35410518 | 0.25 |
ENSDART00000048430
|
gabrr3a
|
gamma-aminobutyric acid (GABA) A receptor, rho 3a |
| chr24_-_30275735 | 0.25 |
ENSDART00000168723
|
snx7
|
sorting nexin 7 |
| chr21_-_2415808 | 0.25 |
ENSDART00000171179
|
si:ch211-241b2.5
|
si:ch211-241b2.5 |
| chr8_-_32803227 | 0.25 |
ENSDART00000110079
|
zgc:194839
|
zgc:194839 |
| chr20_+_26880668 | 0.24 |
ENSDART00000077769
|
serpinb1
|
serpin peptidase inhibitor, clade B (ovalbumin), member 1 |
| chr19_+_19747430 | 0.24 |
ENSDART00000166129
|
hoxa9a
|
homeobox A9a |
| chr3_-_4501026 | 0.24 |
ENSDART00000163052
|
zgc:162198
|
zgc:162198 |
| chr3_-_53091946 | 0.24 |
ENSDART00000187297
|
lpar2a
|
lysophosphatidic acid receptor 2a |
| chr1_-_56213723 | 0.24 |
ENSDART00000142505
ENSDART00000137237 |
si:dkey-76b14.2
|
si:dkey-76b14.2 |
| chr8_-_6897619 | 0.24 |
ENSDART00000133606
|
si:ch211-255g12.8
|
si:ch211-255g12.8 |
| chr16_+_42471455 | 0.24 |
ENSDART00000166640
|
si:ch211-215k15.5
|
si:ch211-215k15.5 |
| chr2_+_36112273 | 0.24 |
ENSDART00000191315
|
traj35
|
T-cell receptor alpha joining 35 |
| chr15_-_563877 | 0.23 |
ENSDART00000128032
|
cbln18
|
cerebellin 18 |
| chr8_+_30709685 | 0.23 |
ENSDART00000133989
|
upb1
|
ureidopropionase, beta |
| chr20_-_43741159 | 0.23 |
ENSDART00000192621
|
si:dkeyp-50f7.2
|
si:dkeyp-50f7.2 |
| chr5_+_64856666 | 0.23 |
ENSDART00000050863
|
zgc:101858
|
zgc:101858 |
| chr3_+_32526263 | 0.23 |
ENSDART00000150897
|
si:ch73-367p23.2
|
si:ch73-367p23.2 |
| chr12_+_20552190 | 0.23 |
ENSDART00000113224
|
gna13a
|
guanine nucleotide binding protein (G protein), alpha 13a |
| chr21_+_11969603 | 0.23 |
ENSDART00000142247
ENSDART00000140652 |
mlnl
|
motilin-like |
| chr5_+_38631119 | 0.23 |
ENSDART00000131832
ENSDART00000162215 |
si:ch211-271e10.6
|
si:ch211-271e10.6 |
| chr10_-_1625080 | 0.22 |
ENSDART00000137285
|
nup155
|
nucleoporin 155 |
| chr20_-_34670236 | 0.22 |
ENSDART00000033325
|
slc25a24
|
solute carrier family 25 (mitochondrial carrier; phosphate carrier), member 24 |
| chr1_-_9249943 | 0.22 |
ENSDART00000055011
|
zgc:136472
|
zgc:136472 |
| chr5_-_30984271 | 0.22 |
ENSDART00000051392
|
spns3
|
spinster homolog 3 (Drosophila) |
| chr24_-_17029374 | 0.21 |
ENSDART00000039267
|
ptgdsb.1
|
prostaglandin D2 synthase b, tandem duplicate 1 |
| chr5_-_57723929 | 0.21 |
ENSDART00000144237
|
gig2p
|
grass carp reovirus (GCRV)-induced gene 2p |
| chr9_+_54290896 | 0.21 |
ENSDART00000149175
|
pou4f3
|
POU class 4 homeobox 3 |
| chr10_-_15849027 | 0.21 |
ENSDART00000184682
|
tjp2a
|
tight junction protein 2a (zona occludens 2) |
| chr20_-_29499363 | 0.21 |
ENSDART00000152889
ENSDART00000153252 ENSDART00000170972 ENSDART00000166420 ENSDART00000163079 |
rrm2
|
ribonucleotide reductase M2 polypeptide |
| chr16_+_20934353 | 0.20 |
ENSDART00000052660
|
skap2
|
src kinase associated phosphoprotein 2 |
| chr25_-_26758253 | 0.20 |
ENSDART00000123004
|
si:dkeyp-73b11.8
|
si:dkeyp-73b11.8 |
| chr14_+_35369979 | 0.20 |
ENSDART00000144702
ENSDART00000169712 |
clint1a
|
clathrin interactor 1a |
| chr23_+_43718115 | 0.20 |
ENSDART00000149266
ENSDART00000149503 |
anapc10
|
anaphase promoting complex subunit 10 |
| chr6_-_46768040 | 0.20 |
ENSDART00000154071
|
igfn1.2
|
immunoglobulin-like and fibronectin type III domain containing 1, tandem duplicate 2 |
| chr19_-_7225060 | 0.20 |
ENSDART00000133179
ENSDART00000151138 ENSDART00000104799 ENSDART00000128331 |
col11a2
|
collagen, type XI, alpha 2 |
| chr7_+_1473929 | 0.20 |
ENSDART00000050687
|
lpcat4
|
lysophosphatidylcholine acyltransferase 4 |
| chr23_+_44349252 | 0.19 |
ENSDART00000097644
|
ephb4b
|
eph receptor B4b |
| chr20_+_11800063 | 0.19 |
ENSDART00000152230
|
si:ch211-155o21.4
|
si:ch211-155o21.4 |
| chr11_-_22361306 | 0.19 |
ENSDART00000180688
ENSDART00000182200 ENSDART00000006580 |
tfeb
|
transcription factor EB |
| chr15_+_9297340 | 0.19 |
ENSDART00000055554
|
slc37a4a
|
solute carrier family 37 (glucose-6-phosphate transporter), member 4a |
| chr20_-_2641233 | 0.19 |
ENSDART00000145335
ENSDART00000133121 |
bub1
|
BUB1 mitotic checkpoint serine/threonine kinase |
| chr24_-_36723116 | 0.19 |
ENSDART00000088204
|
si:ch73-334d15.1
|
si:ch73-334d15.1 |
| chr21_+_20396858 | 0.19 |
ENSDART00000003299
ENSDART00000146615 |
zgc:103482
|
zgc:103482 |
| chr2_+_5927255 | 0.19 |
ENSDART00000152866
|
si:ch211-168b3.2
|
si:ch211-168b3.2 |
| chr11_+_24314148 | 0.19 |
ENSDART00000171491
|
rem1
|
RAS (RAD and GEM)-like GTP-binding 1 |
| chr21_-_3700334 | 0.19 |
ENSDART00000137844
|
atp8b1
|
ATPase phospholipid transporting 8B1 |
| chr10_+_35526528 | 0.19 |
ENSDART00000184110
|
phldb2a
|
pleckstrin homology-like domain, family B, member 2a |
| chr13_+_13668991 | 0.18 |
ENSDART00000148266
|
pimr48
|
Pim proto-oncogene, serine/threonine kinase, related 48 |
| chr15_-_4580763 | 0.18 |
ENSDART00000008170
|
rffl
|
ring finger and FYVE-like domain containing E3 ubiquitin protein ligase |
| chr18_+_9171778 | 0.18 |
ENSDART00000101192
|
sema3d
|
sema domain, immunoglobulin domain (Ig), short basic domain, secreted, (semaphorin) 3D |
| chr25_-_35047838 | 0.18 |
ENSDART00000155060
|
CU302436.3
|
|
| chr16_-_16225260 | 0.18 |
ENSDART00000165790
|
gra
|
granulito |
| chr11_+_44622472 | 0.18 |
ENSDART00000159068
ENSDART00000166323 ENSDART00000187753 |
rbm34
|
RNA binding motif protein 34 |
| chr7_-_18508815 | 0.18 |
ENSDART00000173539
|
rgs12a
|
regulator of G protein signaling 12a |
| chr10_-_21545091 | 0.18 |
ENSDART00000029122
ENSDART00000132207 |
zgc:165539
|
zgc:165539 |
| chr14_+_32918172 | 0.18 |
ENSDART00000182867
|
lnx2b
|
ligand of numb-protein X 2b |
| chr6_-_25165693 | 0.18 |
ENSDART00000167259
|
znf326
|
zinc finger protein 326 |
| chr15_+_37950556 | 0.18 |
ENSDART00000153947
|
si:dkey-238d18.12
|
si:dkey-238d18.12 |
| chr10_+_44700103 | 0.17 |
ENSDART00000165999
|
scarb1
|
scavenger receptor class B, member 1 |
| chr7_+_66822229 | 0.17 |
ENSDART00000112109
|
lyve1a
|
lymphatic vessel endothelial hyaluronic receptor 1a |
| chr5_+_38662125 | 0.17 |
ENSDART00000136949
|
si:dkey-58f10.13
|
si:dkey-58f10.13 |
| chr25_+_29161609 | 0.17 |
ENSDART00000180752
|
pkmb
|
pyruvate kinase M1/2b |
| chr12_+_20567382 | 0.17 |
ENSDART00000123737
|
FP885542.1
|
|
| chr2_+_42715995 | 0.17 |
ENSDART00000143419
ENSDART00000183914 ENSDART00000184371 ENSDART00000185485 ENSDART00000037332 |
ftr12
|
finTRIM family, member 12 |
| chr16_+_13993285 | 0.17 |
ENSDART00000139130
ENSDART00000130353 |
si:dkey-85k15.7
fdps
|
si:dkey-85k15.7 farnesyl diphosphate synthase (farnesyl pyrophosphate synthetase, dimethylallyltranstransferase, geranyltranstransferase) |
| chr8_-_39739056 | 0.17 |
ENSDART00000147992
|
si:ch211-170d8.5
|
si:ch211-170d8.5 |
| chr2_-_17235891 | 0.17 |
ENSDART00000144251
|
artnb
|
artemin b |
| chr9_+_3282369 | 0.17 |
ENSDART00000044128
|
hat1
|
histone acetyltransferase 1 |
| chr2_+_19522082 | 0.17 |
ENSDART00000146098
|
pimr49
|
Pim proto-oncogene, serine/threonine kinase, related 49 |
| chr22_+_30496360 | 0.16 |
ENSDART00000189145
|
BX649490.4
|
|
| chr14_+_22447662 | 0.16 |
ENSDART00000161776
|
sowahab
|
sosondowah ankyrin repeat domain family member Ab |
| chr1_+_34496855 | 0.16 |
ENSDART00000012873
|
klf12a
|
Kruppel-like factor 12a |
| chr21_-_11970199 | 0.15 |
ENSDART00000114524
|
nop56
|
NOP56 ribonucleoprotein homolog |
| chr7_-_29292206 | 0.15 |
ENSDART00000086753
|
dapk2a
|
death-associated protein kinase 2a |
| chr16_-_14534724 | 0.15 |
ENSDART00000148126
|
si:dkey-237j11.3
|
si:dkey-237j11.3 |
| chr2_+_52847049 | 0.15 |
ENSDART00000121980
|
creb3l3b
|
cAMP responsive element binding protein 3-like 3b |
| chr5_-_16351306 | 0.15 |
ENSDART00000168643
|
CABZ01088700.1
|
|
| chr23_-_39959173 | 0.15 |
ENSDART00000077122
|
xcr1a.1
|
chemokine (C motif) receptor 1a, duplicate 1 |
| chr3_-_60589292 | 0.15 |
ENSDART00000157822
|
jmjd6
|
jumonji domain containing 6 |
| chr19_+_9097013 | 0.15 |
ENSDART00000139781
|
si:ch211-81a5.5
|
si:ch211-81a5.5 |
| chr25_-_27633115 | 0.15 |
ENSDART00000131539
|
hyal6
|
hyaluronoglucosaminidase 6 |
| chr3_+_49074008 | 0.14 |
ENSDART00000168864
|
zgc:112146
|
zgc:112146 |
| chr24_+_1042594 | 0.14 |
ENSDART00000109117
|
si:dkey-192l18.9
|
si:dkey-192l18.9 |
| chr5_+_38679623 | 0.14 |
ENSDART00000131879
ENSDART00000170010 |
si:dkey-58f10.11
si:dkey-58f10.10
|
si:dkey-58f10.11 si:dkey-58f10.10 |
| chr3_+_36646054 | 0.14 |
ENSDART00000170013
ENSDART00000159948 |
gspt1l
|
G1 to S phase transition 1, like |
| chr21_-_41838284 | 0.14 |
ENSDART00000141067
|
cct6a
|
chaperonin containing TCP1, subunit 6A (zeta 1) |
| chr14_+_14841685 | 0.14 |
ENSDART00000158291
ENSDART00000162039 |
slbp
|
stem-loop binding protein |
| chr2_+_22409249 | 0.14 |
ENSDART00000182915
|
zgc:56628
|
zgc:56628 |
| chr1_+_44780849 | 0.14 |
ENSDART00000145895
|
si:dkey-9i23.5
|
si:dkey-9i23.5 |
| chr7_+_18075504 | 0.13 |
ENSDART00000173689
|
si:ch73-40a2.1
|
si:ch73-40a2.1 |
| chr8_-_39739627 | 0.13 |
ENSDART00000135422
ENSDART00000067844 |
si:ch211-170d8.5
|
si:ch211-170d8.5 |
| chr9_+_20526472 | 0.13 |
ENSDART00000166457
ENSDART00000141775 |
trim45
|
tripartite motif containing 45 |
| chr12_+_10053852 | 0.13 |
ENSDART00000078497
|
slc4a1b
|
solute carrier family 4 (anion exchanger), member 1b (Diego blood group) |
| chr1_-_9486214 | 0.13 |
ENSDART00000137821
|
micall2b
|
mical-like 2b |
| chr18_+_36664654 | 0.13 |
ENSDART00000128707
ENSDART00000098972 |
capn12
|
calpain 12 |
| chr2_-_36914281 | 0.13 |
ENSDART00000008322
|
si:dkey-193b15.5
|
si:dkey-193b15.5 |
| chr10_-_3332362 | 0.13 |
ENSDART00000007577
ENSDART00000055140 |
tor4aa
|
torsin family 4, member Aa |
| chr20_-_49889111 | 0.13 |
ENSDART00000058858
|
kif13bb
|
kinesin family member 13Bb |
| chr4_-_5775507 | 0.13 |
ENSDART00000021753
|
ccnc
|
cyclin C |
| chr22_+_2111120 | 0.13 |
ENSDART00000165525
|
znf1144
|
zinc finger protein 1144 |
| chr5_-_9073433 | 0.12 |
ENSDART00000099891
|
atp5meb
|
ATP synthase membrane subunit eb |
| chr22_-_10440688 | 0.12 |
ENSDART00000111962
|
nol8
|
nucleolar protein 8 |
| chr9_+_53637932 | 0.12 |
ENSDART00000188962
|
CABZ01084081.1
|
|
| chr13_+_45431660 | 0.12 |
ENSDART00000099950
|
syf2
|
SYF2 pre-mRNA-splicing factor |
| chr18_+_14684115 | 0.12 |
ENSDART00000108469
|
spata2l
|
spermatogenesis associated 2-like |
| chr10_+_44924684 | 0.12 |
ENSDART00000181360
ENSDART00000170418 ENSDART00000170327 |
sec14l7
|
SEC14-like lipid binding 7 |
| chr5_-_23715861 | 0.12 |
ENSDART00000019992
|
gbgt1l1
|
globoside alpha-1,3-N-acetylgalactosaminyltransferase 1, like 1 |
| chr7_+_73827805 | 0.12 |
ENSDART00000109316
|
zgc:173587
|
zgc:173587 |
| chr12_-_6172154 | 0.11 |
ENSDART00000185434
|
a1cf
|
apobec1 complementation factor |
| chr2_-_5728843 | 0.11 |
ENSDART00000014020
|
sst2
|
somatostatin 2 |
| chr19_+_27970292 | 0.11 |
ENSDART00000103887
ENSDART00000145549 |
med10
|
mediator complex subunit 10 |
| chr1_+_54683655 | 0.11 |
ENSDART00000132785
|
knop1
|
lysine-rich nucleolar protein 1 |
| chr19_+_1873059 | 0.11 |
ENSDART00000145246
|
snrpd1
|
small nuclear ribonucleoprotein D1 polypeptide |
| chr4_+_33574463 | 0.11 |
ENSDART00000150255
|
si:dkey-84h14.1
|
si:dkey-84h14.1 |
| chr13_-_25719628 | 0.11 |
ENSDART00000135383
|
si:dkey-192p21.6
|
si:dkey-192p21.6 |
| chr21_+_20356458 | 0.11 |
ENSDART00000135816
|
pde6b
|
phosphodiesterase 6B, cGMP-specific, rod, beta |
| chr5_-_9625459 | 0.11 |
ENSDART00000143347
|
sh2b3
|
SH2B adaptor protein 3 |
| chr19_+_3840955 | 0.11 |
ENSDART00000172305
|
lsm10
|
LSM10, U7 small nuclear RNA associated |
| chr11_-_19097746 | 0.11 |
ENSDART00000103973
|
rhol
|
rhodopsin, like |
| chr20_+_38458084 | 0.11 |
ENSDART00000020153
ENSDART00000135912 |
coq8a
|
coenzyme Q8A |
| chr16_+_38940758 | 0.11 |
ENSDART00000102482
ENSDART00000136215 |
eny2
|
enhancer of yellow 2 homolog (Drosophila) |
| chr6_-_7107868 | 0.11 |
ENSDART00000171123
|
nhej1
|
nonhomologous end-joining factor 1 |
| chr10_+_8481660 | 0.11 |
ENSDART00000109559
|
BX682234.1
|
|
| chr3_-_34624745 | 0.11 |
ENSDART00000151091
|
tac4
|
tachykinin 4 (hemokinin) |
| chr9_-_22355391 | 0.10 |
ENSDART00000009115
|
crygm3
|
crystallin, gamma M3 |
| chr2_+_14992879 | 0.10 |
ENSDART00000137546
|
pimr55
|
Pim proto-oncogene, serine/threonine kinase, related 55 |
| chr1_-_53468160 | 0.10 |
ENSDART00000143349
|
zgc:66455
|
zgc:66455 |
| chr21_-_8153165 | 0.10 |
ENSDART00000182580
|
CABZ01074363.1
|
|
| chr2_-_19520690 | 0.10 |
ENSDART00000133559
|
pimr52
|
Pim proto-oncogene, serine/threonine kinase, related 52 |
| chr16_+_54829574 | 0.10 |
ENSDART00000148392
|
pabpc1a
|
poly(A) binding protein, cytoplasmic 1a |
| chr7_+_18989241 | 0.10 |
ENSDART00000053213
|
zgc:194252
|
zgc:194252 |
| chr1_-_43897831 | 0.10 |
ENSDART00000048225
|
si:dkey-22i16.2
|
si:dkey-22i16.2 |
| chr1_-_14332283 | 0.09 |
ENSDART00000090025
|
wfs1a
|
Wolfram syndrome 1a (wolframin) |
| chr13_+_18520738 | 0.09 |
ENSDART00000113952
|
tlr4al
|
toll-like receptor 4a, like |
| chr4_-_20135919 | 0.09 |
ENSDART00000172230
|
cep83
|
centrosomal protein 83 |
| chr9_-_48370645 | 0.09 |
ENSDART00000140185
|
col28a2a
|
collagen, type XXVIII, alpha 2a |
| chr22_+_2512154 | 0.09 |
ENSDART00000097363
|
zgc:173726
|
zgc:173726 |
| chr11_+_25257022 | 0.09 |
ENSDART00000156052
|
tp53inp2
|
tumor protein p53 inducible nuclear protein 2 |
Network of associatons between targets according to the STRING database.
First level regulatory network of cdx1a+cdx1b
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.5 | GO:0022009 | central nervous system vasculogenesis(GO:0022009) |
| 0.1 | 0.5 | GO:0048618 | post-embryonic foregut morphogenesis(GO:0048618) |
| 0.1 | 0.4 | GO:0002184 | cytoplasmic translational termination(GO:0002184) |
| 0.1 | 0.3 | GO:0033313 | meiotic cell cycle checkpoint(GO:0033313) |
| 0.1 | 0.2 | GO:0043654 | recognition of apoptotic cell(GO:0043654) |
| 0.1 | 0.2 | GO:0036228 | protein targeting to nuclear inner membrane(GO:0036228) protein localization to nuclear envelope(GO:0090435) |
| 0.1 | 0.4 | GO:0051148 | smooth muscle cell differentiation(GO:0051145) negative regulation of muscle cell differentiation(GO:0051148) |
| 0.1 | 0.3 | GO:0007250 | activation of NF-kappaB-inducing kinase activity(GO:0007250) |
| 0.1 | 0.2 | GO:2000048 | activation of phospholipase D activity(GO:0031584) negative regulation of cell-cell adhesion mediated by cadherin(GO:2000048) |
| 0.1 | 0.3 | GO:0010719 | photoreceptor cell morphogenesis(GO:0008594) negative regulation of epithelial to mesenchymal transition(GO:0010719) |
| 0.1 | 0.3 | GO:0010481 | ventriculo bulbo valve development(GO:0003173) epidermal cell division(GO:0010481) regulation of epidermal cell division(GO:0010482) |
| 0.1 | 1.1 | GO:0008078 | mesodermal cell migration(GO:0008078) |
| 0.0 | 0.2 | GO:0045647 | negative regulation of erythrocyte differentiation(GO:0045647) |
| 0.0 | 0.3 | GO:0010269 | response to selenium ion(GO:0010269) |
| 0.0 | 0.2 | GO:0019482 | beta-alanine metabolic process(GO:0019482) beta-alanine biosynthetic process(GO:0019483) |
| 0.0 | 0.3 | GO:0061641 | chromatin remodeling at centromere(GO:0031055) CENP-A containing nucleosome assembly(GO:0034080) CENP-A containing chromatin organization(GO:0061641) |
| 0.0 | 0.5 | GO:0009263 | deoxyribonucleotide biosynthetic process(GO:0009263) |
| 0.0 | 0.1 | GO:1905072 | cardiac jelly development(GO:1905072) |
| 0.0 | 0.2 | GO:1901842 | regulation of high voltage-gated calcium channel activity(GO:1901841) negative regulation of high voltage-gated calcium channel activity(GO:1901842) |
| 0.0 | 0.2 | GO:0002164 | larval development(GO:0002164) larval heart development(GO:0007508) |
| 0.0 | 0.2 | GO:0014707 | branchiomeric skeletal muscle development(GO:0014707) |
| 0.0 | 0.3 | GO:0006857 | oligopeptide transport(GO:0006857) |
| 0.0 | 0.2 | GO:0045337 | farnesyl diphosphate biosynthetic process(GO:0045337) |
| 0.0 | 0.2 | GO:0051754 | meiotic sister chromatid cohesion, centromeric(GO:0051754) |
| 0.0 | 0.2 | GO:2001270 | negative regulation of execution phase of apoptosis(GO:1900118) regulation of cysteine-type endopeptidase activity involved in execution phase of apoptosis(GO:2001270) negative regulation of cysteine-type endopeptidase activity involved in execution phase of apoptosis(GO:2001271) |
| 0.0 | 0.1 | GO:0017185 | peptidyl-lysine hydroxylation(GO:0017185) |
| 0.0 | 0.2 | GO:0045947 | negative regulation of translational initiation(GO:0045947) |
| 0.0 | 0.1 | GO:0003250 | cell proliferation involved in heart valve morphogenesis(GO:0003249) regulation of cell proliferation involved in heart valve morphogenesis(GO:0003250) endocardial cell development(GO:0060958) cell proliferation involved in heart valve development(GO:2000793) |
| 0.0 | 0.1 | GO:2000471 | regulation of hematopoietic stem cell migration(GO:2000471) |
| 0.0 | 0.2 | GO:0009954 | proximal/distal pattern formation(GO:0009954) |
| 0.0 | 0.2 | GO:0031295 | lymphocyte costimulation(GO:0031294) T cell costimulation(GO:0031295) |
| 0.0 | 0.1 | GO:0007217 | tachykinin receptor signaling pathway(GO:0007217) |
| 0.0 | 0.1 | GO:0006398 | mRNA 3'-end processing by stem-loop binding and cleavage(GO:0006398) |
| 0.0 | 0.2 | GO:0045948 | positive regulation of translational initiation(GO:0045948) |
| 0.0 | 0.2 | GO:0006348 | chromatin silencing at telomere(GO:0006348) |
| 0.0 | 0.2 | GO:0045682 | regulation of epidermis development(GO:0045682) |
| 0.0 | 0.2 | GO:0070979 | protein K11-linked ubiquitination(GO:0070979) |
| 0.0 | 0.4 | GO:0000027 | ribosomal large subunit assembly(GO:0000027) |
| 0.0 | 0.1 | GO:0038093 | Fc receptor signaling pathway(GO:0038093) |
| 0.0 | 0.1 | GO:0002495 | antigen processing and presentation of peptide antigen via MHC class II(GO:0002495) |
| 0.0 | 0.4 | GO:0033627 | cell adhesion mediated by integrin(GO:0033627) |
| 0.0 | 0.3 | GO:0008354 | germ cell migration(GO:0008354) |
| 0.0 | 0.1 | GO:0072386 | plus-end-directed vesicle transport along microtubule(GO:0072383) plus-end-directed organelle transport along microtubule(GO:0072386) |
| 0.0 | 0.3 | GO:0007168 | receptor guanylyl cyclase signaling pathway(GO:0007168) |
| 0.0 | 0.1 | GO:0030214 | hyaluronan catabolic process(GO:0030214) |
| 0.0 | 0.3 | GO:0019835 | cytolysis(GO:0019835) |
| 0.0 | 0.1 | GO:0030216 | keratinocyte differentiation(GO:0030216) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.5 | GO:1990909 | Wnt signalosome(GO:1990909) |
| 0.1 | 0.4 | GO:0018444 | translation release factor complex(GO:0018444) |
| 0.1 | 0.5 | GO:0005853 | eukaryotic translation elongation factor 1 complex(GO:0005853) |
| 0.1 | 0.2 | GO:0044611 | nuclear pore inner ring(GO:0044611) |
| 0.0 | 0.2 | GO:0044609 | DBIRD complex(GO:0044609) |
| 0.0 | 0.2 | GO:0031526 | brush border membrane(GO:0031526) |
| 0.0 | 0.2 | GO:0031428 | box C/D snoRNP complex(GO:0031428) |
| 0.0 | 0.2 | GO:0034715 | pICln-Sm protein complex(GO:0034715) |
| 0.0 | 0.1 | GO:0070390 | transcription export complex 2(GO:0070390) |
| 0.0 | 0.1 | GO:0032807 | DNA ligase IV complex(GO:0032807) |
| 0.0 | 0.3 | GO:0005579 | membrane attack complex(GO:0005579) |
| 0.0 | 0.1 | GO:0042622 | photoreceptor outer segment membrane(GO:0042622) |
| 0.0 | 0.1 | GO:0000974 | Prp19 complex(GO:0000974) |
| 0.0 | 0.1 | GO:0005832 | chaperonin-containing T-complex(GO:0005832) |
| 0.0 | 0.5 | GO:0055038 | recycling endosome membrane(GO:0055038) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 1.1 | GO:0051059 | NF-kappaB binding(GO:0051059) |
| 0.1 | 0.3 | GO:1990825 | sequence-specific mRNA binding(GO:1990825) |
| 0.1 | 0.2 | GO:0031752 | D5 dopamine receptor binding(GO:0031752) |
| 0.1 | 0.4 | GO:0070180 | large ribosomal subunit rRNA binding(GO:0070180) |
| 0.1 | 0.5 | GO:0004748 | ribonucleoside-diphosphate reductase activity, thioredoxin disulfide as acceptor(GO:0004748) oxidoreductase activity, acting on CH or CH2 groups, disulfide as acceptor(GO:0016728) ribonucleoside-diphosphate reductase activity(GO:0061731) |
| 0.1 | 0.2 | GO:0004337 | dimethylallyltranstransferase activity(GO:0004161) geranyltranstransferase activity(GO:0004337) |
| 0.0 | 0.2 | GO:0030169 | low-density lipoprotein particle binding(GO:0030169) |
| 0.0 | 0.3 | GO:0071916 | dipeptide transporter activity(GO:0042936) dipeptide transmembrane transporter activity(GO:0071916) |
| 0.0 | 0.3 | GO:0016176 | superoxide-generating NADPH oxidase activator activity(GO:0016176) |
| 0.0 | 0.1 | GO:0031835 | neurokinin receptor binding(GO:0031834) substance P receptor binding(GO:0031835) |
| 0.0 | 0.1 | GO:0070815 | peptidyl-lysine 5-dioxygenase activity(GO:0070815) |
| 0.0 | 0.2 | GO:0004743 | pyruvate kinase activity(GO:0004743) |
| 0.0 | 0.2 | GO:0001972 | retinoic acid binding(GO:0001972) |
| 0.0 | 0.2 | GO:0008190 | eukaryotic initiation factor 4E binding(GO:0008190) |
| 0.0 | 0.1 | GO:0001875 | lipopolysaccharide receptor activity(GO:0001875) |
| 0.0 | 0.3 | GO:0005332 | gamma-aminobutyric acid:sodium symporter activity(GO:0005332) |
| 0.0 | 0.1 | GO:0005068 | transmembrane receptor protein tyrosine kinase adaptor activity(GO:0005068) |
| 0.0 | 0.1 | GO:0008079 | translation release factor activity(GO:0003747) translation termination factor activity(GO:0008079) |
| 0.0 | 0.2 | GO:0070915 | lysophosphatidic acid receptor activity(GO:0070915) |
| 0.0 | 0.2 | GO:0010485 | H4 histone acetyltransferase activity(GO:0010485) |
| 0.0 | 0.3 | GO:0045159 | myosin II binding(GO:0045159) |
| 0.0 | 0.3 | GO:0004602 | glutathione peroxidase activity(GO:0004602) |
| 0.0 | 0.2 | GO:0038191 | neuropilin binding(GO:0038191) |
| 0.0 | 0.1 | GO:0004415 | hyalurononglucosaminidase activity(GO:0004415) |
| 0.0 | 0.3 | GO:0097153 | cysteine-type endopeptidase activity involved in apoptotic process(GO:0097153) |
| 0.0 | 0.0 | GO:0008442 | 3-hydroxyisobutyrate dehydrogenase activity(GO:0008442) |
| 0.0 | 0.1 | GO:0016918 | retinal binding(GO:0016918) |
| 0.0 | 0.5 | GO:0003746 | translation elongation factor activity(GO:0003746) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.4 | PID INTEGRIN5 PATHWAY | Beta5 beta6 beta7 and beta8 integrin cell surface interactions |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.8 | REACTOME TRAF6 MEDIATED IRF7 ACTIVATION | Genes involved in TRAF6 mediated IRF7 activation |
| 0.0 | 0.2 | REACTOME ACYL CHAIN REMODELLING OF PS | Genes involved in Acyl chain remodelling of PS |
| 0.0 | 0.2 | REACTOME SIGNAL REGULATORY PROTEIN SIRP FAMILY INTERACTIONS | Genes involved in Signal regulatory protein (SIRP) family interactions |
| 0.0 | 0.2 | REACTOME PYRIMIDINE CATABOLISM | Genes involved in Pyrimidine catabolism |
| 0.0 | 0.4 | REACTOME IMMUNOREGULATORY INTERACTIONS BETWEEN A LYMPHOID AND A NON LYMPHOID CELL | Genes involved in Immunoregulatory interactions between a Lymphoid and a non-Lymphoid cell |
| 0.0 | 0.5 | REACTOME SYNTHESIS AND INTERCONVERSION OF NUCLEOTIDE DI AND TRIPHOSPHATES | Genes involved in Synthesis and interconversion of nucleotide di- and triphosphates |
| 0.0 | 0.2 | REACTOME SLBP DEPENDENT PROCESSING OF REPLICATION DEPENDENT HISTONE PRE MRNAS | Genes involved in SLBP Dependent Processing of Replication-Dependent Histone Pre-mRNAs |
| 0.0 | 0.2 | REACTOME HDL MEDIATED LIPID TRANSPORT | Genes involved in HDL-mediated lipid transport |
| 0.0 | 0.3 | REACTOME DEPOSITION OF NEW CENPA CONTAINING NUCLEOSOMES AT THE CENTROMERE | Genes involved in Deposition of New CENPA-containing Nucleosomes at the Centromere |
| 0.0 | 0.3 | REACTOME COMPLEMENT CASCADE | Genes involved in Complement cascade |