Project
PRJNA207719: Tissue specific transcriptome profiling
Navigation
Downloads
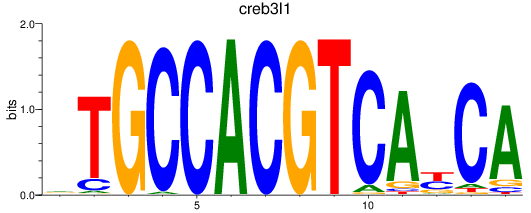
Results for creb3l1
Z-value: 0.44
Transcription factors associated with creb3l1
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
creb3l1
|
ENSDARG00000015793 | cAMP responsive element binding protein 3-like 1 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| creb3l1 | dr11_v1_chr7_+_38897836_38897836 | -0.81 | 9.6e-02 | Click! |
Activity profile of creb3l1 motif
Sorted Z-values of creb3l1 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr4_+_68562464 | 0.43 |
ENSDART00000192954
|
BX548011.4
|
|
| chr22_-_2886937 | 0.38 |
ENSDART00000063533
|
aqp12
|
aquaporin 12 |
| chr21_-_43636595 | 0.25 |
ENSDART00000151115
ENSDART00000151486 ENSDART00000151778 |
si:ch1073-263o8.2
|
si:ch1073-263o8.2 |
| chr1_-_7930679 | 0.24 |
ENSDART00000146090
|
si:dkey-79f11.10
|
si:dkey-79f11.10 |
| chr15_-_41245962 | 0.22 |
ENSDART00000155359
|
smco4
|
single-pass membrane protein with coiled-coil domains 4 |
| chr4_-_26095755 | 0.19 |
ENSDART00000100611
ENSDART00000191266 |
si:ch211-244b2.3
|
si:ch211-244b2.3 |
| chr18_-_34549721 | 0.19 |
ENSDART00000137101
ENSDART00000021880 |
ssr3
|
signal sequence receptor, gamma |
| chr18_-_7481036 | 0.18 |
ENSDART00000101292
|
si:dkey-238c7.16
|
si:dkey-238c7.16 |
| chr5_+_32924669 | 0.17 |
ENSDART00000085219
|
lmo4a
|
LIM domain only 4a |
| chr4_-_17812131 | 0.16 |
ENSDART00000025731
|
spic
|
Spi-C transcription factor (Spi-1/PU.1 related) |
| chr20_+_43083745 | 0.16 |
ENSDART00000139014
ENSDART00000153438 |
moxd1l
|
monooxygenase, DBH-like 1, like |
| chr22_-_31059670 | 0.16 |
ENSDART00000022445
|
cand2
|
cullin-associated and neddylation-dissociated 2 (putative) |
| chr22_+_30446987 | 0.15 |
ENSDART00000146471
|
si:dkey-169i5.4
|
si:dkey-169i5.4 |
| chr24_+_854877 | 0.15 |
ENSDART00000188408
|
piezo2a.2
|
piezo-type mechanosensitive ion channel component 2a, tandem duplicate 2 |
| chr23_-_31266586 | 0.14 |
ENSDART00000139746
|
si:dkey-261l7.2
|
si:dkey-261l7.2 |
| chr6_-_50685862 | 0.14 |
ENSDART00000134146
|
mtss1
|
metastasis suppressor 1 |
| chr21_+_10701834 | 0.14 |
ENSDART00000192473
|
lman1
|
lectin, mannose-binding, 1 |
| chr21_+_10702031 | 0.13 |
ENSDART00000102304
|
lman1
|
lectin, mannose-binding, 1 |
| chr4_+_58576146 | 0.13 |
ENSDART00000164911
|
si:ch211-212k5.4
|
si:ch211-212k5.4 |
| chr4_+_11464255 | 0.12 |
ENSDART00000008584
|
gdi2
|
GDP dissociation inhibitor 2 |
| chr4_+_25651720 | 0.12 |
ENSDART00000100693
ENSDART00000100717 |
acot16
|
acyl-CoA thioesterase 16 |
| chr16_+_19637384 | 0.12 |
ENSDART00000184773
ENSDART00000191895 ENSDART00000182020 ENSDART00000135359 |
macc1
|
metastasis associated in colon cancer 1 |
| chr4_+_13568469 | 0.11 |
ENSDART00000171235
ENSDART00000136152 |
calua
|
calumenin a |
| chr18_+_7553950 | 0.11 |
ENSDART00000193420
ENSDART00000062143 |
zgc:77650
|
zgc:77650 |
| chr11_-_2456656 | 0.11 |
ENSDART00000171219
ENSDART00000167938 |
slc2a10
|
solute carrier family 2 (facilitated glucose transporter), member 10 |
| chr18_-_21097663 | 0.11 |
ENSDART00000060196
|
anpepa
|
alanyl (membrane) aminopeptidase a |
| chr1_+_277731 | 0.10 |
ENSDART00000133431
|
cenpe
|
centromere protein E |
| chr3_+_32862730 | 0.10 |
ENSDART00000144939
ENSDART00000125126 ENSDART00000144531 ENSDART00000141717 ENSDART00000137599 |
zgc:162613
|
zgc:162613 |
| chr4_+_2637947 | 0.10 |
ENSDART00000130623
|
dus4l
|
dihydrouridine synthase 4-like (S. cerevisiae) |
| chr18_-_7031409 | 0.10 |
ENSDART00000148485
ENSDART00000005405 |
calub
|
calumenin b |
| chr18_+_2153530 | 0.09 |
ENSDART00000158255
|
pygo1
|
pygopus family PHD finger 1 |
| chr3_-_180860 | 0.09 |
ENSDART00000059956
ENSDART00000192506 |
kdelr3
|
KDEL (Lys-Asp-Glu-Leu) endoplasmic reticulum protein retention receptor 3 |
| chr25_+_37443194 | 0.09 |
ENSDART00000163178
ENSDART00000190262 |
slc10a3
|
solute carrier family 10, member 3 |
| chr10_-_3427589 | 0.09 |
ENSDART00000133452
ENSDART00000037183 |
tmed2
|
transmembrane p24 trafficking protein 2 |
| chr22_-_10482253 | 0.09 |
ENSDART00000143164
|
aspn
|
asporin (LRR class 1) |
| chr5_-_48680580 | 0.08 |
ENSDART00000031194
|
lysmd3
|
LysM, putative peptidoglycan-binding, domain containing 3 |
| chr25_-_3034668 | 0.08 |
ENSDART00000053405
|
scamp2
|
secretory carrier membrane protein 2 |
| chr12_+_30789611 | 0.08 |
ENSDART00000181501
|
aldh18a1
|
aldehyde dehydrogenase 18 family, member A1 |
| chr20_+_34511678 | 0.08 |
ENSDART00000061588
|
prrx1b
|
paired related homeobox 1b |
| chr14_-_45464112 | 0.08 |
ENSDART00000173209
|
ehd1a
|
EH-domain containing 1a |
| chr6_+_13506841 | 0.08 |
ENSDART00000032331
|
gmppab
|
GDP-mannose pyrophosphorylase Ab |
| chr7_-_45990386 | 0.07 |
ENSDART00000186008
|
si:ch211-260e23.7
|
si:ch211-260e23.7 |
| chr7_-_45990681 | 0.07 |
ENSDART00000165441
|
si:ch211-260e23.7
|
si:ch211-260e23.7 |
| chr22_+_31059919 | 0.07 |
ENSDART00000077063
|
sec13
|
SEC13 homolog, nuclear pore and COPII coat complex component |
| chrM_+_9735 | 0.07 |
ENSDART00000093613
|
mt-co3
|
cytochrome c oxidase III, mitochondrial |
| chr17_-_36818176 | 0.07 |
ENSDART00000061762
|
myo6b
|
myosin VIb |
| chr25_-_37331513 | 0.06 |
ENSDART00000111862
|
ldlrad3
|
low density lipoprotein receptor class A domain containing 3 |
| chr10_+_31646020 | 0.06 |
ENSDART00000115251
|
esama
|
endothelial cell adhesion molecule a |
| chr1_-_18592068 | 0.06 |
ENSDART00000082063
|
fam114a1
|
family with sequence similarity 114, member A1 |
| chr7_-_58826164 | 0.06 |
ENSDART00000171095
|
sox32
|
SRY (sex determining region Y)-box 32 |
| chr20_+_22799857 | 0.06 |
ENSDART00000058527
|
scfd2
|
sec1 family domain containing 2 |
| chr15_-_29387446 | 0.06 |
ENSDART00000145976
ENSDART00000035096 |
serpinh1b
|
serpin peptidase inhibitor, clade H (heat shock protein 47), member 1b |
| chr3_+_26123736 | 0.06 |
ENSDART00000010477
|
hsp70.3
|
heat shock cognate 70-kd protein, tandem duplicate 3 |
| chr18_-_18587745 | 0.06 |
ENSDART00000191973
|
sf3b3
|
splicing factor 3b, subunit 3 |
| chr22_-_14346575 | 0.05 |
ENSDART00000136326
|
si:dkeyp-122a9.1
|
si:dkeyp-122a9.1 |
| chr19_+_44039849 | 0.05 |
ENSDART00000086040
|
lrrc14b
|
leucine rich repeat containing 14B |
| chr22_-_1168291 | 0.05 |
ENSDART00000167724
|
si:ch1073-181h11.4
|
si:ch1073-181h11.4 |
| chr9_-_42730672 | 0.05 |
ENSDART00000136728
|
fkbp7
|
FK506 binding protein 7 |
| chr5_-_39474235 | 0.05 |
ENSDART00000171557
|
antxr2a
|
anthrax toxin receptor 2a |
| chr21_+_5932140 | 0.05 |
ENSDART00000193767
|
rexo4
|
REX4 homolog, 3'-5' exonuclease |
| chr9_-_29497916 | 0.04 |
ENSDART00000060246
|
dnajc3a
|
DnaJ (Hsp40) homolog, subfamily C, member 3a |
| chr22_+_31295791 | 0.04 |
ENSDART00000092447
|
grip2b
|
glutamate receptor interacting protein 2b |
| chr17_-_11417551 | 0.04 |
ENSDART00000128291
|
arid4a
|
AT rich interactive domain 4A (RBP1-like) |
| chr3_-_26184018 | 0.04 |
ENSDART00000191604
|
si:ch211-11k18.4
|
si:ch211-11k18.4 |
| chr17_-_39880725 | 0.03 |
ENSDART00000184030
|
zmp:0000000545
|
zmp:0000000545 |
| chr5_+_58996765 | 0.03 |
ENSDART00000062175
|
derl2
|
derlin 2 |
| chr23_-_25798099 | 0.03 |
ENSDART00000041833
|
fitm2
|
fat storage-inducing transmembrane protein 2 |
| chr4_-_2637689 | 0.02 |
ENSDART00000192550
ENSDART00000021953 ENSDART00000150344 |
cog5
|
component of oligomeric golgi complex 5 |
| chr20_-_14875308 | 0.02 |
ENSDART00000141290
|
dnm3a
|
dynamin 3a |
| chr17_-_20849879 | 0.02 |
ENSDART00000088100
ENSDART00000149630 |
ank3b
|
ankyrin 3b |
| chr5_-_28968964 | 0.02 |
ENSDART00000184936
ENSDART00000016628 |
fam129bb
|
family with sequence similarity 129, member Bb |
| chr5_-_25066780 | 0.02 |
ENSDART00000002118
ENSDART00000182575 |
pnpla7b
|
patatin-like phospholipase domain containing 7b |
| chr4_+_58016732 | 0.02 |
ENSDART00000165777
|
CT027609.1
|
|
| chr20_-_22464250 | 0.02 |
ENSDART00000165904
|
pdgfra
|
platelet-derived growth factor receptor, alpha polypeptide |
| chr7_+_13491452 | 0.02 |
ENSDART00000053535
|
arih1l
|
ariadne homolog, ubiquitin-conjugating enzyme E2 binding protein, 1 like |
| chr23_-_306796 | 0.02 |
ENSDART00000143125
|
anks1aa
|
ankyrin repeat and sterile alpha motif domain containing 1Aa |
| chr3_-_26183699 | 0.01 |
ENSDART00000147517
ENSDART00000140731 |
si:ch211-11k18.4
|
si:ch211-11k18.4 |
| chr6_+_3864040 | 0.01 |
ENSDART00000013743
|
gorasp2
|
golgi reassembly stacking protein 2 |
| chr20_-_38617766 | 0.00 |
ENSDART00000050474
|
slc30a2
|
solute carrier family 30 (zinc transporter), member 2 |
| chr2_-_23172708 | 0.00 |
ENSDART00000041365
|
prrx1a
|
paired related homeobox 1a |
| chr9_-_49464993 | 0.00 |
ENSDART00000181471
|
xirp2b
|
xin actin binding repeat containing 2b |
| chr8_+_47219107 | 0.00 |
ENSDART00000146018
ENSDART00000075068 |
mthfr
|
methylenetetrahydrofolate reductase (NAD(P)H) |
Network of associatons between targets according to the STRING database.
First level regulatory network of creb3l1
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.2 | GO:0006589 | octopamine biosynthetic process(GO:0006589) dopamine catabolic process(GO:0042420) norepinephrine biosynthetic process(GO:0042421) octopamine metabolic process(GO:0046333) |
| 0.1 | 0.2 | GO:0010265 | SCF complex assembly(GO:0010265) |
| 0.0 | 0.1 | GO:0070278 | extracellular matrix constituent secretion(GO:0070278) |
| 0.0 | 0.1 | GO:0071260 | cellular response to mechanical stimulus(GO:0071260) |
| 0.0 | 0.1 | GO:0006621 | protein retention in ER lumen(GO:0006621) |
| 0.0 | 0.7 | GO:0071391 | cellular response to estrogen stimulus(GO:0071391) |
| 0.0 | 0.1 | GO:0009298 | GDP-mannose biosynthetic process(GO:0009298) |
| 0.0 | 0.2 | GO:0006614 | SRP-dependent cotranslational protein targeting to membrane(GO:0006614) |
| 0.0 | 0.1 | GO:1905066 | regulation of canonical Wnt signaling pathway involved in heart development(GO:1905066) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.2 | GO:0033018 | sarcoplasmic reticulum lumen(GO:0033018) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.2 | GO:0004500 | dopamine beta-monooxygenase activity(GO:0004500) |
| 0.0 | 0.1 | GO:0005093 | Rab GDP-dissociation inhibitor activity(GO:0005093) |
| 0.0 | 0.1 | GO:0046923 | ER retention sequence binding(GO:0046923) |
| 0.0 | 0.3 | GO:0005537 | mannose binding(GO:0005537) |
| 0.0 | 0.1 | GO:0004475 | mannose-1-phosphate guanylyltransferase activity(GO:0004475) mannose-phosphate guanylyltransferase activity(GO:0008905) |
| 0.0 | 0.1 | GO:0016774 | phosphotransferase activity, carboxyl group as acceptor(GO:0016774) |
| 0.0 | 0.1 | GO:0017150 | tRNA dihydrouridine synthase activity(GO:0017150) |
| 0.0 | 0.1 | GO:0008508 | bile acid:sodium symporter activity(GO:0008508) |
| 0.0 | 0.1 | GO:0030620 | U2 snRNA binding(GO:0030620) |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | REACTOME FACILITATIVE NA INDEPENDENT GLUCOSE TRANSPORTERS | Genes involved in Facilitative Na+-independent glucose transporters |