Project
PRJNA207719: Tissue specific transcriptome profiling
Navigation
Downloads
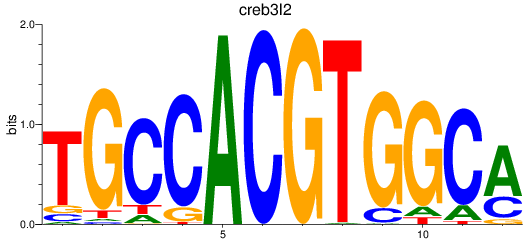
Results for creb3l2
Z-value: 0.76
Transcription factors associated with creb3l2
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
creb3l2
|
ENSDARG00000063563 | cAMP responsive element binding protein 3-like 2 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| creb3l2 | dr11_v1_chr4_-_4751981_4751983 | -0.31 | 6.1e-01 | Click! |
Activity profile of creb3l2 motif
Sorted Z-values of creb3l2 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr23_-_10175898 | 1.15 |
ENSDART00000146185
|
krt5
|
keratin 5 |
| chr16_-_45178430 | 0.88 |
ENSDART00000165186
|
si:dkey-33i11.9
|
si:dkey-33i11.9 |
| chr16_+_49249669 | 0.79 |
ENSDART00000181899
|
LO018128.1
|
|
| chr2_+_50999675 | 0.75 |
ENSDART00000158064
ENSDART00000165746 ENSDART00000163917 ENSDART00000172038 ENSDART00000169048 ENSDART00000164775 |
eef1da
|
eukaryotic translation elongation factor 1 delta a (guanine nucleotide exchange protein) |
| chr3_-_55092051 | 0.71 |
ENSDART00000053077
|
hbba2
|
hemoglobin, beta adult 2 |
| chr2_+_50999477 | 0.64 |
ENSDART00000190111
|
eef1da
|
eukaryotic translation elongation factor 1 delta a (guanine nucleotide exchange protein) |
| chr2_+_39618951 | 0.60 |
ENSDART00000077108
|
zgc:136870
|
zgc:136870 |
| chr18_+_7553950 | 0.59 |
ENSDART00000193420
ENSDART00000062143 |
zgc:77650
|
zgc:77650 |
| chr8_+_44783424 | 0.51 |
ENSDART00000025875
|
si:ch1073-459j12.1
|
si:ch1073-459j12.1 |
| chr5_-_1962500 | 0.50 |
ENSDART00000150163
|
rplp0
|
ribosomal protein, large, P0 |
| chr22_-_817479 | 0.48 |
ENSDART00000123487
|
zgc:153675
|
zgc:153675 |
| chr12_-_9132682 | 0.43 |
ENSDART00000066471
|
adam8b
|
ADAM metallopeptidase domain 8b |
| chr19_+_47405867 | 0.42 |
ENSDART00000041114
|
psmb2
|
proteasome subunit beta 2 |
| chr2_+_3452590 | 0.40 |
ENSDART00000153935
|
cyp8b2
|
cytochrome P450, family 8, subfamily B, polypeptide 2 |
| chr6_+_269204 | 0.40 |
ENSDART00000191678
|
atf4a
|
activating transcription factor 4a |
| chr8_+_7316568 | 0.38 |
ENSDART00000140874
|
selenoh
|
selenoprotein H |
| chr22_+_9871238 | 0.38 |
ENSDART00000141085
ENSDART00000105939 |
si:dkey-253d23.4
|
si:dkey-253d23.4 |
| chr14_+_22076596 | 0.37 |
ENSDART00000106147
ENSDART00000100278 ENSDART00000131489 |
slc43a1a
|
solute carrier family 43 (amino acid system L transporter), member 1a |
| chr8_-_15292197 | 0.37 |
ENSDART00000140867
|
spata6
|
spermatogenesis associated 6 |
| chr16_-_17200120 | 0.36 |
ENSDART00000147739
|
gapdh
|
glyceraldehyde-3-phosphate dehydrogenase |
| chr6_+_12968101 | 0.35 |
ENSDART00000013781
|
mcm6
|
minichromosome maintenance complex component 6 |
| chr22_-_31059670 | 0.34 |
ENSDART00000022445
|
cand2
|
cullin-associated and neddylation-dissociated 2 (putative) |
| chr12_+_13282797 | 0.34 |
ENSDART00000137757
ENSDART00000152397 |
irf9
|
interferon regulatory factor 9 |
| chr21_-_5881344 | 0.32 |
ENSDART00000009241
|
rpl35
|
ribosomal protein L35 |
| chr25_+_5035343 | 0.31 |
ENSDART00000011751
|
parvb
|
parvin, beta |
| chr5_-_54714789 | 0.29 |
ENSDART00000063357
|
ccnb1
|
cyclin B1 |
| chr20_+_32406011 | 0.29 |
ENSDART00000018640
ENSDART00000137910 |
snx3
|
sorting nexin 3 |
| chr18_-_37252036 | 0.28 |
ENSDART00000132230
|
six5
|
SIX homeobox 5 |
| chr22_+_31059919 | 0.27 |
ENSDART00000077063
|
sec13
|
SEC13 homolog, nuclear pore and COPII coat complex component |
| chr5_-_54714525 | 0.26 |
ENSDART00000150138
ENSDART00000150070 |
ccnb1
|
cyclin B1 |
| chr7_-_45990386 | 0.26 |
ENSDART00000186008
|
si:ch211-260e23.7
|
si:ch211-260e23.7 |
| chr3_+_22335030 | 0.25 |
ENSDART00000055676
|
zgc:103564
|
zgc:103564 |
| chr8_-_46386024 | 0.25 |
ENSDART00000136602
ENSDART00000060919 ENSDART00000137472 |
qars
|
glutaminyl-tRNA synthetase |
| chr9_+_6587364 | 0.25 |
ENSDART00000122279
|
fhl2a
|
four and a half LIM domains 2a |
| chr25_-_7670616 | 0.24 |
ENSDART00000131583
ENSDART00000142794 |
bet1l
|
Bet1 golgi vesicular membrane trafficking protein-like |
| chr24_+_12913329 | 0.22 |
ENSDART00000141829
|
si:dkeyp-28d2.4
|
si:dkeyp-28d2.4 |
| chr4_-_17409533 | 0.22 |
ENSDART00000011943
|
pah
|
phenylalanine hydroxylase |
| chr7_-_45990681 | 0.22 |
ENSDART00000165441
|
si:ch211-260e23.7
|
si:ch211-260e23.7 |
| chr19_-_42607451 | 0.21 |
ENSDART00000004392
|
fkbp9
|
FK506 binding protein 9 |
| chr3_+_44062576 | 0.21 |
ENSDART00000161277
ENSDART00000168784 |
nsmce1
|
NSE1 homolog, SMC5-SMC6 complex component |
| chr2_-_10386738 | 0.20 |
ENSDART00000016369
|
wls
|
wntless Wnt ligand secretion mediator |
| chr5_-_28968964 | 0.20 |
ENSDART00000184936
ENSDART00000016628 |
fam129bb
|
family with sequence similarity 129, member Bb |
| chr8_+_8699085 | 0.20 |
ENSDART00000021209
|
uxt
|
ubiquitously-expressed, prefoldin-like chaperone |
| chr17_-_11417904 | 0.20 |
ENSDART00000103228
|
arid4a
|
AT rich interactive domain 4A (RBP1-like) |
| chr8_-_51578926 | 0.19 |
ENSDART00000190625
|
ankrd39
|
ankyrin repeat domain 39 |
| chr8_-_51579286 | 0.18 |
ENSDART00000147878
|
ankrd39
|
ankyrin repeat domain 39 |
| chr16_+_813780 | 0.18 |
ENSDART00000162474
ENSDART00000161774 |
irx1a
|
iroquois homeobox 1a |
| chr22_-_94352 | 0.18 |
ENSDART00000184883
|
ndufaf3
|
NADH dehydrogenase (ubiquinone) complex I, assembly factor 3 |
| chr25_-_37084032 | 0.17 |
ENSDART00000025494
|
hprt1l
|
hypoxanthine phosphoribosyltransferase 1, like |
| chr21_-_27413294 | 0.17 |
ENSDART00000131646
|
slc29a2
|
solute carrier family 29 (equilibrative nucleoside transporter), member 2 |
| chr17_-_15229787 | 0.17 |
ENSDART00000039165
|
styx
|
serine/threonine/tyrosine interacting protein |
| chr20_+_15565926 | 0.16 |
ENSDART00000063917
|
si:dkey-86e18.1
|
si:dkey-86e18.1 |
| chr22_-_25043103 | 0.16 |
ENSDART00000015512
|
polr3f
|
polymerase (RNA) III (DNA directed) polypeptide F |
| chr4_-_17391091 | 0.15 |
ENSDART00000056002
|
th2
|
tyrosine hydroxylase 2 |
| chr9_-_7683799 | 0.15 |
ENSDART00000102713
|
si:ch73-199e17.1
|
si:ch73-199e17.1 |
| chr19_-_3303995 | 0.15 |
ENSDART00000105150
|
si:ch211-133n4.9
|
si:ch211-133n4.9 |
| chr4_-_22749553 | 0.15 |
ENSDART00000040033
|
nup107
|
nucleoporin 107 |
| chr18_-_43857089 | 0.15 |
ENSDART00000150170
|
treh
|
trehalase (brush-border membrane glycoprotein) |
| chr16_+_20294976 | 0.14 |
ENSDART00000059619
|
fkbp14
|
FK506 binding protein 14 |
| chr8_+_49065348 | 0.14 |
ENSDART00000032277
|
ddx51
|
DEAD (Asp-Glu-Ala-Asp) box polypeptide 51 |
| chr22_+_25841786 | 0.14 |
ENSDART00000180863
|
vasna
|
vasorin a |
| chr14_+_16287968 | 0.14 |
ENSDART00000106593
|
prpf19
|
pre-mRNA processing factor 19 |
| chr16_+_26824691 | 0.14 |
ENSDART00000135053
|
zmp:0000001316
|
zmp:0000001316 |
| chr22_-_14128716 | 0.13 |
ENSDART00000140323
|
si:ch211-246m6.4
|
si:ch211-246m6.4 |
| chr17_-_11417551 | 0.13 |
ENSDART00000128291
|
arid4a
|
AT rich interactive domain 4A (RBP1-like) |
| chr5_-_32338866 | 0.13 |
ENSDART00000017956
ENSDART00000047670 |
dab2
|
Dab, mitogen-responsive phosphoprotein, homolog 2 (Drosophila) |
| chr25_+_37443194 | 0.11 |
ENSDART00000163178
ENSDART00000190262 |
slc10a3
|
solute carrier family 10, member 3 |
| chr15_-_43978141 | 0.11 |
ENSDART00000041249
|
chordc1a
|
cysteine and histidine-rich domain (CHORD) containing 1a |
| chr17_+_28706946 | 0.11 |
ENSDART00000126967
|
strn3
|
striatin, calmodulin binding protein 3 |
| chr7_-_15830251 | 0.11 |
ENSDART00000102363
|
rcn1
|
reticulocalbin 1, EF-hand calcium binding domain |
| chr3_+_42923275 | 0.11 |
ENSDART00000168228
|
tmem184a
|
transmembrane protein 184a |
| chr16_-_31622777 | 0.11 |
ENSDART00000137311
ENSDART00000002930 |
phf20l1
|
PHD finger protein 20 like 1 |
| chr12_-_44010532 | 0.10 |
ENSDART00000183875
|
si:ch211-182p11.1
|
si:ch211-182p11.1 |
| chr4_+_17844013 | 0.10 |
ENSDART00000019165
|
apaf1
|
apoptotic peptidase activating factor 1 |
| chr4_+_357810 | 0.10 |
ENSDART00000163436
ENSDART00000103645 |
tmem181
|
transmembrane protein 181 |
| chr22_-_5099824 | 0.09 |
ENSDART00000122341
ENSDART00000161345 |
zfr2
|
zinc finger RNA binding protein 2 |
| chr3_+_39568290 | 0.08 |
ENSDART00000020741
|
aldoaa
|
aldolase a, fructose-bisphosphate, a |
| chr24_-_982443 | 0.08 |
ENSDART00000063151
|
napga
|
N-ethylmaleimide-sensitive factor attachment protein, gamma a |
| chr5_-_22130937 | 0.07 |
ENSDART00000138606
|
las1l
|
LAS1-like, ribosome biogenesis factor |
| chr17_-_28811747 | 0.07 |
ENSDART00000001444
|
g2e3
|
G2/M-phase specific E3 ubiquitin protein ligase |
| chr7_-_47148440 | 0.07 |
ENSDART00000185435
ENSDART00000189719 |
BX005073.3
|
|
| chr4_-_16883051 | 0.07 |
ENSDART00000124627
|
strap
|
serine/threonine kinase receptor associated protein |
| chr14_-_26436760 | 0.06 |
ENSDART00000088677
|
si:dkeyp-110e4.6
|
si:dkeyp-110e4.6 |
| chr17_+_44441042 | 0.06 |
ENSDART00000142123
|
ap5m1
|
adaptor-related protein complex 5, mu 1 subunit |
| chr2_-_44971551 | 0.06 |
ENSDART00000018818
|
mul1a
|
mitochondrial E3 ubiquitin protein ligase 1a |
| chr20_+_29209926 | 0.05 |
ENSDART00000152949
ENSDART00000153016 |
katnbl1
|
katanin p80 subunit B-like 1 |
| chr3_-_18384501 | 0.05 |
ENSDART00000027630
|
kdelr2a
|
KDEL (Lys-Asp-Glu-Leu) endoplasmic reticulum protein retention receptor 2a |
| chr14_-_26436951 | 0.05 |
ENSDART00000140173
|
si:dkeyp-110e4.6
|
si:dkeyp-110e4.6 |
| chr7_-_28058442 | 0.05 |
ENSDART00000173842
|
si:ch211-235p24.2
|
si:ch211-235p24.2 |
| chr17_-_28797395 | 0.05 |
ENSDART00000134735
|
scfd1
|
sec1 family domain containing 1 |
| chr1_+_17593392 | 0.05 |
ENSDART00000078889
|
helt
|
helt bHLH transcription factor |
| chr4_+_77957611 | 0.05 |
ENSDART00000156692
|
arfgap3
|
ADP-ribosylation factor GTPase activating protein 3 |
| chr4_-_48578324 | 0.05 |
ENSDART00000122707
|
znf980
|
zinc finger protein 980 |
| chr2_-_55298075 | 0.04 |
ENSDART00000186404
ENSDART00000149062 |
rab8a
|
RAB8A, member RAS oncogene family |
| chr1_-_26444075 | 0.04 |
ENSDART00000125690
|
ints12
|
integrator complex subunit 12 |
| chr15_-_29387446 | 0.04 |
ENSDART00000145976
ENSDART00000035096 |
serpinh1b
|
serpin peptidase inhibitor, clade H (heat shock protein 47), member 1b |
| chr8_+_23355484 | 0.04 |
ENSDART00000085361
ENSDART00000125729 |
dnmt3ba
|
DNA (cytosine-5-)-methyltransferase 3 beta, duplicate a |
| chr17_+_1514711 | 0.04 |
ENSDART00000165641
|
akt1
|
v-akt murine thymoma viral oncogene homolog 1 |
| chr7_+_15308042 | 0.03 |
ENSDART00000185170
|
mespaa
|
mesoderm posterior aa |
| chr19_+_7173613 | 0.03 |
ENSDART00000001331
|
hsd17b8
|
hydroxysteroid (17-beta) dehydrogenase 8 |
| chr5_-_67757188 | 0.03 |
ENSDART00000167168
|
b4galt4
|
UDP-Gal:betaGlcNAc beta 1,4- galactosyltransferase, polypeptide 4 |
| chr8_+_46386601 | 0.03 |
ENSDART00000129661
ENSDART00000084081 |
ogg1
|
8-oxoguanine DNA glycosylase |
| chr21_+_45685757 | 0.03 |
ENSDART00000160530
|
sec24a
|
SEC24 homolog A, COPII coat complex component |
| chr20_+_29209767 | 0.03 |
ENSDART00000141252
|
katnbl1
|
katanin p80 subunit B-like 1 |
| chr20_-_7069612 | 0.03 |
ENSDART00000040793
|
sirt5
|
sirtuin 5 |
| chr9_+_31222026 | 0.03 |
ENSDART00000145573
|
clybl
|
citrate lyase beta like |
| chr8_-_25980694 | 0.03 |
ENSDART00000135456
|
si:dkey-72l14.3
|
si:dkey-72l14.3 |
| chr4_+_17843717 | 0.02 |
ENSDART00000113507
|
apaf1
|
apoptotic peptidase activating factor 1 |
| chr11_-_43104475 | 0.02 |
ENSDART00000125368
|
acyp2
|
acylphosphatase 2, muscle type |
| chr4_-_12718367 | 0.02 |
ENSDART00000035259
|
mgst1.1
|
microsomal glutathione S-transferase 1.1 |
| chr2_-_11504347 | 0.02 |
ENSDART00000019392
|
sdr16c5a
|
short chain dehydrogenase/reductase family 16C, member 5a |
| chr19_-_7540821 | 0.01 |
ENSDART00000143958
|
lix1l
|
limb and CNS expressed 1 like |
| chr4_-_12718103 | 0.01 |
ENSDART00000144388
|
mgst1.1
|
microsomal glutathione S-transferase 1.1 |
| chr15_-_29388012 | 0.01 |
ENSDART00000115032
|
serpinh1b
|
serpin peptidase inhibitor, clade H (heat shock protein 47), member 1b |
| chr20_+_51061695 | 0.01 |
ENSDART00000134416
|
im:7140055
|
im:7140055 |
| chr23_-_46034609 | 0.01 |
ENSDART00000158712
|
zgc:65873
|
zgc:65873 |
| chr21_+_40589770 | 0.00 |
ENSDART00000164650
ENSDART00000161584 ENSDART00000161108 |
pdk3b
|
pyruvate dehydrogenase kinase, isozyme 3b |
| chr11_-_7156620 | 0.00 |
ENSDART00000172823
ENSDART00000172879 ENSDART00000078916 |
smim7
|
small integral membrane protein 7 |
Network of associatons between targets according to the STRING database.
First level regulatory network of creb3l2
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.4 | GO:0051044 | positive regulation of membrane protein ectodomain proteolysis(GO:0051044) |
| 0.1 | 0.3 | GO:0010265 | SCF complex assembly(GO:0010265) |
| 0.1 | 0.4 | GO:1904353 | positive regulation of telomere maintenance(GO:0032206) regulation of telomere capping(GO:1904353) positive regulation of telomere capping(GO:1904355) |
| 0.1 | 0.3 | GO:0070278 | extracellular matrix constituent secretion(GO:0070278) |
| 0.1 | 0.2 | GO:0061355 | Wnt protein secretion(GO:0061355) |
| 0.1 | 0.6 | GO:0007080 | mitotic metaphase plate congression(GO:0007080) |
| 0.1 | 0.2 | GO:0006178 | guanine salvage(GO:0006178) GMP salvage(GO:0032263) guanine biosynthetic process(GO:0046099) |
| 0.1 | 0.2 | GO:0000350 | generation of catalytic spliceosome for second transesterification step(GO:0000350) |
| 0.1 | 0.2 | GO:0042416 | dopamine biosynthetic process from tyrosine(GO:0006585) dopamine biosynthetic process(GO:0042416) |
| 0.0 | 0.1 | GO:0000973 | posttranscriptional tethering of RNA polymerase II gene DNA at nuclear periphery(GO:0000973) |
| 0.0 | 0.1 | GO:0005991 | trehalose metabolic process(GO:0005991) |
| 0.0 | 0.1 | GO:0000393 | spliceosomal conformational changes to generate catalytic conformation(GO:0000393) |
| 0.0 | 0.7 | GO:0015671 | oxygen transport(GO:0015671) |
| 0.0 | 0.4 | GO:0070309 | intrinsic apoptotic signaling pathway in response to endoplasmic reticulum stress(GO:0070059) lens fiber cell development(GO:0070307) lens fiber cell morphogenesis(GO:0070309) |
| 0.0 | 0.2 | GO:0043393 | regulation of protein binding(GO:0043393) |
| 0.0 | 0.4 | GO:1902299 | pre-replicative complex assembly involved in nuclear cell cycle DNA replication(GO:0006267) pre-replicative complex assembly(GO:0036388) pre-replicative complex assembly involved in cell cycle DNA replication(GO:1902299) |
| 0.0 | 0.2 | GO:0015862 | uridine transport(GO:0015862) pyrimidine nucleoside transport(GO:0015864) |
| 0.0 | 1.4 | GO:0006414 | translational elongation(GO:0006414) |
| 0.0 | 0.5 | GO:0000027 | ribosomal large subunit assembly(GO:0000027) |
| 0.0 | 0.3 | GO:0000463 | maturation of LSU-rRNA from tricistronic rRNA transcript (SSU-rRNA, 5.8S rRNA, LSU-rRNA)(GO:0000463) |
| 0.0 | 0.2 | GO:0006570 | L-phenylalanine metabolic process(GO:0006558) L-phenylalanine catabolic process(GO:0006559) tyrosine metabolic process(GO:0006570) erythrose 4-phosphate/phosphoenolpyruvate family amino acid metabolic process(GO:1902221) erythrose 4-phosphate/phosphoenolpyruvate family amino acid catabolic process(GO:1902222) |
| 0.0 | 0.1 | GO:2000725 | regulation of cardiocyte differentiation(GO:1905207) regulation of cardiac muscle cell differentiation(GO:2000725) |
| 0.0 | 0.0 | GO:0035964 | COPI-coated vesicle budding(GO:0035964) Golgi transport vesicle coating(GO:0048200) COPI coating of Golgi vesicle(GO:0048205) |
| 0.0 | 0.4 | GO:0045454 | cell redox homeostasis(GO:0045454) |
| 0.0 | 0.4 | GO:0010499 | proteasomal ubiquitin-independent protein catabolic process(GO:0010499) |
| 0.0 | 0.3 | GO:0030032 | lamellipodium assembly(GO:0030032) |
| 0.0 | 0.3 | GO:0042541 | hemoglobin biosynthetic process(GO:0042541) |
| 0.0 | 0.4 | GO:0015804 | neutral amino acid transport(GO:0015804) |
| 0.0 | 0.4 | GO:0050821 | protein stabilization(GO:0050821) |
| 0.0 | 1.2 | GO:0031101 | fin regeneration(GO:0031101) |
| 0.0 | 0.0 | GO:0048210 | Golgi vesicle fusion to target membrane(GO:0048210) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 1.4 | GO:0005853 | eukaryotic translation elongation factor 1 complex(GO:0005853) |
| 0.2 | 0.6 | GO:0097125 | cyclin B1-CDK1 complex(GO:0097125) |
| 0.1 | 0.4 | GO:0071065 | alpha9-beta1 integrin-vascular cell adhesion molecule-1 complex(GO:0071065) |
| 0.1 | 1.1 | GO:0045095 | keratin filament(GO:0045095) |
| 0.1 | 0.4 | GO:0097224 | sperm connecting piece(GO:0097224) |
| 0.1 | 0.7 | GO:0031838 | haptoglobin-hemoglobin complex(GO:0031838) |
| 0.0 | 0.4 | GO:0031080 | nuclear pore outer ring(GO:0031080) |
| 0.0 | 0.2 | GO:0000138 | Golgi trans cisterna(GO:0000138) |
| 0.0 | 0.4 | GO:1990589 | ATF4-CREB1 transcription factor complex(GO:1990589) |
| 0.0 | 0.2 | GO:0030915 | Smc5-Smc6 complex(GO:0030915) |
| 0.0 | 0.3 | GO:0030904 | retromer complex(GO:0030904) |
| 0.0 | 0.1 | GO:0071006 | U2-type catalytic step 1 spliceosome(GO:0071006) catalytic step 1 spliceosome(GO:0071012) |
| 0.0 | 0.2 | GO:0016272 | prefoldin complex(GO:0016272) |
| 0.0 | 0.4 | GO:0005839 | proteasome core complex(GO:0005839) |
| 0.0 | 0.8 | GO:0022625 | cytosolic large ribosomal subunit(GO:0022625) |
| 0.0 | 0.2 | GO:0042555 | MCM complex(GO:0042555) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.4 | GO:0008397 | sterol 12-alpha-hydroxylase activity(GO:0008397) |
| 0.1 | 0.4 | GO:0032027 | myosin light chain binding(GO:0032027) |
| 0.1 | 0.4 | GO:0004365 | glyceraldehyde-3-phosphate dehydrogenase (NAD+) (phosphorylating) activity(GO:0004365) glyceraldehyde-3-phosphate dehydrogenase (NAD(P)+) (phosphorylating) activity(GO:0043891) |
| 0.1 | 0.5 | GO:0070180 | large ribosomal subunit rRNA binding(GO:0070180) |
| 0.1 | 0.4 | GO:0043140 | ATP-dependent 3'-5' DNA helicase activity(GO:0043140) single-stranded DNA-dependent ATP-dependent 3'-5' DNA helicase activity(GO:1990518) |
| 0.1 | 0.7 | GO:0031720 | haptoglobin binding(GO:0031720) |
| 0.1 | 0.2 | GO:0004422 | hypoxanthine phosphoribosyltransferase activity(GO:0004422) |
| 0.1 | 0.4 | GO:0008140 | cAMP response element binding protein binding(GO:0008140) |
| 0.0 | 0.1 | GO:0015927 | alpha,alpha-trehalase activity(GO:0004555) trehalase activity(GO:0015927) |
| 0.0 | 0.4 | GO:0016714 | oxidoreductase activity, acting on paired donors, with incorporation or reduction of molecular oxygen, reduced pteridine as one donor, and incorporation of one atom of oxygen(GO:0016714) |
| 0.0 | 1.4 | GO:0003746 | translation elongation factor activity(GO:0003746) |
| 0.0 | 0.1 | GO:0043531 | ADP binding(GO:0043531) |
| 0.0 | 0.1 | GO:0008508 | bile acid:sodium symporter activity(GO:0008508) |
| 0.0 | 0.1 | GO:0070016 | armadillo repeat domain binding(GO:0070016) |
| 0.0 | 0.1 | GO:0046923 | ER retention sequence binding(GO:0046923) |
| 0.0 | 0.0 | GO:0033130 | acetylcholine receptor binding(GO:0033130) |
| 0.0 | 0.1 | GO:0005483 | soluble NSF attachment protein activity(GO:0005483) |
| 0.0 | 0.2 | GO:0070003 | threonine-type endopeptidase activity(GO:0004298) threonine-type peptidase activity(GO:0070003) |
| 0.0 | 0.1 | GO:0004332 | fructose-bisphosphate aldolase activity(GO:0004332) |
| 0.0 | 0.0 | GO:0000035 | acyl binding(GO:0000035) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.3 | ST TYPE I INTERFERON PATHWAY | Type I Interferon (alpha/beta IFN) Pathway |
| 0.0 | 1.1 | PID DELTA NP63 PATHWAY | Validated transcriptional targets of deltaNp63 isoforms |
| 0.0 | 0.1 | SA PROGRAMMED CELL DEATH | Programmed cell death, or apoptosis, eliminates damaged or unneeded cells. |
| 0.0 | 0.9 | PID MYC ACTIV PATHWAY | Validated targets of C-MYC transcriptional activation |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.6 | REACTOME RECRUITMENT OF NUMA TO MITOTIC CENTROSOMES | Genes involved in Recruitment of NuMA to mitotic centrosomes |
| 0.0 | 0.1 | REACTOME DIGESTION OF DIETARY CARBOHYDRATE | Genes involved in Digestion of dietary carbohydrate |
| 0.0 | 0.4 | REACTOME UNWINDING OF DNA | Genes involved in Unwinding of DNA |
| 0.0 | 0.3 | REACTOME CELL EXTRACELLULAR MATRIX INTERACTIONS | Genes involved in Cell-extracellular matrix interactions |
| 0.0 | 0.1 | REACTOME GAP JUNCTION DEGRADATION | Genes involved in Gap junction degradation |
| 0.0 | 0.4 | REACTOME GLYCOLYSIS | Genes involved in Glycolysis |
| 0.0 | 0.3 | REACTOME ANTIGEN PRESENTATION FOLDING ASSEMBLY AND PEPTIDE LOADING OF CLASS I MHC | Genes involved in Antigen Presentation: Folding, assembly and peptide loading of class I MHC |
| 0.0 | 0.3 | REACTOME INTERFERON GAMMA SIGNALING | Genes involved in Interferon gamma signaling |
| 0.0 | 0.3 | REACTOME MITOCHONDRIAL TRNA AMINOACYLATION | Genes involved in Mitochondrial tRNA aminoacylation |
| 0.0 | 0.4 | REACTOME CROSS PRESENTATION OF SOLUBLE EXOGENOUS ANTIGENS ENDOSOMES | Genes involved in Cross-presentation of soluble exogenous antigens (endosomes) |