Project
PRJNA207719: Tissue specific transcriptome profiling
Navigation
Downloads
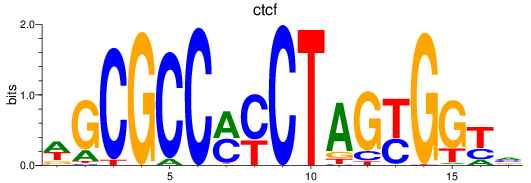
Results for ctcf
Z-value: 1.05
Transcription factors associated with ctcf
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
ctcf
|
ENSDARG00000056621 | CCCTC-binding factor (zinc finger protein) |
|
ctcf
|
ENSDARG00000113285 | CCCTC-binding factor (zinc finger protein) |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| ctcf | dr11_v1_chr18_+_22285992_22286029 | 0.94 | 1.8e-02 | Click! |
Activity profile of ctcf motif
Sorted Z-values of ctcf motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr2_+_56463167 | 0.53 |
ENSDART00000123392
|
rab11bb
|
RAB11B, member RAS oncogene family, b |
| chr21_+_45626136 | 0.42 |
ENSDART00000158742
|
irf1b
|
interferon regulatory factor 1b |
| chr9_-_12594759 | 0.40 |
ENSDART00000021266
|
tra2b
|
transformer 2 beta homolog |
| chr18_+_907266 | 0.40 |
ENSDART00000171729
|
pkma
|
pyruvate kinase M1/2a |
| chr20_-_29864390 | 0.39 |
ENSDART00000161834
ENSDART00000132278 |
rnf144ab
|
ring finger protein 144ab |
| chr21_+_17024002 | 0.39 |
ENSDART00000080628
|
arpc3
|
actin related protein 2/3 complex, subunit 3 |
| chr9_-_12595003 | 0.38 |
ENSDART00000184900
|
tra2b
|
transformer 2 beta homolog |
| chr17_+_6276559 | 0.37 |
ENSDART00000131075
|
dusp23b
|
dual specificity phosphatase 23b |
| chr5_+_57320113 | 0.36 |
ENSDART00000036331
|
atp6v1g1
|
ATPase H+ transporting V1 subunit G1 |
| chr22_-_57177 | 0.36 |
ENSDART00000163959
|
CABZ01085139.1
|
|
| chr6_+_39836474 | 0.36 |
ENSDART00000112637
|
smarcc2
|
SWI/SNF related, matrix associated, actin dependent regulator of chromatin, subfamily c, member 2 |
| chr20_-_30931139 | 0.36 |
ENSDART00000006778
ENSDART00000146376 |
acat2
|
acetyl-CoA acetyltransferase 2 |
| chr19_-_3916075 | 0.35 |
ENSDART00000161799
|
si:ch73-281i18.7
|
si:ch73-281i18.7 |
| chr25_+_31122806 | 0.34 |
ENSDART00000067039
|
rassf8a
|
Ras association (RalGDS/AF-6) domain family (N-terminal) member 8a |
| chr18_-_14677936 | 0.34 |
ENSDART00000111995
|
si:dkey-238o13.4
|
si:dkey-238o13.4 |
| chr8_+_7144066 | 0.32 |
ENSDART00000146306
|
slc6a6a
|
solute carrier family 6 (neurotransmitter transporter), member 6a |
| chr3_-_21280373 | 0.32 |
ENSDART00000003939
|
syngr1a
|
synaptogyrin 1a |
| chr19_-_3896514 | 0.32 |
ENSDART00000161346
|
si:ch73-281i18.3
|
si:ch73-281i18.3 |
| chr5_-_6377865 | 0.31 |
ENSDART00000031775
|
zgc:73226
|
zgc:73226 |
| chr8_-_11324143 | 0.31 |
ENSDART00000008215
|
pip5k1bb
|
phosphatidylinositol-4-phosphate 5-kinase, type I, beta b |
| chr12_-_31794908 | 0.31 |
ENSDART00000105583
ENSDART00000153449 |
nat9
|
N-acetyltransferase 9 (GCN5-related, putative) |
| chr19_-_3906369 | 0.30 |
ENSDART00000160299
ENSDART00000169205 |
si:ch73-281i18.4
|
si:ch73-281i18.4 |
| chr19_-_3925801 | 0.30 |
ENSDART00000129570
ENSDART00000163138 |
si:ch73-281i18.6
|
si:ch73-281i18.6 |
| chr14_-_44790817 | 0.29 |
ENSDART00000098640
|
grhpra
|
glyoxylate reductase/hydroxypyruvate reductase a |
| chr19_-_7070691 | 0.28 |
ENSDART00000168755
|
tapbp.2
|
TAP binding protein (tapasin), tandem duplicate 2 |
| chr9_-_29643628 | 0.28 |
ENSDART00000101177
|
spryd7b
|
SPRY domain containing 7b |
| chr10_+_26926654 | 0.27 |
ENSDART00000078980
ENSDART00000100289 |
rab1bb
|
RAB1B, member RAS oncogene family b |
| chr2_-_40135942 | 0.27 |
ENSDART00000176951
ENSDART00000098632 ENSDART00000148563 ENSDART00000149895 |
epha4a
|
eph receptor A4a |
| chr13_-_46429220 | 0.27 |
ENSDART00000149125
ENSDART00000098269 ENSDART00000150061 ENSDART00000080916 |
fgfr2
|
fibroblast growth factor receptor 2 |
| chr12_-_48374728 | 0.27 |
ENSDART00000153403
ENSDART00000188117 |
dnajb12b
|
DnaJ (Hsp40) homolog, subfamily B, member 12b |
| chr3_+_24050043 | 0.27 |
ENSDART00000151788
|
cbx1a
|
chromobox homolog 1a (HP1 beta homolog Drosophila) |
| chr22_-_372723 | 0.26 |
ENSDART00000112895
|
FNDC10
|
si:zfos-1324h11.5 |
| chr12_-_3077395 | 0.26 |
ENSDART00000002867
ENSDART00000126315 |
rfng
|
RFNG O-fucosylpeptide 3-beta-N-acetylglucosaminyltransferase |
| chr12_-_7807298 | 0.26 |
ENSDART00000191355
|
ank3b
|
ankyrin 3b |
| chr1_-_8651718 | 0.26 |
ENSDART00000133319
|
actb1
|
actin, beta 1 |
| chr7_+_24049776 | 0.25 |
ENSDART00000166559
|
efs
|
embryonal Fyn-associated substrate |
| chr7_+_30787903 | 0.25 |
ENSDART00000174000
|
apba2b
|
amyloid beta (A4) precursor protein-binding, family A, member 2b |
| chr19_-_33274978 | 0.25 |
ENSDART00000020301
ENSDART00000114714 |
fam92a1
|
family with sequence similarity 92, member A1 |
| chr9_-_30243393 | 0.25 |
ENSDART00000089539
|
si:dkey-100n23.3
|
si:dkey-100n23.3 |
| chr20_+_49895220 | 0.25 |
ENSDART00000097865
|
ints9
|
integrator complex subunit 9 |
| chr1_+_218524 | 0.25 |
ENSDART00000109529
|
tmco3
|
transmembrane and coiled-coil domains 3 |
| chr3_-_45777226 | 0.25 |
ENSDART00000192849
|
h3f3b.1
|
H3 histone, family 3B.1 |
| chr11_+_25157374 | 0.25 |
ENSDART00000019450
|
trpc4apa
|
transient receptor potential cation channel, subfamily C, member 4 associated protein a |
| chr19_+_20785724 | 0.24 |
ENSDART00000038942
|
adnp2b
|
ADNP homeobox 2b |
| chr17_+_17955063 | 0.24 |
ENSDART00000104999
|
ccdc85ca
|
coiled-coil domain containing 85C, a |
| chr1_+_45958904 | 0.23 |
ENSDART00000108528
|
arhgef7b
|
Rho guanine nucleotide exchange factor (GEF) 7b |
| chr21_+_261490 | 0.23 |
ENSDART00000177919
|
jak2a
|
Janus kinase 2a |
| chr19_-_29437108 | 0.23 |
ENSDART00000052108
ENSDART00000074497 |
fndc5b
|
fibronectin type III domain containing 5b |
| chr1_-_45577980 | 0.23 |
ENSDART00000160961
|
atf7ip
|
activating transcription factor 7 interacting protein |
| chr25_+_28823952 | 0.23 |
ENSDART00000067072
|
nfybb
|
nuclear transcription factor Y, beta b |
| chr6_+_59967994 | 0.23 |
ENSDART00000050457
|
zgc:65895
|
zgc:65895 |
| chr7_-_50917255 | 0.22 |
ENSDART00000022918
|
ankrd46b
|
ankyrin repeat domain 46b |
| chr1_-_22851481 | 0.22 |
ENSDART00000054386
|
qdprb1
|
quinoid dihydropteridine reductase b1 |
| chr13_-_40499296 | 0.22 |
ENSDART00000158338
|
CNNM1
|
Danio rerio cyclin and CBS domain divalent metal cation transport mediator 1 (cnnm1), mRNA. |
| chr4_+_12931763 | 0.22 |
ENSDART00000016382
|
wif1
|
wnt inhibitory factor 1 |
| chr1_+_22851261 | 0.21 |
ENSDART00000193925
|
gtf2e2
|
general transcription factor IIE, polypeptide 2, beta |
| chr5_+_52039067 | 0.21 |
ENSDART00000143276
|
setbp1
|
SET binding protein 1 |
| chr6_+_48041759 | 0.21 |
ENSDART00000140086
|
si:dkey-92f12.2
|
si:dkey-92f12.2 |
| chr5_+_55934129 | 0.21 |
ENSDART00000050969
|
tmem150ab
|
transmembrane protein 150Ab |
| chr1_+_22851745 | 0.21 |
ENSDART00000138235
ENSDART00000016488 |
gtf2e2
|
general transcription factor IIE, polypeptide 2, beta |
| chr3_-_33417826 | 0.21 |
ENSDART00000084284
|
abi3a
|
ABI family, member 3a |
| chr19_+_13420884 | 0.20 |
ENSDART00000160209
|
si:ch211-204a13.2
|
si:ch211-204a13.2 |
| chr16_-_32672883 | 0.20 |
ENSDART00000124515
ENSDART00000190920 ENSDART00000188776 |
pnisr
|
PNN-interacting serine/arginine-rich protein |
| chr15_-_16323750 | 0.20 |
ENSDART00000028500
|
nxn
|
nucleoredoxin |
| chr19_+_232536 | 0.20 |
ENSDART00000137880
|
syngap1a
|
synaptic Ras GTPase activating protein 1a |
| chr13_-_16191674 | 0.20 |
ENSDART00000147868
|
vwc2
|
von Willebrand factor C domain containing 2 |
| chr21_+_17005737 | 0.20 |
ENSDART00000101246
|
vps29
|
vacuolar protein sorting 29 homolog (S. cerevisiae) |
| chr2_+_42724404 | 0.19 |
ENSDART00000075392
|
basp1
|
brain abundant, membrane attached signal protein 1 |
| chr20_-_52338782 | 0.19 |
ENSDART00000109735
ENSDART00000132941 |
si:ch1073-287p18.1
|
si:ch1073-287p18.1 |
| chr20_+_26002534 | 0.19 |
ENSDART00000147845
|
nrbp1
|
nuclear receptor binding protein 1 |
| chr5_-_29514689 | 0.19 |
ENSDART00000126018
ENSDART00000125175 |
ehmt1a
|
euchromatic histone-lysine N-methyltransferase 1a |
| chr13_-_14908664 | 0.19 |
ENSDART00000138867
|
ap5s1
|
adaptor-related protein complex 5, sigma 1 subunit |
| chr10_+_6340478 | 0.19 |
ENSDART00000163182
ENSDART00000163722 |
tpm2
|
tropomyosin 2 (beta) |
| chr1_+_34763539 | 0.19 |
ENSDART00000077725
ENSDART00000113808 |
zgc:172122
|
zgc:172122 |
| chr12_-_7639120 | 0.18 |
ENSDART00000126712
ENSDART00000126219 |
ccdc6b
|
coiled-coil domain containing 6b |
| chr1_+_41690402 | 0.18 |
ENSDART00000177298
|
fbxo41
|
F-box protein 41 |
| chr4_+_9177997 | 0.18 |
ENSDART00000057254
ENSDART00000154614 |
nfyba
|
nuclear transcription factor Y, beta a |
| chr3_+_22057109 | 0.18 |
ENSDART00000153709
|
kansl1b
|
KAT8 regulatory NSL complex subunit 1b |
| chr15_+_22435460 | 0.18 |
ENSDART00000031976
|
tmem136a
|
transmembrane protein 136a |
| chr8_+_2757821 | 0.18 |
ENSDART00000051403
ENSDART00000160551 |
sh3glb2a
|
SH3-domain GRB2-like endophilin B2a |
| chr4_-_18512550 | 0.18 |
ENSDART00000045639
|
rint1
|
RAD50 interactor 1 |
| chr3_-_22216442 | 0.18 |
ENSDART00000153613
|
maptb
|
microtubule-associated protein tau b |
| chr8_-_13364879 | 0.18 |
ENSDART00000063863
|
ccdc124
|
coiled-coil domain containing 124 |
| chr25_-_554142 | 0.17 |
ENSDART00000028997
|
myo9ab
|
myosin IXAb |
| chr9_-_11655031 | 0.17 |
ENSDART00000044314
|
itgav
|
integrin, alpha V |
| chr19_-_7043355 | 0.17 |
ENSDART00000104845
|
tapbp.1
|
TAP binding protein (tapasin), tandem duplicate 1 |
| chr13_+_24834199 | 0.17 |
ENSDART00000101274
|
zgc:153981
|
zgc:153981 |
| chr10_+_33626344 | 0.17 |
ENSDART00000109563
|
b3glctb
|
beta 3-glucosyltransferase b |
| chr4_-_9667380 | 0.17 |
ENSDART00000189671
ENSDART00000133214 |
dmtf1
|
cyclin D binding myb-like transcription factor 1 |
| chr8_+_25900049 | 0.17 |
ENSDART00000124300
ENSDART00000127618 ENSDART00000024009 |
rhoab
|
ras homolog gene family, member Ab |
| chr21_+_15713097 | 0.17 |
ENSDART00000015841
|
gstt1b
|
glutathione S-transferase theta 1b |
| chr2_+_25019387 | 0.17 |
ENSDART00000142601
|
stag1a
|
stromal antigen 1a |
| chr25_+_37130450 | 0.17 |
ENSDART00000183358
|
si:ch1073-174d20.1
|
si:ch1073-174d20.1 |
| chr19_+_18903533 | 0.17 |
ENSDART00000157523
ENSDART00000166562 |
slc39a7
|
solute carrier family 39 (zinc transporter), member 7 |
| chr9_-_41507712 | 0.17 |
ENSDART00000135821
|
mfsd6b
|
major facilitator superfamily domain containing 6b |
| chr25_-_12788370 | 0.17 |
ENSDART00000158551
|
slc7a5
|
solute carrier family 7 (amino acid transporter light chain, L system), member 5 |
| chr24_-_41478917 | 0.17 |
ENSDART00000192192
|
CABZ01084131.1
|
|
| chr11_+_40032790 | 0.17 |
ENSDART00000158809
|
si:dkey-264d12.1
|
si:dkey-264d12.1 |
| chr5_-_32396929 | 0.17 |
ENSDART00000023977
|
fbxw2
|
F-box and WD repeat domain containing 2 |
| chr13_-_27038407 | 0.16 |
ENSDART00000146712
|
ccdc85a
|
coiled-coil domain containing 85A |
| chr16_-_8132742 | 0.16 |
ENSDART00000104323
|
snrka
|
SNF related kinase a |
| chr1_+_35194454 | 0.16 |
ENSDART00000165389
|
sc:d189
|
sc:d189 |
| chr13_-_14908999 | 0.16 |
ENSDART00000112771
|
ap5s1
|
adaptor-related protein complex 5, sigma 1 subunit |
| chr22_-_3595439 | 0.16 |
ENSDART00000083308
|
ptprsa
|
protein tyrosine phosphatase, receptor type, s, a |
| chr19_+_24882845 | 0.16 |
ENSDART00000010580
|
si:ch211-195b13.1
|
si:ch211-195b13.1 |
| chr13_-_33822550 | 0.16 |
ENSDART00000143703
|
flrt3
|
fibronectin leucine rich transmembrane 3 |
| chr19_+_233143 | 0.16 |
ENSDART00000175273
|
syngap1a
|
synaptic Ras GTPase activating protein 1a |
| chr15_+_17441734 | 0.16 |
ENSDART00000153729
|
snx19b
|
sorting nexin 19b |
| chr8_-_34427364 | 0.16 |
ENSDART00000112854
ENSDART00000161282 ENSDART00000113230 |
gapvd1
|
GTPase activating protein and VPS9 domains 1 |
| chr21_-_18824434 | 0.16 |
ENSDART00000156333
|
si:dkey-112m2.1
|
si:dkey-112m2.1 |
| chr12_-_33582382 | 0.16 |
ENSDART00000009794
ENSDART00000136617 |
tdrkh
|
tudor and KH domain containing |
| chr6_+_13779857 | 0.15 |
ENSDART00000154793
|
tmem198b
|
transmembrane protein 198b |
| chr3_+_40407352 | 0.15 |
ENSDART00000155112
|
tnrc18
|
trinucleotide repeat containing 18 |
| chr5_-_54497319 | 0.15 |
ENSDART00000160492
|
alad
|
aminolevulinate dehydratase |
| chr6_-_35046735 | 0.15 |
ENSDART00000143649
|
uap1
|
UDP-N-acetylglucosamine pyrophosphorylase 1 |
| chr14_+_6954579 | 0.15 |
ENSDART00000060998
|
nme5
|
NME/NM23 family member 5 |
| chr1_+_14283692 | 0.15 |
ENSDART00000017679
|
ppp2r2ca
|
protein phosphatase 2, regulatory subunit B, gamma a |
| chr1_+_59154521 | 0.15 |
ENSDART00000130089
ENSDART00000152456 |
soul5l
|
heme-binding protein soul5, like |
| chr25_-_13490744 | 0.15 |
ENSDART00000056721
|
ldhd
|
lactate dehydrogenase D |
| chr25_+_19734038 | 0.14 |
ENSDART00000067354
|
zgc:101783
|
zgc:101783 |
| chr25_-_20730800 | 0.14 |
ENSDART00000172170
|
si:ch211-127m7.2
|
si:ch211-127m7.2 |
| chr3_-_61354239 | 0.14 |
ENSDART00000154312
|
znf1134
|
zinc finger protein 1134 |
| chr9_+_219124 | 0.14 |
ENSDART00000161484
|
map3k12
|
mitogen-activated protein kinase kinase kinase 12 |
| chr23_-_20429741 | 0.14 |
ENSDART00000104334
|
taf13
|
TATA-box binding protein associated factor 13 |
| chr10_-_7974155 | 0.14 |
ENSDART00000147368
ENSDART00000075524 |
osbp2
|
oxysterol binding protein 2 |
| chr11_+_22623284 | 0.14 |
ENSDART00000111850
|
si:ch211-86h15.1
|
si:ch211-86h15.1 |
| chr20_+_19512727 | 0.14 |
ENSDART00000063696
|
atraid
|
all-trans retinoic acid-induced differentiation factor |
| chr23_+_43954809 | 0.14 |
ENSDART00000164080
|
corin
|
corin, serine peptidase |
| chr22_+_737211 | 0.14 |
ENSDART00000017305
|
znf76
|
zinc finger protein 76 |
| chr22_+_35131890 | 0.13 |
ENSDART00000003303
ENSDART00000130581 |
rnf13
|
ring finger protein 13 |
| chr4_+_3478049 | 0.13 |
ENSDART00000153944
|
grm8a
|
glutamate receptor, metabotropic 8a |
| chr24_+_37533728 | 0.13 |
ENSDART00000061203
|
rhot2
|
ras homolog family member T2 |
| chr7_+_29080684 | 0.13 |
ENSDART00000173709
ENSDART00000173576 |
acd
|
ACD, shelterin complex subunit and telomerase recruitment factor |
| chr21_-_21824285 | 0.13 |
ENSDART00000030984
|
wdr73
|
WD repeat domain 73 |
| chr5_+_61941610 | 0.13 |
ENSDART00000168808
|
si:dkeyp-117b8.4
|
si:dkeyp-117b8.4 |
| chr2_-_50298337 | 0.13 |
ENSDART00000155125
|
cntnap2b
|
contactin associated protein like 2b |
| chr19_+_24068223 | 0.13 |
ENSDART00000141351
ENSDART00000100420 |
pex11b
|
peroxisomal biogenesis factor 11 beta |
| chr9_+_22929675 | 0.13 |
ENSDART00000061299
|
tsn
|
translin |
| chr22_+_18786797 | 0.13 |
ENSDART00000141864
|
cbarpb
|
calcium channel, voltage-dependent, beta subunit associated regulatory protein b |
| chr13_+_51853716 | 0.13 |
ENSDART00000175341
ENSDART00000187855 |
LT631684.1
|
|
| chr25_+_26921480 | 0.13 |
ENSDART00000155949
|
grm8b
|
glutamate receptor, metabotropic 8b |
| chr15_+_6786757 | 0.12 |
ENSDART00000013278
|
sirt2
|
sirtuin 2 (silent mating type information regulation 2, homolog) 2 (S. cerevisiae) |
| chr6_+_47846366 | 0.12 |
ENSDART00000064842
|
padi2
|
peptidyl arginine deiminase, type II |
| chr19_-_3886991 | 0.12 |
ENSDART00000162085
|
thrap3b
|
thyroid hormone receptor associated protein 3b |
| chr22_-_20403194 | 0.12 |
ENSDART00000010048
|
map2k2a
|
mitogen-activated protein kinase kinase 2a |
| chr10_+_2842923 | 0.12 |
ENSDART00000181895
|
ykt6
|
YKT6 v-SNARE homolog (S. cerevisiae) |
| chr14_-_17588345 | 0.12 |
ENSDART00000143486
|
selenot2
|
selenoprotein T, 2 |
| chr10_+_11355841 | 0.12 |
ENSDART00000193067
ENSDART00000064215 |
cops4
|
COP9 constitutive photomorphogenic homolog subunit 4 (Arabidopsis) |
| chr16_+_23799622 | 0.11 |
ENSDART00000046922
|
rab13
|
RAB13, member RAS oncogene family |
| chr16_+_12812214 | 0.11 |
ENSDART00000124875
|
u2af2a
|
U2 small nuclear RNA auxiliary factor 2a |
| chr1_+_49878000 | 0.11 |
ENSDART00000047876
|
lef1
|
lymphoid enhancer-binding factor 1 |
| chr12_+_18681477 | 0.11 |
ENSDART00000127981
ENSDART00000143979 |
rgs9b
|
regulator of G protein signaling 9b |
| chr5_+_58687541 | 0.11 |
ENSDART00000083015
ENSDART00000181902 |
ccdc84
|
coiled-coil domain containing 84 |
| chr23_+_43950674 | 0.11 |
ENSDART00000167813
|
corin
|
corin, serine peptidase |
| chr24_-_30096666 | 0.11 |
ENSDART00000183285
|
plppr4b
|
phospholipid phosphatase related 4b |
| chr13_-_35765028 | 0.11 |
ENSDART00000157391
|
erlec1
|
endoplasmic reticulum lectin 1 |
| chr19_-_7144548 | 0.11 |
ENSDART00000147177
ENSDART00000134850 |
psmb8a
psmb13a
|
proteasome subunit beta 8A proteasome subunit beta 13a |
| chr5_+_41496490 | 0.11 |
ENSDART00000039369
|
fancg
|
Fanconi anemia, complementation group G |
| chr20_+_12702923 | 0.11 |
ENSDART00000163499
|
zgc:153383
|
zgc:153383 |
| chr18_+_44649804 | 0.11 |
ENSDART00000059063
|
ehd2b
|
EH-domain containing 2b |
| chr8_+_13364950 | 0.11 |
ENSDART00000159760
|
slc5a5
|
solute carrier family 5 (sodium/iodide cotransporter), member 5 |
| chr22_+_5135884 | 0.11 |
ENSDART00000141276
|
mydgf
|
myeloid-derived growth factor |
| chr1_+_2301961 | 0.11 |
ENSDART00000108919
ENSDART00000143361 ENSDART00000142944 |
farp1
|
FERM, RhoGEF (ARHGEF) and pleckstrin domain protein 1 (chondrocyte-derived) |
| chr3_-_30248277 | 0.11 |
ENSDART00000133516
ENSDART00000077029 |
emc10
|
ER membrane protein complex subunit 10 |
| chr6_+_45934682 | 0.11 |
ENSDART00000103489
|
cenps
|
centromere protein S |
| chr1_+_21937201 | 0.11 |
ENSDART00000087729
|
kdm4c
|
lysine (K)-specific demethylase 4C |
| chr7_+_39758714 | 0.11 |
ENSDART00000111278
|
sorcs2
|
sortilin-related VPS10 domain containing receptor 2 |
| chr10_-_40826657 | 0.11 |
ENSDART00000076304
|
pcna
|
proliferating cell nuclear antigen |
| chr14_-_30557127 | 0.11 |
ENSDART00000053936
|
pfdn6
|
prefoldin subunit 6 |
| chr24_+_28935200 | 0.11 |
ENSDART00000017427
|
rnpc3
|
RNA-binding region (RNP1, RRM) containing 3 |
| chr18_+_35229115 | 0.11 |
ENSDART00000129624
ENSDART00000184596 |
tbrg1
|
transforming growth factor beta regulator 1 |
| chr3_-_22216771 | 0.11 |
ENSDART00000130546
|
maptb
|
microtubule-associated protein tau b |
| chr11_-_2270069 | 0.11 |
ENSDART00000189005
|
znf740a
|
zinc finger protein 740a |
| chr20_-_54192130 | 0.10 |
ENSDART00000020834
|
CABZ01111496.1
|
|
| chr16_+_12812472 | 0.10 |
ENSDART00000008535
|
u2af2a
|
U2 small nuclear RNA auxiliary factor 2a |
| chr18_+_8402129 | 0.10 |
ENSDART00000081154
ENSDART00000171974 |
prpf18
|
PRP18 pre-mRNA processing factor 18 homolog (yeast) |
| chr8_+_26007988 | 0.10 |
ENSDART00000193948
ENSDART00000058100 |
xpc
|
xeroderma pigmentosum, complementation group C |
| chr19_+_40337457 | 0.10 |
ENSDART00000089547
ENSDART00000140857 |
fam133b
|
family with sequence similarity 133, member B |
| chr4_-_75158035 | 0.10 |
ENSDART00000174353
|
CABZ01066312.1
|
|
| chr3_-_26524934 | 0.10 |
ENSDART00000087118
|
xylt1
|
xylosyltransferase I |
| chr21_+_34976600 | 0.10 |
ENSDART00000191672
ENSDART00000139635 |
rbm11
|
RNA binding motif protein 11 |
| chr6_+_23935045 | 0.10 |
ENSDART00000162993
|
gadd45ab
|
growth arrest and DNA-damage-inducible, alpha, b |
| chr5_+_32076109 | 0.10 |
ENSDART00000051357
ENSDART00000144510 |
zmat5
|
zinc finger, matrin-type 5 |
| chr24_+_39027481 | 0.09 |
ENSDART00000085565
|
capn15
|
calpain 15 |
| chr16_+_34493987 | 0.09 |
ENSDART00000138374
|
si:ch211-255i3.4
|
si:ch211-255i3.4 |
| chr17_-_51829310 | 0.09 |
ENSDART00000154544
|
numb
|
numb homolog (Drosophila) |
| chr13_-_44630111 | 0.09 |
ENSDART00000110092
|
mdga1
|
MAM domain containing glycosylphosphatidylinositol anchor 1 |
| chr9_-_16877456 | 0.09 |
ENSDART00000161105
ENSDART00000160869 |
fbxl3a
|
F-box and leucine-rich repeat protein 3a |
| chr23_-_19486571 | 0.09 |
ENSDART00000009092
|
fam208ab
|
family with sequence similarity 208, member Ab |
| chr6_+_22337081 | 0.09 |
ENSDART00000128047
ENSDART00000138930 |
uqcrc1
|
ubiquinol-cytochrome c reductase core protein 1 |
| chr1_+_34181581 | 0.09 |
ENSDART00000146042
|
epha6
|
eph receptor A6 |
| chr7_-_34256374 | 0.09 |
ENSDART00000075176
|
dis3l
|
DIS3 like exosome 3'-5' exoribonuclease |
| chr16_+_2843428 | 0.08 |
ENSDART00000149485
|
clec3ba
|
C-type lectin domain family 3, member Ba |
| chr22_-_718615 | 0.08 |
ENSDART00000149320
|
arl8a
|
ADP-ribosylation factor-like 8A |
| chr17_+_25677458 | 0.08 |
ENSDART00000029703
|
kcnh1a
|
potassium voltage-gated channel, subfamily H (eag-related), member 1a |
| chr2_-_9744081 | 0.08 |
ENSDART00000097732
|
dvl3a
|
dishevelled segment polarity protein 3a |
| chr14_+_904850 | 0.08 |
ENSDART00000161847
|
si:ch73-208h1.1
|
si:ch73-208h1.1 |
| chr9_+_38043337 | 0.08 |
ENSDART00000022574
|
stam2
|
signal transducing adaptor molecule (SH3 domain and ITAM motif) 2 |
| chr11_+_575665 | 0.08 |
ENSDART00000122133
|
mkrn2os.1
|
MKRN2 opposite strand, tandem duplicate 1 |
| chr5_+_54497475 | 0.08 |
ENSDART00000158149
ENSDART00000163968 ENSDART00000160446 |
tmem203
|
transmembrane protein 203 |
Network of associatons between targets according to the STRING database.
First level regulatory network of ctcf
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.2 | GO:0071896 | protein localization to adherens junction(GO:0071896) |
| 0.1 | 0.3 | GO:0090527 | actin filament reorganization(GO:0090527) |
| 0.0 | 0.4 | GO:0097345 | mitochondrial outer membrane permeabilization(GO:0097345) |
| 0.0 | 0.1 | GO:1902175 | regulation of oxidative stress-induced intrinsic apoptotic signaling pathway(GO:1902175) negative regulation of oxidative stress-induced intrinsic apoptotic signaling pathway(GO:1902176) |
| 0.0 | 0.1 | GO:0006089 | lactate metabolic process(GO:0006089) |
| 0.0 | 0.8 | GO:0048026 | positive regulation of mRNA splicing, via spliceosome(GO:0048026) |
| 0.0 | 0.1 | GO:0051973 | positive regulation of telomerase activity(GO:0051973) |
| 0.0 | 0.3 | GO:0034334 | adherens junction maintenance(GO:0034334) |
| 0.0 | 0.2 | GO:0042590 | antigen processing and presentation of exogenous peptide antigen(GO:0002478) antigen processing and presentation of exogenous antigen(GO:0019884) antigen processing and presentation of exogenous peptide antigen via MHC class I(GO:0042590) |
| 0.0 | 0.1 | GO:0043388 | negative regulation of gliogenesis(GO:0014014) positive regulation of DNA binding(GO:0043388) negative regulation of reactive oxygen species metabolic process(GO:2000378) |
| 0.0 | 0.3 | GO:0003404 | optic vesicle morphogenesis(GO:0003404) |
| 0.0 | 0.3 | GO:0033278 | cell proliferation in midbrain(GO:0033278) |
| 0.0 | 0.2 | GO:0098529 | neuromuscular junction development, skeletal muscle fiber(GO:0098529) |
| 0.0 | 0.4 | GO:0050688 | regulation of defense response to virus(GO:0050688) |
| 0.0 | 0.1 | GO:0000350 | generation of catalytic spliceosome for second transesterification step(GO:0000350) |
| 0.0 | 0.1 | GO:0045669 | positive regulation of osteoblast differentiation(GO:0045669) |
| 0.0 | 0.2 | GO:0060628 | regulation of ER to Golgi vesicle-mediated transport(GO:0060628) |
| 0.0 | 0.2 | GO:0090245 | axis elongation involved in somitogenesis(GO:0090245) |
| 0.0 | 0.1 | GO:0048210 | Golgi vesicle fusion to target membrane(GO:0048210) |
| 0.0 | 0.1 | GO:0006272 | leading strand elongation(GO:0006272) |
| 0.0 | 0.1 | GO:0099612 | protein localization to juxtaparanode region of axon(GO:0071205) protein localization to axon(GO:0099612) |
| 0.0 | 0.1 | GO:0019240 | citrulline biosynthetic process(GO:0019240) |
| 0.0 | 0.2 | GO:1902766 | skeletal muscle satellite cell migration(GO:1902766) |
| 0.0 | 0.3 | GO:0021592 | fourth ventricle development(GO:0021592) central nervous system segmentation(GO:0035283) brain segmentation(GO:0035284) |
| 0.0 | 0.1 | GO:0060784 | regulation of cell proliferation involved in tissue homeostasis(GO:0060784) |
| 0.0 | 0.1 | GO:0016559 | peroxisome fission(GO:0016559) |
| 0.0 | 0.1 | GO:0045721 | negative regulation of gluconeogenesis(GO:0045721) |
| 0.0 | 0.1 | GO:1903306 | negative regulation of calcium ion-dependent exocytosis(GO:0045955) negative regulation of regulated secretory pathway(GO:1903306) |
| 0.0 | 0.1 | GO:2001032 | regulation of double-strand break repair via nonhomologous end joining(GO:2001032) |
| 0.0 | 0.2 | GO:0006346 | methylation-dependent chromatin silencing(GO:0006346) positive regulation of chromatin silencing(GO:0031937) regulation of methylation-dependent chromatin silencing(GO:0090308) positive regulation of methylation-dependent chromatin silencing(GO:0090309) |
| 0.0 | 0.1 | GO:0071236 | cellular response to antibiotic(GO:0071236) cellular response to toxic substance(GO:0097237) |
| 0.0 | 0.2 | GO:0019883 | antigen processing and presentation of endogenous peptide antigen(GO:0002483) antigen processing and presentation of endogenous antigen(GO:0019883) antigen processing and presentation of endogenous peptide antigen via MHC class I(GO:0019885) |
| 0.0 | 0.0 | GO:0042373 | vitamin K metabolic process(GO:0042373) |
| 0.0 | 0.2 | GO:0016926 | protein desumoylation(GO:0016926) |
| 0.0 | 0.1 | GO:0045050 | protein insertion into ER membrane by stop-transfer membrane-anchor sequence(GO:0045050) |
| 0.0 | 0.1 | GO:2000480 | negative regulation of cAMP-dependent protein kinase activity(GO:2000480) |
| 0.0 | 0.1 | GO:0070262 | peptidyl-serine dephosphorylation(GO:0070262) |
| 0.0 | 0.0 | GO:0000379 | tRNA-type intron splice site recognition and cleavage(GO:0000379) |
| 0.0 | 0.1 | GO:0042264 | peptidyl-aspartic acid modification(GO:0018197) peptidyl-aspartic acid hydroxylation(GO:0042264) |
| 0.0 | 0.3 | GO:0051788 | response to misfolded protein(GO:0051788) cellular response to misfolded protein(GO:0071218) |
| 0.0 | 0.2 | GO:0099560 | synaptic membrane adhesion(GO:0099560) |
| 0.0 | 0.1 | GO:0006048 | UDP-N-acetylglucosamine biosynthetic process(GO:0006048) |
| 0.0 | 0.1 | GO:0021694 | cerebellar Purkinje cell layer formation(GO:0021694) cerebellar Purkinje cell differentiation(GO:0021702) |
| 0.0 | 0.1 | GO:0000185 | activation of MAPKKK activity(GO:0000185) |
| 0.0 | 0.2 | GO:0031397 | negative regulation of protein ubiquitination(GO:0031397) |
| 0.0 | 0.4 | GO:0034314 | Arp2/3 complex-mediated actin nucleation(GO:0034314) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.4 | GO:0005673 | transcription factor TFIIE complex(GO:0005673) |
| 0.1 | 0.3 | GO:0097433 | dense body(GO:0097433) |
| 0.0 | 0.1 | GO:0043218 | compact myelin(GO:0043218) Schmidt-Lanterman incisure(GO:0043220) |
| 0.0 | 0.4 | GO:0016602 | CCAAT-binding factor complex(GO:0016602) |
| 0.0 | 0.2 | GO:0031464 | Cul4A-RING E3 ubiquitin ligase complex(GO:0031464) |
| 0.0 | 0.2 | GO:0089701 | U2AF(GO:0089701) |
| 0.0 | 0.4 | GO:0071564 | npBAF complex(GO:0071564) |
| 0.0 | 0.4 | GO:0005885 | Arp2/3 protein complex(GO:0005885) |
| 0.0 | 0.1 | GO:0034709 | methylosome(GO:0034709) |
| 0.0 | 0.2 | GO:0043240 | Fanconi anaemia nuclear complex(GO:0043240) |
| 0.0 | 0.1 | GO:0033010 | paranodal junction(GO:0033010) |
| 0.0 | 0.4 | GO:0016471 | vacuolar proton-transporting V-type ATPase complex(GO:0016471) |
| 0.0 | 0.2 | GO:0030904 | retromer complex(GO:0030904) |
| 0.0 | 0.1 | GO:0005779 | integral component of peroxisomal membrane(GO:0005779) intrinsic component of peroxisomal membrane(GO:0031231) |
| 0.0 | 0.1 | GO:0016272 | prefoldin complex(GO:0016272) |
| 0.0 | 0.2 | GO:0044545 | NSL complex(GO:0044545) |
| 0.0 | 0.2 | GO:0032039 | integrator complex(GO:0032039) |
| 0.0 | 0.1 | GO:0032593 | insulin-responsive compartment(GO:0032593) |
| 0.0 | 0.3 | GO:0005689 | U12-type spliceosomal complex(GO:0005689) |
| 0.0 | 0.2 | GO:0034993 | microtubule organizing center attachment site(GO:0034992) LINC complex(GO:0034993) |
| 0.0 | 0.1 | GO:0031307 | integral component of mitochondrial outer membrane(GO:0031307) |
| 0.0 | 0.0 | GO:0019185 | snRNA-activating protein complex(GO:0019185) |
| 0.0 | 0.3 | GO:0031594 | neuromuscular junction(GO:0031594) |
| 0.0 | 0.1 | GO:0008180 | COP9 signalosome(GO:0008180) |
| 0.0 | 0.0 | GO:0000214 | tRNA-intron endonuclease complex(GO:0000214) |
| 0.0 | 0.1 | GO:0005682 | U5 snRNP(GO:0005682) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.4 | GO:0003985 | acetyl-CoA C-acetyltransferase activity(GO:0003985) |
| 0.1 | 0.3 | GO:0030267 | hydroxypyruvate reductase activity(GO:0016618) glyoxylate reductase (NADP) activity(GO:0030267) |
| 0.1 | 0.4 | GO:0004743 | pyruvate kinase activity(GO:0004743) |
| 0.1 | 0.3 | GO:0033829 | O-fucosylpeptide 3-beta-N-acetylglucosaminyltransferase activity(GO:0033829) |
| 0.1 | 0.3 | GO:0098973 | structural constituent of postsynaptic actin cytoskeleton(GO:0098973) |
| 0.1 | 0.2 | GO:0047134 | protein-disulfide reductase activity(GO:0047134) |
| 0.0 | 0.2 | GO:0030626 | U12 snRNA binding(GO:0030626) |
| 0.0 | 0.3 | GO:0001642 | group III metabotropic glutamate receptor activity(GO:0001642) |
| 0.0 | 0.2 | GO:0070139 | ubiquitin-like protein-specific endopeptidase activity(GO:0070137) SUMO-specific endopeptidase activity(GO:0070139) |
| 0.0 | 0.1 | GO:0042903 | tubulin deacetylase activity(GO:0042903) |
| 0.0 | 0.1 | GO:0003977 | UDP-N-acetylglucosamine diphosphorylase activity(GO:0003977) |
| 0.0 | 0.2 | GO:0035035 | histone acetyltransferase binding(GO:0035035) |
| 0.0 | 0.2 | GO:0030628 | pre-mRNA 3'-splice site binding(GO:0030628) |
| 0.0 | 0.4 | GO:0005005 | transmembrane-ephrin receptor activity(GO:0005005) |
| 0.0 | 0.3 | GO:0005007 | fibroblast growth factor-activated receptor activity(GO:0005007) |
| 0.0 | 0.1 | GO:0004457 | lactate dehydrogenase activity(GO:0004457) |
| 0.0 | 0.3 | GO:0030544 | Hsp70 protein binding(GO:0030544) |
| 0.0 | 0.2 | GO:0043495 | protein anchor(GO:0043495) |
| 0.0 | 0.1 | GO:0004706 | JUN kinase kinase kinase activity(GO:0004706) |
| 0.0 | 0.1 | GO:0004597 | peptide-aspartate beta-dioxygenase activity(GO:0004597) |
| 0.0 | 0.1 | GO:0004862 | cAMP-dependent protein kinase inhibitor activity(GO:0004862) |
| 0.0 | 0.1 | GO:0035252 | UDP-xylosyltransferase activity(GO:0035252) |
| 0.0 | 0.1 | GO:0016813 | hydrolase activity, acting on carbon-nitrogen (but not peptide) bonds, in linear amidines(GO:0016813) |
| 0.0 | 0.1 | GO:0004791 | thioredoxin-disulfide reductase activity(GO:0004791) |
| 0.0 | 0.1 | GO:0032977 | membrane insertase activity(GO:0032977) |
| 0.0 | 0.3 | GO:0001540 | beta-amyloid binding(GO:0001540) |
| 0.0 | 0.2 | GO:0002039 | p53 binding(GO:0002039) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.2 | PID INTEGRIN5 PATHWAY | Beta5 beta6 beta7 and beta8 integrin cell surface interactions |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.3 | REACTOME FGFR2C LIGAND BINDING AND ACTIVATION | Genes involved in FGFR2c ligand binding and activation |
| 0.0 | 0.4 | REACTOME INSULIN RECEPTOR RECYCLING | Genes involved in Insulin receptor recycling |
| 0.0 | 0.2 | REACTOME PECAM1 INTERACTIONS | Genes involved in PECAM1 interactions |
| 0.0 | 0.3 | REACTOME PRE NOTCH PROCESSING IN GOLGI | Genes involved in Pre-NOTCH Processing in Golgi |
| 0.0 | 0.6 | REACTOME RNA POL II TRANSCRIPTION PRE INITIATION AND PROMOTER OPENING | Genes involved in RNA Polymerase II Transcription Pre-Initiation And Promoter Opening |
| 0.0 | 0.2 | REACTOME ZINC TRANSPORTERS | Genes involved in Zinc transporters |
| 0.0 | 0.1 | REACTOME METABOLISM OF PORPHYRINS | Genes involved in Metabolism of porphyrins |
| 0.0 | 0.1 | REACTOME AMINE DERIVED HORMONES | Genes involved in Amine-derived hormones |