Project
PRJNA207719: Tissue specific transcriptome profiling
Navigation
Downloads
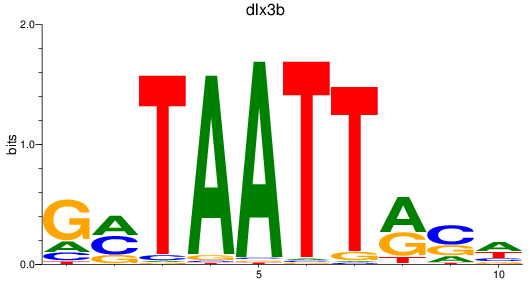
Results for dlx3b
Z-value: 0.43
Transcription factors associated with dlx3b
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
dlx3b
|
ENSDARG00000014626 | distal-less homeobox 3b |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| dlx3b | dr11_v1_chr12_+_5708400_5708400 | 0.62 | 2.7e-01 | Click! |
Activity profile of dlx3b motif
Sorted Z-values of dlx3b motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr24_-_17023392 | 0.49 |
ENSDART00000106058
|
ptgdsb.2
|
prostaglandin D2 synthase b, tandem duplicate 2 |
| chr24_-_17029374 | 0.45 |
ENSDART00000039267
|
ptgdsb.1
|
prostaglandin D2 synthase b, tandem duplicate 1 |
| chr14_-_34044369 | 0.24 |
ENSDART00000149396
ENSDART00000123607 ENSDART00000190746 |
cyfip2
|
cytoplasmic FMR1 interacting protein 2 |
| chr7_-_25895189 | 0.23 |
ENSDART00000173599
ENSDART00000079235 ENSDART00000173786 ENSDART00000173602 ENSDART00000079245 ENSDART00000187568 ENSDART00000173505 |
cd99l2
|
CD99 molecule-like 2 |
| chr14_-_30587814 | 0.23 |
ENSDART00000144912
ENSDART00000149714 |
tmem265
|
transmembrane protein 265 |
| chr4_+_14727018 | 0.22 |
ENSDART00000124189
|
cmasa
|
cytidine monophosphate N-acetylneuraminic acid synthetase a |
| chr12_+_22580579 | 0.20 |
ENSDART00000171725
ENSDART00000192290 |
capgb
|
capping protein (actin filament), gelsolin-like b |
| chr3_+_13624815 | 0.19 |
ENSDART00000161451
|
pglyrp6
|
peptidoglycan recognition protein 6 |
| chr8_-_31107537 | 0.18 |
ENSDART00000098925
|
vgll4l
|
vestigial like 4 like |
| chr14_-_24410673 | 0.18 |
ENSDART00000125923
|
cxcl14
|
chemokine (C-X-C motif) ligand 14 |
| chr4_+_14727212 | 0.17 |
ENSDART00000158094
|
cmasa
|
cytidine monophosphate N-acetylneuraminic acid synthetase a |
| chr25_-_32869794 | 0.16 |
ENSDART00000162784
|
tmem266
|
transmembrane protein 266 |
| chr25_+_10410620 | 0.15 |
ENSDART00000151886
|
ehf
|
ets homologous factor |
| chr9_-_21976670 | 0.15 |
ENSDART00000104322
|
uchl3
|
ubiquitin carboxyl-terminal esterase L3 (ubiquitin thiolesterase) |
| chr18_-_1185772 | 0.14 |
ENSDART00000143245
|
nptnb
|
neuroplastin b |
| chr12_-_35386910 | 0.14 |
ENSDART00000153453
|
camk2g1
|
calcium/calmodulin-dependent protein kinase (CaM kinase) II gamma 1 |
| chr11_-_25418856 | 0.13 |
ENSDART00000013714
|
gata1a
|
GATA binding protein 1a |
| chr6_-_35472923 | 0.13 |
ENSDART00000185907
|
rgs8
|
regulator of G protein signaling 8 |
| chr23_-_18130264 | 0.13 |
ENSDART00000016976
|
nucks1b
|
nuclear casein kinase and cyclin-dependent kinase substrate 1b |
| chr16_-_19890303 | 0.13 |
ENSDART00000147161
ENSDART00000079159 |
hdac9b
|
histone deacetylase 9b |
| chr10_-_21955231 | 0.13 |
ENSDART00000183695
|
FO744833.2
|
|
| chr19_+_20793237 | 0.13 |
ENSDART00000014774
|
txnl4a
|
thioredoxin-like 4A |
| chr3_-_21280373 | 0.12 |
ENSDART00000003939
|
syngr1a
|
synaptogyrin 1a |
| chr23_+_17839187 | 0.12 |
ENSDART00000104647
|
prim1
|
DNA primase subunit 1 |
| chr21_-_5856050 | 0.12 |
ENSDART00000115367
|
CABZ01071020.1
|
|
| chr15_-_9272328 | 0.12 |
ENSDART00000172114
|
calm2a
|
calmodulin 2a (phosphorylase kinase, delta) |
| chr21_-_10488379 | 0.12 |
ENSDART00000163878
|
hcn1
|
hyperpolarization activated cyclic nucleotide-gated potassium channel 1 |
| chr19_-_32641725 | 0.12 |
ENSDART00000165006
ENSDART00000188185 |
hpca
|
hippocalcin |
| chr19_+_20793388 | 0.12 |
ENSDART00000142463
|
txnl4a
|
thioredoxin-like 4A |
| chr4_+_9177997 | 0.12 |
ENSDART00000057254
ENSDART00000154614 |
nfyba
|
nuclear transcription factor Y, beta a |
| chr16_+_5774977 | 0.11 |
ENSDART00000134202
|
ccka
|
cholecystokinin a |
| chr1_-_58036509 | 0.11 |
ENSDART00000081122
|
COLGALT1
|
si:ch211-114l13.7 |
| chr1_+_10318089 | 0.11 |
ENSDART00000029774
|
pip4p1b
|
phosphatidylinositol-4,5-bisphosphate 4-phosphatase 1b |
| chr14_+_23717165 | 0.10 |
ENSDART00000006373
|
ndfip1
|
Nedd4 family interacting protein 1 |
| chr17_+_16046314 | 0.10 |
ENSDART00000154554
ENSDART00000154338 ENSDART00000155336 |
si:ch73-204p21.2
|
si:ch73-204p21.2 |
| chr21_-_43949208 | 0.10 |
ENSDART00000150983
|
camk2a
|
calcium/calmodulin-dependent protein kinase II alpha |
| chr4_-_17629444 | 0.10 |
ENSDART00000108814
|
nrip2
|
nuclear receptor interacting protein 2 |
| chr4_+_3980247 | 0.10 |
ENSDART00000049194
|
gpr37b
|
G protein-coupled receptor 37b |
| chr7_+_30787903 | 0.10 |
ENSDART00000174000
|
apba2b
|
amyloid beta (A4) precursor protein-binding, family A, member 2b |
| chr2_-_23004286 | 0.10 |
ENSDART00000134664
ENSDART00000110373 ENSDART00000185833 ENSDART00000187235 |
znf414
mllt1b
|
zinc finger protein 414 MLLT1, super elongation complex subunit b |
| chr16_-_21620947 | 0.10 |
ENSDART00000115011
ENSDART00000183125 ENSDART00000188856 ENSDART00000189460 |
ddr1
|
discoidin domain receptor tyrosine kinase 1 |
| chr21_-_26490186 | 0.09 |
ENSDART00000009889
|
zgc:110540
|
zgc:110540 |
| chr23_+_26946744 | 0.09 |
ENSDART00000115141
|
cacnb3b
|
calcium channel, voltage-dependent, beta 3b |
| chr3_-_28428198 | 0.09 |
ENSDART00000151546
|
rbfox1
|
RNA binding fox-1 homolog 1 |
| chr23_-_16666583 | 0.09 |
ENSDART00000189804
ENSDART00000136496 |
sdcbp2
|
syndecan binding protein (syntenin) 2 |
| chr19_-_42607451 | 0.09 |
ENSDART00000004392
|
fkbp9
|
FK506 binding protein 9 |
| chr5_+_59234421 | 0.09 |
ENSDART00000045518
|
prkrip1
|
PRKR interacting protein 1 |
| chr15_-_42736433 | 0.09 |
ENSDART00000154379
|
si:ch211-181d7.1
|
si:ch211-181d7.1 |
| chr23_-_16682186 | 0.09 |
ENSDART00000020810
|
sdcbp2
|
syndecan binding protein (syntenin) 2 |
| chr14_-_36378494 | 0.08 |
ENSDART00000058503
|
gpm6aa
|
glycoprotein M6Aa |
| chr21_+_11244068 | 0.08 |
ENSDART00000163432
|
arid6
|
AT-rich interaction domain 6 |
| chr10_-_34871737 | 0.08 |
ENSDART00000138755
|
dclk1a
|
doublecortin-like kinase 1a |
| chr17_-_20717845 | 0.08 |
ENSDART00000150037
|
ank3b
|
ankyrin 3b |
| chr10_+_22381802 | 0.08 |
ENSDART00000112484
|
nlgn2b
|
neuroligin 2b |
| chr1_+_37752171 | 0.08 |
ENSDART00000183247
ENSDART00000189756 ENSDART00000139448 |
GALNTL6
|
si:ch211-15e22.3 |
| chr18_-_2433011 | 0.08 |
ENSDART00000181922
ENSDART00000193276 |
CR769778.1
|
|
| chr19_+_7735157 | 0.08 |
ENSDART00000186717
|
tuft1b
|
tuftelin 1b |
| chr16_-_12953739 | 0.08 |
ENSDART00000103894
|
cacng8b
|
calcium channel, voltage-dependent, gamma subunit 8b |
| chr11_-_29768054 | 0.08 |
ENSDART00000079117
|
plekha3
|
pleckstrin homology domain containing, family A (phosphoinositide binding specific) member 3 |
| chr15_-_755023 | 0.08 |
ENSDART00000155594
|
znf1011
|
zinc finger protein 1011 |
| chr11_+_38280454 | 0.08 |
ENSDART00000171496
|
CDK18
|
si:dkey-166c18.1 |
| chr4_+_12612145 | 0.07 |
ENSDART00000181201
|
lmo3
|
LIM domain only 3 |
| chr20_-_37813863 | 0.07 |
ENSDART00000147529
|
batf3
|
basic leucine zipper transcription factor, ATF-like 3 |
| chr8_+_28452738 | 0.07 |
ENSDART00000062706
|
tmem189
|
transmembrane protein 189 |
| chr3_+_32832538 | 0.07 |
ENSDART00000139410
|
cd2bp2
|
CD2 (cytoplasmic tail) binding protein 2 |
| chr20_-_32270866 | 0.07 |
ENSDART00000153140
|
armc2
|
armadillo repeat containing 2 |
| chr2_-_28671139 | 0.07 |
ENSDART00000165272
ENSDART00000164657 |
dhcr7
|
7-dehydrocholesterol reductase |
| chr5_+_26213874 | 0.07 |
ENSDART00000193816
ENSDART00000098514 |
oclnb
|
occludin b |
| chr9_+_11532025 | 0.07 |
ENSDART00000109037
|
cdk5r2b
|
cyclin-dependent kinase 5, regulatory subunit 2b (p39) |
| chr20_+_30490682 | 0.07 |
ENSDART00000184871
|
myt1la
|
myelin transcription factor 1-like, a |
| chr16_-_13623659 | 0.06 |
ENSDART00000168978
|
si:dkeyp-69b9.6
|
si:dkeyp-69b9.6 |
| chr17_+_44780166 | 0.06 |
ENSDART00000156260
|
tmem63c
|
transmembrane protein 63C |
| chr13_+_31205439 | 0.06 |
ENSDART00000132326
|
ptpn20
|
protein tyrosine phosphatase, non-receptor type 20 |
| chr6_-_6487876 | 0.06 |
ENSDART00000137642
|
cep170ab
|
centrosomal protein 170Ab |
| chr19_+_31904836 | 0.06 |
ENSDART00000162297
ENSDART00000088340 ENSDART00000151280 ENSDART00000151218 |
tpd52
|
tumor protein D52 |
| chr18_+_9171778 | 0.06 |
ENSDART00000101192
|
sema3d
|
sema domain, immunoglobulin domain (Ig), short basic domain, secreted, (semaphorin) 3D |
| chr17_-_15546862 | 0.06 |
ENSDART00000091021
|
col10a1a
|
collagen, type X, alpha 1a |
| chr11_+_33818179 | 0.06 |
ENSDART00000109418
|
spoplb
|
speckle-type POZ protein-like b |
| chr23_+_28582865 | 0.06 |
ENSDART00000020296
|
l1cama
|
L1 cell adhesion molecule, paralog a |
| chr17_+_16046132 | 0.06 |
ENSDART00000155005
|
si:ch73-204p21.2
|
si:ch73-204p21.2 |
| chr3_-_16719244 | 0.06 |
ENSDART00000055859
|
pold1
|
polymerase (DNA directed), delta 1, catalytic subunit |
| chr3_-_46811611 | 0.05 |
ENSDART00000134092
|
elavl3
|
ELAV like neuron-specific RNA binding protein 3 |
| chr10_+_32104305 | 0.05 |
ENSDART00000099880
|
wnt11r
|
wingless-type MMTV integration site family, member 11, related |
| chr15_-_33172246 | 0.05 |
ENSDART00000158666
|
nbeab
|
neurobeachin b |
| chr4_-_15420452 | 0.05 |
ENSDART00000016230
|
plxna4
|
plexin A4 |
| chr11_-_42554290 | 0.05 |
ENSDART00000130573
|
atp6ap1la
|
ATPase H+ transporting accessory protein 1 like a |
| chr20_-_9462433 | 0.05 |
ENSDART00000152674
ENSDART00000040557 |
zgc:101840
|
zgc:101840 |
| chr13_+_35339182 | 0.05 |
ENSDART00000019323
|
jag1b
|
jagged 1b |
| chr16_+_39159752 | 0.05 |
ENSDART00000122081
|
sybu
|
syntabulin (syntaxin-interacting) |
| chr9_-_32730487 | 0.05 |
ENSDART00000147795
|
olig1
|
oligodendrocyte transcription factor 1 |
| chr13_+_22295905 | 0.05 |
ENSDART00000180133
ENSDART00000181125 |
usp54a
|
ubiquitin specific peptidase 54a |
| chr5_+_27525477 | 0.05 |
ENSDART00000051491
|
sfrp1a
|
secreted frizzled-related protein 1a |
| chr15_+_11825366 | 0.04 |
ENSDART00000190042
|
prkd2
|
protein kinase D2 |
| chr23_-_7799184 | 0.04 |
ENSDART00000190946
ENSDART00000165427 |
myt1b
|
myelin transcription factor 1b |
| chr11_-_42418374 | 0.04 |
ENSDART00000160704
|
slmapa
|
sarcolemma associated protein a |
| chr1_-_44701313 | 0.04 |
ENSDART00000193926
|
si:dkey-28b4.8
|
si:dkey-28b4.8 |
| chr12_+_25432627 | 0.04 |
ENSDART00000011662
|
ppm1bb
|
protein phosphatase, Mg2+/Mn2+ dependent, 1Bb |
| chr19_-_38830582 | 0.04 |
ENSDART00000189966
ENSDART00000183055 |
adgrb2
|
adhesion G protein-coupled receptor B2 |
| chr23_+_43489182 | 0.04 |
ENSDART00000162062
|
znf341
|
zinc finger protein 341 |
| chr3_-_31079186 | 0.04 |
ENSDART00000145636
ENSDART00000140569 |
ELOB (1 of many)
elob
|
elongin B elongin B |
| chr9_-_3934963 | 0.04 |
ENSDART00000062336
|
ubr3
|
ubiquitin protein ligase E3 component n-recognin 3 |
| chr10_+_3049636 | 0.04 |
ENSDART00000081794
ENSDART00000183167 ENSDART00000191634 ENSDART00000183514 |
rasgrf2a
|
Ras protein-specific guanine nucleotide-releasing factor 2a |
| chr15_-_18115540 | 0.03 |
ENSDART00000131639
ENSDART00000047902 |
arcn1b
|
archain 1b |
| chr16_-_13623928 | 0.03 |
ENSDART00000164344
|
si:dkeyp-69b9.6
|
si:dkeyp-69b9.6 |
| chr4_+_11464255 | 0.03 |
ENSDART00000008584
|
gdi2
|
GDP dissociation inhibitor 2 |
| chr2_+_2168547 | 0.03 |
ENSDART00000029347
|
higd1a
|
HIG1 hypoxia inducible domain family, member 1A |
| chr1_+_23783349 | 0.03 |
ENSDART00000007531
|
slit2
|
slit homolog 2 (Drosophila) |
| chr2_-_26720854 | 0.03 |
ENSDART00000148110
|
si:dkey-181m9.8
|
si:dkey-181m9.8 |
| chr8_-_21142550 | 0.03 |
ENSDART00000143192
ENSDART00000186820 ENSDART00000135938 |
cpt2
|
carnitine palmitoyltransferase 2 |
| chr11_+_18873619 | 0.03 |
ENSDART00000176141
|
magi1b
|
membrane associated guanylate kinase, WW and PDZ domain containing 1b |
| chr22_-_7129631 | 0.03 |
ENSDART00000171359
|
asic1b
|
acid-sensing (proton-gated) ion channel 1b |
| chr16_-_29194517 | 0.03 |
ENSDART00000046114
ENSDART00000148899 |
mef2d
|
myocyte enhancer factor 2d |
| chr14_+_25817628 | 0.03 |
ENSDART00000047680
|
glra1
|
glycine receptor, alpha 1 |
| chr20_-_35578435 | 0.03 |
ENSDART00000142444
|
adgrf6
|
adhesion G protein-coupled receptor F6 |
| chr11_-_39118882 | 0.02 |
ENSDART00000113185
ENSDART00000156526 |
ap5b1
|
adaptor-related protein complex 5, beta 1 subunit |
| chr1_-_19402802 | 0.02 |
ENSDART00000135552
|
rbm47
|
RNA binding motif protein 47 |
| chr10_-_1788376 | 0.02 |
ENSDART00000123842
|
epb41l4a
|
erythrocyte membrane protein band 4.1 like 4A |
| chr3_+_22059066 | 0.02 |
ENSDART00000155739
|
kansl1b
|
KAT8 regulatory NSL complex subunit 1b |
| chr6_-_40713183 | 0.02 |
ENSDART00000157113
ENSDART00000154810 ENSDART00000153702 |
si:ch211-157b11.12
|
si:ch211-157b11.12 |
| chr4_-_8058214 | 0.02 |
ENSDART00000132228
|
wnk1b
|
WNK lysine deficient protein kinase 1b |
| chr8_-_24252933 | 0.02 |
ENSDART00000057624
|
zgc:110353
|
zgc:110353 |
| chr21_-_11654422 | 0.02 |
ENSDART00000081614
ENSDART00000132699 |
cast
|
calpastatin |
| chr11_+_31864921 | 0.02 |
ENSDART00000180252
|
diaph3
|
diaphanous-related formin 3 |
| chr9_+_11034314 | 0.02 |
ENSDART00000032695
|
asic4a
|
acid-sensing (proton-gated) ion channel family member 4a |
| chr8_-_23776399 | 0.02 |
ENSDART00000114800
|
INAVA
|
si:ch211-163l21.4 |
| chr3_-_26183699 | 0.02 |
ENSDART00000147517
ENSDART00000140731 |
si:ch211-11k18.4
|
si:ch211-11k18.4 |
| chr7_-_12464412 | 0.02 |
ENSDART00000178723
|
adamtsl3
|
ADAMTS-like 3 |
| chr22_-_3564563 | 0.02 |
ENSDART00000145114
|
ptprsa
|
protein tyrosine phosphatase, receptor type, s, a |
| chrM_+_9052 | 0.01 |
ENSDART00000093612
|
mt-atp6
|
ATP synthase 6, mitochondrial |
| chr9_+_21165484 | 0.01 |
ENSDART00000177286
|
si:rp71-68n21.9
|
si:rp71-68n21.9 |
| chr2_+_56213694 | 0.01 |
ENSDART00000162582
|
upf1
|
upf1 regulator of nonsense transcripts homolog (yeast) |
| chr7_+_15872357 | 0.01 |
ENSDART00000165757
|
pax6b
|
paired box 6b |
| chr1_-_47071979 | 0.01 |
ENSDART00000160817
|
itsn1
|
intersectin 1 (SH3 domain protein) |
| chr18_-_46258612 | 0.01 |
ENSDART00000153930
|
si:dkey-244a7.1
|
si:dkey-244a7.1 |
| chr18_-_37007061 | 0.01 |
ENSDART00000136432
|
map3k10
|
mitogen-activated protein kinase kinase kinase 10 |
| chr21_-_37790727 | 0.01 |
ENSDART00000162907
|
gabrb4
|
gamma-aminobutyric acid (GABA) A receptor, beta 4 |
| chr16_-_20312146 | 0.01 |
ENSDART00000134980
|
si:dkeyp-86h10.3
|
si:dkeyp-86h10.3 |
| chr18_-_37007294 | 0.01 |
ENSDART00000088309
|
map3k10
|
mitogen-activated protein kinase kinase kinase 10 |
| chr4_+_12612723 | 0.01 |
ENSDART00000133767
|
lmo3
|
LIM domain only 3 |
| chr6_+_9289802 | 0.01 |
ENSDART00000188650
|
kalrnb
|
kalirin RhoGEF kinase b |
| chr12_-_6818676 | 0.01 |
ENSDART00000106391
|
pcdh15b
|
protocadherin-related 15b |
| chr2_+_32826235 | 0.01 |
ENSDART00000143127
|
si:dkey-154p10.3
|
si:dkey-154p10.3 |
| chr4_+_57580303 | 0.01 |
ENSDART00000166492
ENSDART00000103025 ENSDART00000170786 |
il17ra1b
il17ra2
|
interleukin 17 receptor A1b interleukin 17 receptor A2 |
| chr8_-_5847533 | 0.01 |
ENSDART00000192489
|
CABZ01102147.1
|
|
| chr5_-_48268049 | 0.01 |
ENSDART00000187454
|
mef2cb
|
myocyte enhancer factor 2cb |
| chr2_-_55298075 | 0.01 |
ENSDART00000186404
ENSDART00000149062 |
rab8a
|
RAB8A, member RAS oncogene family |
| chr20_+_4060839 | 0.01 |
ENSDART00000178565
|
TRIM67
|
tripartite motif containing 67 |
| chr7_-_69647988 | 0.00 |
ENSDART00000169943
|
LO018231.1
|
|
| chr16_-_16120941 | 0.00 |
ENSDART00000131227
|
ankib1b
|
ankyrin repeat and IBR domain containing 1b |
| chr17_+_26352372 | 0.00 |
ENSDART00000155177
|
grid1a
|
glutamate receptor, ionotropic, delta 1a |
| chr14_+_36220479 | 0.00 |
ENSDART00000148319
|
pitx2
|
paired-like homeodomain 2 |
| chr9_+_1313418 | 0.00 |
ENSDART00000191361
|
casp8l2
|
caspase 8, apoptosis-related cysteine peptidase, like 2 |
| chr12_-_18577983 | 0.00 |
ENSDART00000193262
|
zdhhc4
|
zinc finger, DHHC-type containing 4 |
Network of associatons between targets according to the STRING database.
First level regulatory network of dlx3b
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.2 | GO:0016045 | detection of biotic stimulus(GO:0009595) detection of bacterium(GO:0016045) detection of other organism(GO:0098543) detection of external biotic stimulus(GO:0098581) |
| 0.0 | 0.4 | GO:0006054 | N-acetylneuraminate metabolic process(GO:0006054) |
| 0.0 | 0.1 | GO:0030224 | monocyte differentiation(GO:0030224) |
| 0.0 | 0.1 | GO:0060159 | regulation of dopamine receptor signaling pathway(GO:0060159) |
| 0.0 | 0.1 | GO:0006269 | DNA replication, synthesis of RNA primer(GO:0006269) |
| 0.0 | 0.1 | GO:0007414 | axonal defasciculation(GO:0007414) |
| 0.0 | 0.2 | GO:0001964 | startle response(GO:0001964) |
| 0.0 | 0.1 | GO:0035627 | ceramide transport(GO:0035627) |
| 0.0 | 0.1 | GO:0006297 | nucleotide-excision repair, DNA gap filling(GO:0006297) |
| 0.0 | 0.0 | GO:0060074 | synapse maturation(GO:0060074) |
| 0.0 | 0.1 | GO:0007508 | larval development(GO:0002164) larval heart development(GO:0007508) |
| 0.0 | 0.1 | GO:0035677 | posterior lateral line neuromast hair cell development(GO:0035677) |
| 0.0 | 0.0 | GO:0003322 | pancreatic A cell development(GO:0003322) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | GO:1990077 | primosome complex(GO:1990077) |
| 0.0 | 0.1 | GO:0060077 | inhibitory synapse(GO:0060077) |
| 0.0 | 0.3 | GO:0005682 | U5 snRNP(GO:0005682) |
| 0.0 | 0.2 | GO:0031209 | SCAR complex(GO:0031209) |
| 0.0 | 0.1 | GO:0016533 | cyclin-dependent protein kinase 5 holoenzyme complex(GO:0016533) |
| 0.0 | 0.1 | GO:0044295 | axonal growth cone(GO:0044295) |
| 0.0 | 0.1 | GO:0043625 | delta DNA polymerase complex(GO:0043625) |
| 0.0 | 0.1 | GO:0016602 | CCAAT-binding factor complex(GO:0016602) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.2 | GO:0016019 | peptidoglycan receptor activity(GO:0016019) |
| 0.1 | 0.2 | GO:0045545 | syndecan binding(GO:0045545) |
| 0.1 | 0.4 | GO:0001972 | retinoic acid binding(GO:0001972) |
| 0.0 | 0.4 | GO:0070567 | cytidylyltransferase activity(GO:0070567) |
| 0.0 | 0.2 | GO:0001223 | transcription coactivator binding(GO:0001223) |
| 0.0 | 0.1 | GO:1902387 | ceramide transporter activity(GO:0035620) ceramide 1-phosphate binding(GO:1902387) ceramide 1-phosphate transporter activity(GO:1902388) |
| 0.0 | 0.1 | GO:0050699 | WW domain binding(GO:0050699) |
| 0.0 | 0.2 | GO:0000340 | RNA 7-methylguanosine cap binding(GO:0000340) |
| 0.0 | 0.1 | GO:0034597 | phosphatidylinositol-4,5-bisphosphate 4-phosphatase activity(GO:0034597) |
| 0.0 | 0.1 | GO:0019136 | deoxynucleoside kinase activity(GO:0019136) |
| 0.0 | 0.1 | GO:0008046 | axon guidance receptor activity(GO:0008046) |
| 0.0 | 0.0 | GO:0005093 | Rab GDP-dissociation inhibitor activity(GO:0005093) |
| 0.0 | 0.1 | GO:0038064 | protein tyrosine kinase collagen receptor activity(GO:0038062) collagen receptor activity(GO:0038064) |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | REACTOME UNBLOCKING OF NMDA RECEPTOR GLUTAMATE BINDING AND ACTIVATION | Genes involved in Unblocking of NMDA receptor, glutamate binding and activation |
| 0.0 | 0.2 | REACTOME POL SWITCHING | Genes involved in Polymerase switching |