Project
PRJNA207719: Tissue specific transcriptome profiling
Navigation
Downloads
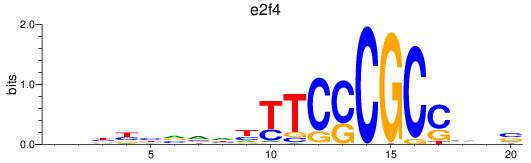
Results for e2f4
Z-value: 0.67
Transcription factors associated with e2f4
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
e2f4
|
ENSDARG00000101578 | E2F transcription factor 4 |
|
e2f4
|
ENSDARG00000109320 | E2F transcription factor 4 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| e2f4 | dr11_v1_chr25_+_17405201_17405201 | -0.73 | 1.7e-01 | Click! |
Activity profile of e2f4 motif
Sorted Z-values of e2f4 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr19_-_48391415 | 0.58 |
ENSDART00000170726
ENSDART00000169577 |
si:ch73-359m17.2
|
si:ch73-359m17.2 |
| chr7_+_24814866 | 0.58 |
ENSDART00000173581
|
si:ch211-226l4.6
|
si:ch211-226l4.6 |
| chr7_+_24881680 | 0.48 |
ENSDART00000058843
|
krcp
|
kelch repeat-containing protein |
| chr20_-_29498178 | 0.40 |
ENSDART00000152986
ENSDART00000027851 ENSDART00000152954 |
rrm2
|
ribonucleotide reductase M2 polypeptide |
| chr19_-_47570672 | 0.39 |
ENSDART00000112155
|
rrm2
|
ribonucleotide reductase M2 polypeptide |
| chr3_+_29641181 | 0.38 |
ENSDART00000151517
|
eif3d
|
eukaryotic translation initiation factor 3, subunit D |
| chr22_+_3153876 | 0.36 |
ENSDART00000163327
|
rpl36
|
ribosomal protein L36 |
| chr7_-_6429031 | 0.33 |
ENSDART00000173413
|
zgc:112234
|
zgc:112234 |
| chr25_-_774350 | 0.31 |
ENSDART00000166321
ENSDART00000160386 |
IRAK4
|
interleukin 1 receptor associated kinase 4 |
| chr25_+_192116 | 0.30 |
ENSDART00000153983
|
zgc:114188
|
zgc:114188 |
| chr24_+_21621654 | 0.30 |
ENSDART00000002595
|
rpl21
|
ribosomal protein L21 |
| chr17_-_15189397 | 0.28 |
ENSDART00000133710
ENSDART00000110507 |
wdhd1
|
WD repeat and HMG-box DNA binding protein 1 |
| chr10_+_6884123 | 0.26 |
ENSDART00000149095
ENSDART00000148772 ENSDART00000149334 |
ddx4
|
DEAD (Asp-Glu-Ala-Asp) box polypeptide 4 |
| chr25_-_35124576 | 0.25 |
ENSDART00000128017
|
zgc:171759
|
zgc:171759 |
| chr4_-_17725008 | 0.22 |
ENSDART00000016658
|
chpt1
|
choline phosphotransferase 1 |
| chr20_-_33462961 | 0.21 |
ENSDART00000135927
|
si:dkey-65b13.1
|
si:dkey-65b13.1 |
| chr7_+_5960491 | 0.21 |
ENSDART00000145370
|
zgc:112234
|
zgc:112234 |
| chr20_+_35445462 | 0.21 |
ENSDART00000124497
|
tdrd6
|
tudor domain containing 6 |
| chr7_-_6459481 | 0.20 |
ENSDART00000173158
|
zgc:112234
|
zgc:112234 |
| chr25_-_35109536 | 0.19 |
ENSDART00000185229
|
CU302436.5
|
|
| chr7_-_6415991 | 0.18 |
ENSDART00000173349
|
CU457819.3
|
Histone H3.2 |
| chr6_-_425378 | 0.18 |
ENSDART00000192190
|
fam83fb
|
family with sequence similarity 83, member Fb |
| chr7_-_6351021 | 0.18 |
ENSDART00000159542
|
zgc:112234
|
zgc:112234 |
| chr7_-_6345507 | 0.17 |
ENSDART00000173032
|
CU457819.4
|
Histone H3.2 |
| chr11_+_31680513 | 0.17 |
ENSDART00000139900
ENSDART00000040305 |
diaph3
|
diaphanous-related formin 3 |
| chr20_-_28433990 | 0.16 |
ENSDART00000182824
ENSDART00000193381 |
wdr21
|
WD repeat domain 21 |
| chr25_-_36370292 | 0.15 |
ENSDART00000152766
|
CR354435.1
|
Histone H2B 1/2 |
| chr6_-_33978622 | 0.14 |
ENSDART00000151725
ENSDART00000124917 |
mpl
|
MPL proto-oncogene, thrombopoietin receptor |
| chr6_-_39653972 | 0.14 |
ENSDART00000155859
|
larp4ab
|
La ribonucleoprotein domain family, member 4Ab |
| chr17_+_43889371 | 0.14 |
ENSDART00000156871
ENSDART00000154702 |
msh4
|
mutS homolog 4 |
| chr14_+_94603 | 0.14 |
ENSDART00000162480
|
mcm7
|
minichromosome maintenance complex component 7 |
| chr25_+_35062353 | 0.13 |
ENSDART00000089844
|
zgc:113983
|
zgc:113983 |
| chr10_-_37487612 | 0.13 |
ENSDART00000167964
ENSDART00000171326 |
si:dkey-65j4.2
|
si:dkey-65j4.2 |
| chr16_+_35595312 | 0.13 |
ENSDART00000170438
|
si:ch211-1i11.3
|
si:ch211-1i11.3 |
| chr13_+_48359573 | 0.13 |
ENSDART00000161959
ENSDART00000165311 |
msh6
|
mutS homolog 6 (E. coli) |
| chr23_-_19831739 | 0.13 |
ENSDART00000125066
|
haus7
|
HAUS augmin-like complex, subunit 7 |
| chr18_+_17020967 | 0.13 |
ENSDART00000189168
|
chtf8
|
CTF8, chromosome transmission fidelity factor 8 homolog (S. cerevisiae) |
| chr3_-_61592417 | 0.12 |
ENSDART00000155082
|
nptx2a
|
neuronal pentraxin 2a |
| chr3_+_58119571 | 0.12 |
ENSDART00000108979
|
si:ch211-256e16.4
|
si:ch211-256e16.4 |
| chr7_+_20393386 | 0.12 |
ENSDART00000173471
|
si:dkey-33c9.6
|
si:dkey-33c9.6 |
| chr4_-_75175407 | 0.11 |
ENSDART00000180125
|
CABZ01043953.1
|
|
| chr20_-_28433616 | 0.11 |
ENSDART00000169289
|
wdr21
|
WD repeat domain 21 |
| chr13_-_45942000 | 0.10 |
ENSDART00000158534
ENSDART00000171822 ENSDART00000166442 |
si:ch211-62a1.4
|
si:ch211-62a1.4 |
| chr13_+_15933168 | 0.10 |
ENSDART00000131390
|
fignl1
|
fidgetin-like 1 |
| chr25_+_35058088 | 0.10 |
ENSDART00000156838
|
zgc:112234
|
zgc:112234 |
| chr7_-_73854476 | 0.10 |
ENSDART00000186481
|
zgc:173552
|
zgc:173552 |
| chr7_+_34453185 | 0.10 |
ENSDART00000173875
ENSDART00000173921 ENSDART00000173995 |
si:cabz01009626.1
|
si:cabz01009626.1 |
| chr2_-_57378748 | 0.09 |
ENSDART00000149235
|
tcf3a
|
transcription factor 3a |
| chr23_-_45568816 | 0.09 |
ENSDART00000076086
|
cyp17a2
|
cytochrome P450, family 17, subfamily A, polypeptide 2 |
| chr3_-_7897305 | 0.08 |
ENSDART00000169757
|
ubn2b
|
ubinuclein 2b |
| chr14_+_94946 | 0.08 |
ENSDART00000165766
ENSDART00000163778 |
mcm7
|
minichromosome maintenance complex component 7 |
| chr16_+_11834516 | 0.07 |
ENSDART00000146611
|
cxcr3.3
|
chemokine (C-X-C motif) receptor 3, tandem duplicate 3 |
| chr17_-_50020672 | 0.06 |
ENSDART00000188942
|
filip1a
|
filamin A interacting protein 1a |
| chr8_+_8947623 | 0.06 |
ENSDART00000131215
|
slc35a2
|
solute carrier family 35 (UDP-galactose transporter), member 2 |
| chr4_+_77740228 | 0.06 |
ENSDART00000193397
|
si:zfos-2131b9.2
|
si:zfos-2131b9.2 |
| chr3_-_7897563 | 0.06 |
ENSDART00000185232
|
ubn2b
|
ubinuclein 2b |
| chr18_+_17021391 | 0.04 |
ENSDART00000100160
|
chtf8
|
CTF8, chromosome transmission fidelity factor 8 homolog (S. cerevisiae) |
| chr25_+_35155676 | 0.03 |
ENSDART00000114598
|
zgc:112234
|
zgc:112234 |
| chr17_-_11368662 | 0.03 |
ENSDART00000159061
ENSDART00000188694 ENSDART00000190932 |
si:ch211-185a18.2
|
si:ch211-185a18.2 |
| chr25_+_35067318 | 0.02 |
ENSDART00000186828
|
CU302436.1
|
|
| chr5_+_20453874 | 0.02 |
ENSDART00000124545
ENSDART00000008402 |
sart3
|
squamous cell carcinoma antigen recognized by T cells 3 |
| chr21_+_30355767 | 0.02 |
ENSDART00000189948
|
CR749164.1
|
|
| chr21_-_27413294 | 0.01 |
ENSDART00000131646
|
slc29a2
|
solute carrier family 29 (equilibrative nucleoside transporter), member 2 |
| chr10_+_6010570 | 0.01 |
ENSDART00000190025
ENSDART00000163680 |
hmgcs1
|
3-hydroxy-3-methylglutaryl-CoA synthase 1 (soluble) |
Network of associatons between targets according to the STRING database.
First level regulatory network of e2f4
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.4 | GO:0002191 | cap-dependent translational initiation(GO:0002191) |
| 0.1 | 0.8 | GO:0009263 | deoxyribonucleotide biosynthetic process(GO:0009263) |
| 0.0 | 0.1 | GO:0002566 | somatic diversification of immune receptors via somatic mutation(GO:0002566) pyrimidine dimer repair(GO:0006290) somatic hypermutation of immunoglobulin genes(GO:0016446) |
| 0.0 | 0.2 | GO:1902299 | pre-replicative complex assembly involved in nuclear cell cycle DNA replication(GO:0006267) pre-replicative complex assembly(GO:0036388) pre-replicative complex assembly involved in cell cycle DNA replication(GO:1902299) |
| 0.0 | 0.2 | GO:0030719 | P granule organization(GO:0030719) |
| 0.0 | 0.1 | GO:0000710 | meiotic mismatch repair(GO:0000710) |
| 0.0 | 0.3 | GO:0051445 | regulation of meiotic cell cycle(GO:0051445) |
| 0.0 | 0.1 | GO:0008207 | C21-steroid hormone metabolic process(GO:0008207) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.2 | GO:0033391 | chromatoid body(GO:0033391) |
| 0.1 | 0.2 | GO:0031390 | Ctf18 RFC-like complex(GO:0031390) |
| 0.0 | 0.1 | GO:0032301 | MutSalpha complex(GO:0032301) |
| 0.0 | 0.4 | GO:0016282 | eukaryotic 43S preinitiation complex(GO:0016282) |
| 0.0 | 0.1 | GO:0070652 | HAUS complex(GO:0070652) |
| 0.0 | 0.2 | GO:0042555 | MCM complex(GO:0042555) |
| 0.0 | 0.7 | GO:0022625 | cytosolic large ribosomal subunit(GO:0022625) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.4 | GO:0098808 | mRNA cap binding(GO:0098808) |
| 0.1 | 0.8 | GO:0004748 | ribonucleoside-diphosphate reductase activity, thioredoxin disulfide as acceptor(GO:0004748) oxidoreductase activity, acting on CH or CH2 groups, disulfide as acceptor(GO:0016728) ribonucleoside-diphosphate reductase activity(GO:0061731) |
| 0.0 | 0.2 | GO:0043140 | ATP-dependent 3'-5' DNA helicase activity(GO:0043140) single-stranded DNA-dependent ATP-dependent 3'-5' DNA helicase activity(GO:1990518) |
| 0.0 | 0.2 | GO:0004142 | diacylglycerol cholinephosphotransferase activity(GO:0004142) |
| 0.0 | 0.1 | GO:0032138 | guanine/thymine mispair binding(GO:0032137) single base insertion or deletion binding(GO:0032138) single thymine insertion binding(GO:0032143) oxidized DNA binding(GO:0032356) oxidized purine DNA binding(GO:0032357) mismatch repair complex binding(GO:0032404) MutLalpha complex binding(GO:0032405) |
| 0.0 | 0.6 | GO:0004129 | cytochrome-c oxidase activity(GO:0004129) heme-copper terminal oxidase activity(GO:0015002) oxidoreductase activity, acting on a heme group of donors(GO:0016675) oxidoreductase activity, acting on a heme group of donors, oxygen as acceptor(GO:0016676) |
| 0.0 | 0.1 | GO:0022850 | serotonin-gated cation channel activity(GO:0022850) |
| 0.0 | 0.1 | GO:0030983 | mismatched DNA binding(GO:0030983) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.3 | PID TOLL ENDOGENOUS PATHWAY | Endogenous TLR signaling |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.8 | REACTOME SYNTHESIS AND INTERCONVERSION OF NUCLEOTIDE DI AND TRIPHOSPHATES | Genes involved in Synthesis and interconversion of nucleotide di- and triphosphates |
| 0.0 | 0.3 | REACTOME TRAF6 MEDIATED IRF7 ACTIVATION IN TLR7 8 OR 9 SIGNALING | Genes involved in TRAF6 mediated IRF7 activation in TLR7/8 or 9 signaling |
| 0.0 | 0.2 | REACTOME UNWINDING OF DNA | Genes involved in Unwinding of DNA |
| 0.0 | 0.2 | REACTOME SYNTHESIS OF PC | Genes involved in Synthesis of PC |