Project
PRJNA207719: Tissue specific transcriptome profiling
Navigation
Downloads
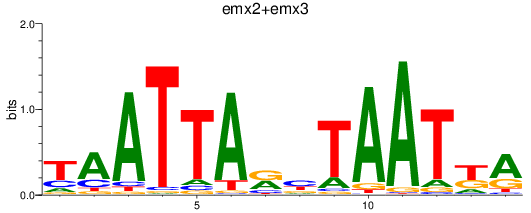
Results for emx2+emx3
Z-value: 0.99
Transcription factors associated with emx2+emx3
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
emx3
|
ENSDARG00000020417 | empty spiracles homeobox 3 |
|
emx2
|
ENSDARG00000039701 | empty spiracles homeobox 2 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| emx2 | dr11_v1_chr13_+_19322686_19322686 | -0.90 | 3.9e-02 | Click! |
| emx3 | dr11_v1_chr14_-_26377044_26377044 | -0.75 | 1.5e-01 | Click! |
Activity profile of emx2+emx3 motif
Sorted Z-values of emx2+emx3 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr5_+_2815021 | 0.91 |
ENSDART00000020472
|
hpda
|
4-hydroxyphenylpyruvate dioxygenase a |
| chr5_+_35458190 | 0.90 |
ENSDART00000051313
|
fbp1b
|
fructose-1,6-bisphosphatase 1b |
| chr4_+_25654686 | 0.78 |
ENSDART00000100714
|
acot16
|
acyl-CoA thioesterase 16 |
| chr2_+_41926707 | 0.71 |
ENSDART00000023208
|
zgc:110183
|
zgc:110183 |
| chr20_-_40755614 | 0.61 |
ENSDART00000061247
|
cx32.3
|
connexin 32.3 |
| chr6_+_49926115 | 0.60 |
ENSDART00000018523
|
ahcy
|
adenosylhomocysteinase |
| chr20_+_54309148 | 0.55 |
ENSDART00000099360
|
zp2.1
|
zona pellucida glycoprotein 2, tandem duplicate 1 |
| chr25_+_22320738 | 0.54 |
ENSDART00000073566
|
cyp11a1
|
cytochrome P450, family 11, subfamily A, polypeptide 1 |
| chr1_-_18811517 | 0.54 |
ENSDART00000142026
|
si:dkey-167i21.2
|
si:dkey-167i21.2 |
| chr8_-_23780334 | 0.54 |
ENSDART00000145179
ENSDART00000145894 |
zgc:195245
|
zgc:195245 |
| chr20_+_54299419 | 0.53 |
ENSDART00000056089
ENSDART00000193107 |
si:zfos-1505d6.3
|
si:zfos-1505d6.3 |
| chr5_-_42083363 | 0.52 |
ENSDART00000162596
|
cxcl11.5
|
chemokine (C-X-C motif) ligand 11, duplicate 5 |
| chr20_-_43663494 | 0.50 |
ENSDART00000144564
|
BX470188.1
|
|
| chr8_-_2616326 | 0.50 |
ENSDART00000027214
|
slc25a25a
|
solute carrier family 25 (mitochondrial carrier; phosphate carrier), member 25a |
| chr15_+_36457888 | 0.49 |
ENSDART00000155100
|
si:dkey-262k9.2
|
si:dkey-262k9.2 |
| chr4_+_7817996 | 0.47 |
ENSDART00000166809
|
si:ch1073-67j19.1
|
si:ch1073-67j19.1 |
| chr22_-_17729778 | 0.46 |
ENSDART00000192132
|
si:ch73-63e15.2
|
si:ch73-63e15.2 |
| chr5_-_42082730 | 0.44 |
ENSDART00000135625
|
cxcl11.5
|
chemokine (C-X-C motif) ligand 11, duplicate 5 |
| chr19_-_10330778 | 0.44 |
ENSDART00000081465
ENSDART00000136653 ENSDART00000171232 |
ccdc106b
|
coiled-coil domain containing 106b |
| chr20_+_54304800 | 0.44 |
ENSDART00000121661
|
zp2.6
|
zona pellucida glycoprotein 2, tandem duplicate 6 |
| chr3_-_59297532 | 0.44 |
ENSDART00000187991
|
CABZ01053748.1
|
|
| chr11_-_6452444 | 0.43 |
ENSDART00000137879
ENSDART00000134957 ENSDART00000004483 |
larp6b
|
La ribonucleoprotein domain family, member 6b |
| chr23_-_44965582 | 0.42 |
ENSDART00000163367
|
tfr2
|
transferrin receptor 2 |
| chr5_+_35458346 | 0.42 |
ENSDART00000141239
|
erlin2
|
ER lipid raft associated 2 |
| chr12_+_20587179 | 0.41 |
ENSDART00000170127
|
arsg
|
arylsulfatase G |
| chr16_-_50897887 | 0.39 |
ENSDART00000156985
|
si:ch73-90p23.1
|
si:ch73-90p23.1 |
| chr19_-_20403845 | 0.39 |
ENSDART00000151265
ENSDART00000147911 ENSDART00000151356 |
dazl
|
deleted in azoospermia-like |
| chr23_+_32101202 | 0.37 |
ENSDART00000000992
|
zgc:56699
|
zgc:56699 |
| chr23_+_26026383 | 0.37 |
ENSDART00000141553
|
pfkfb1
|
6-phosphofructo-2-kinase/fructose-2,6-biphosphatase 1 |
| chr23_+_41679586 | 0.37 |
ENSDART00000067662
|
CU914487.1
|
|
| chr3_+_3681116 | 0.35 |
ENSDART00000109618
|
art4
|
ADP-ribosyltransferase 4 (Dombrock blood group) |
| chr2_+_19522082 | 0.35 |
ENSDART00000146098
|
pimr49
|
Pim proto-oncogene, serine/threonine kinase, related 49 |
| chr10_-_44560165 | 0.34 |
ENSDART00000181217
ENSDART00000076084 |
npm2b
|
nucleophosmin/nucleoplasmin, 2b |
| chr21_+_18997511 | 0.34 |
ENSDART00000145591
|
rpl17
|
ribosomal protein L17 |
| chr6_+_612594 | 0.34 |
ENSDART00000150903
|
kynu
|
kynureninase |
| chr15_-_14625373 | 0.34 |
ENSDART00000171841
|
slc5a2
|
solute carrier family 5 (sodium/glucose cotransporter), member 2 |
| chr9_+_38314466 | 0.33 |
ENSDART00000048753
|
ddx18
|
DEAD (Asp-Glu-Ala-Asp) box polypeptide 18 |
| chr14_+_29941445 | 0.33 |
ENSDART00000181761
|
fam149a
|
family with sequence similarity 149 member A |
| chr13_-_31622195 | 0.33 |
ENSDART00000057432
|
six1a
|
SIX homeobox 1a |
| chr10_+_21867307 | 0.33 |
ENSDART00000126629
|
cbln17
|
cerebellin 17 |
| chr15_-_40157331 | 0.32 |
ENSDART00000187958
|
si:ch211-281l24.3
|
si:ch211-281l24.3 |
| chr18_+_5490668 | 0.32 |
ENSDART00000167035
|
mibp2
|
muscle-specific beta 1 integrin binding protein 2 |
| chr16_-_38118003 | 0.32 |
ENSDART00000058667
|
si:dkey-23o4.6
|
si:dkey-23o4.6 |
| chr6_+_25257728 | 0.32 |
ENSDART00000162581
|
kyat3
|
kynurenine aminotransferase 3 |
| chr7_+_24573721 | 0.31 |
ENSDART00000173938
ENSDART00000173681 |
si:dkeyp-75h12.7
|
si:dkeyp-75h12.7 |
| chr7_+_4694924 | 0.31 |
ENSDART00000144873
|
si:ch211-225k7.3
|
si:ch211-225k7.3 |
| chr22_-_31060579 | 0.31 |
ENSDART00000182376
|
cand2
|
cullin-associated and neddylation-dissociated 2 (putative) |
| chr3_+_54553931 | 0.30 |
ENSDART00000029387
|
ppan
|
peter pan homolog (Drosophila) |
| chr1_-_513762 | 0.30 |
ENSDART00000148162
ENSDART00000144606 |
trmt10c
|
tRNA methyltransferase 10C, mitochondrial RNase P subunit |
| chr2_-_17392799 | 0.30 |
ENSDART00000136470
ENSDART00000141188 |
st3gal3b
|
ST3 beta-galactoside alpha-2,3-sialyltransferase 3b |
| chr15_+_34934568 | 0.30 |
ENSDART00000165210
|
zgc:66024
|
zgc:66024 |
| chr15_-_40157513 | 0.29 |
ENSDART00000184014
|
si:ch211-281l24.3
|
si:ch211-281l24.3 |
| chr23_+_21405201 | 0.29 |
ENSDART00000144409
|
iffo2a
|
intermediate filament family orphan 2a |
| chr21_+_43702016 | 0.29 |
ENSDART00000017176
|
dkc1
|
dyskeratosis congenita 1, dyskerin |
| chr25_+_14092871 | 0.29 |
ENSDART00000067239
|
guca1g
|
guanylate cyclase activator 1g |
| chr10_+_24468922 | 0.29 |
ENSDART00000008248
ENSDART00000183510 |
slc7a1
|
solute carrier family 7 (cationic amino acid transporter, y+ system), member 1 |
| chr19_-_20403507 | 0.29 |
ENSDART00000052603
ENSDART00000137590 |
dazl
|
deleted in azoospermia-like |
| chr7_+_59020972 | 0.29 |
ENSDART00000157873
|
hacd1
|
3-hydroxyacyl-CoA dehydratase 1 |
| chr6_+_39905021 | 0.28 |
ENSDART00000064904
|
endou
|
endonuclease, polyU-specific |
| chr12_-_25294769 | 0.28 |
ENSDART00000153306
|
hcar1-4
|
hydroxycarboxylic acid receptor 1-4 |
| chr7_+_4694762 | 0.28 |
ENSDART00000132862
|
si:ch211-225k7.3
|
si:ch211-225k7.3 |
| chr7_+_4474880 | 0.27 |
ENSDART00000143528
|
si:dkey-83f18.14
|
si:dkey-83f18.14 |
| chr2_+_19633493 | 0.27 |
ENSDART00000147989
|
pimr54
|
Pim proto-oncogene, serine/threonine kinase, related 54 |
| chr5_+_44944778 | 0.26 |
ENSDART00000130428
ENSDART00000044361 ENSDART00000128825 ENSDART00000124637 ENSDART00000126066 ENSDART00000177635 |
dmrt1
|
doublesex and mab-3 related transcription factor 1 |
| chr25_+_31276842 | 0.26 |
ENSDART00000187238
|
tnni2a.4
|
troponin I type 2a (skeletal, fast), tandem duplicate 4 |
| chr11_+_40812590 | 0.26 |
ENSDART00000186690
|
errfi1a
|
ERBB receptor feedback inhibitor 1a |
| chr25_+_31277415 | 0.26 |
ENSDART00000036275
|
tnni2a.4
|
troponin I type 2a (skeletal, fast), tandem duplicate 4 |
| chr15_+_17728180 | 0.26 |
ENSDART00000081105
|
si:ch211-213d14.1
|
si:ch211-213d14.1 |
| chr17_+_8799661 | 0.26 |
ENSDART00000105326
|
tonsl
|
tonsoku-like, DNA repair protein |
| chr9_-_390874 | 0.26 |
ENSDART00000165324
|
si:dkey-11f4.7
|
si:dkey-11f4.7 |
| chr15_-_40157165 | 0.25 |
ENSDART00000192991
|
si:ch211-281l24.3
|
si:ch211-281l24.3 |
| chr23_-_19686791 | 0.25 |
ENSDART00000161973
|
zgc:193598
|
zgc:193598 |
| chr19_+_43780970 | 0.25 |
ENSDART00000063870
|
rpl11
|
ribosomal protein L11 |
| chr18_-_48547564 | 0.25 |
ENSDART00000138607
|
kcnj1a.1
|
potassium inwardly-rectifying channel, subfamily J, member 1a, tandem duplicate 1 |
| chr22_+_19366866 | 0.25 |
ENSDART00000137301
|
si:dkey-21e2.12
|
si:dkey-21e2.12 |
| chr9_-_5318873 | 0.25 |
ENSDART00000129308
|
ACVR1C
|
activin A receptor type 1C |
| chr20_-_33556983 | 0.25 |
ENSDART00000168798
|
rock2bl
|
rho-associated, coiled-coil containing protein kinase 2b, like |
| chr5_+_6954162 | 0.24 |
ENSDART00000086666
|
stpg2
|
sperm-tail PG-rich repeat containing 2 |
| chr16_-_31622777 | 0.24 |
ENSDART00000137311
ENSDART00000002930 |
phf20l1
|
PHD finger protein 20 like 1 |
| chr2_+_30182431 | 0.24 |
ENSDART00000004903
|
rdh10b
|
retinol dehydrogenase 10b |
| chr11_+_11200550 | 0.24 |
ENSDART00000181339
ENSDART00000187116 |
myom2a
|
myomesin 2a |
| chr24_+_42004640 | 0.24 |
ENSDART00000171380
|
top1mt
|
DNA topoisomerase I mitochondrial |
| chr2_+_14992879 | 0.24 |
ENSDART00000137546
|
pimr55
|
Pim proto-oncogene, serine/threonine kinase, related 55 |
| chr1_-_9195629 | 0.24 |
ENSDART00000143587
ENSDART00000192174 |
ern2
|
endoplasmic reticulum to nucleus signaling 2 |
| chr7_-_66868543 | 0.24 |
ENSDART00000149680
|
ampd3a
|
adenosine monophosphate deaminase 3a |
| chr10_+_34010494 | 0.24 |
ENSDART00000186836
ENSDART00000114703 |
kl
|
klotho |
| chr3_+_23029484 | 0.24 |
ENSDART00000187900
|
nags
|
N-acetylglutamate synthase |
| chr1_-_5455498 | 0.23 |
ENSDART00000040368
ENSDART00000114035 |
mnx2b
|
motor neuron and pancreas homeobox 2b |
| chr18_-_977075 | 0.23 |
ENSDART00000032392
|
dhdhl
|
dihydrodiol dehydrogenase (dimeric), like |
| chr2_+_19578446 | 0.23 |
ENSDART00000164758
|
pimr50
|
Pim proto-oncogene, serine/threonine kinase, related 50 |
| chr5_-_25733745 | 0.23 |
ENSDART00000051566
|
zgc:101016
|
zgc:101016 |
| chr20_-_5291012 | 0.23 |
ENSDART00000122892
|
cyp46a1.3
|
cytochrome P450, family 46, subfamily A, polypeptide 1, tandem duplicate 3 |
| chr25_+_3358701 | 0.23 |
ENSDART00000104877
|
chchd3b
|
coiled-coil-helix-coiled-coil-helix domain containing 3b |
| chr2_+_37227011 | 0.23 |
ENSDART00000126587
ENSDART00000084958 |
samd7
|
sterile alpha motif domain containing 7 |
| chr2_+_20406399 | 0.23 |
ENSDART00000006817
ENSDART00000137848 |
palmda
|
palmdelphin a |
| chr20_+_25586099 | 0.23 |
ENSDART00000063122
ENSDART00000134047 |
cyp2p10
|
cytochrome P450, family 2, subfamily P, polypeptide 10 |
| chr3_+_32118670 | 0.23 |
ENSDART00000055287
ENSDART00000111688 |
zgc:109934
|
zgc:109934 |
| chr9_-_22147567 | 0.22 |
ENSDART00000110941
|
crygm2d14
|
crystallin, gamma M2d14 |
| chr18_-_6766354 | 0.22 |
ENSDART00000132611
|
adm2b
|
adrenomedullin 2b |
| chr11_-_10456553 | 0.22 |
ENSDART00000169509
ENSDART00000185574 ENSDART00000188276 |
ect2
|
epithelial cell transforming 2 |
| chr10_-_34916208 | 0.22 |
ENSDART00000187371
|
ccna1
|
cyclin A1 |
| chr13_+_646700 | 0.22 |
ENSDART00000006892
|
tp53bp2a
|
tumor protein p53 binding protein, 2a |
| chr22_-_30770751 | 0.22 |
ENSDART00000172115
|
AL831726.2
|
|
| chr10_-_17550239 | 0.22 |
ENSDART00000057513
|
ska1
|
spindle and kinetochore associated complex subunit 1 |
| chr19_-_20403318 | 0.22 |
ENSDART00000136826
|
dazl
|
deleted in azoospermia-like |
| chr22_-_22337382 | 0.22 |
ENSDART00000144684
|
si:ch211-129c21.1
|
si:ch211-129c21.1 |
| chr12_-_29305533 | 0.22 |
ENSDART00000189410
|
sh2d4bb
|
SH2 domain containing 4Bb |
| chr11_-_10456387 | 0.22 |
ENSDART00000011087
ENSDART00000081827 |
ect2
|
epithelial cell transforming 2 |
| chr19_-_2822372 | 0.21 |
ENSDART00000109130
ENSDART00000122385 |
recql4
|
RecQ helicase-like 4 |
| chr25_+_13157165 | 0.21 |
ENSDART00000155653
|
si:dkeyp-50b9.1
|
si:dkeyp-50b9.1 |
| chr19_-_47323267 | 0.21 |
ENSDART00000190077
|
CU138512.1
|
|
| chr6_+_6828167 | 0.21 |
ENSDART00000181284
|
si:ch211-85n16.4
|
si:ch211-85n16.4 |
| chr15_-_46779934 | 0.21 |
ENSDART00000085136
|
clcn2c
|
chloride channel 2c |
| chr25_-_12937727 | 0.21 |
ENSDART00000172643
|
ccl39.6
|
chemokine (C-C motif) ligand 39, duplicate 6 |
| chr9_+_29548195 | 0.21 |
ENSDART00000176057
|
rnf17
|
ring finger protein 17 |
| chr21_-_1644414 | 0.21 |
ENSDART00000105736
ENSDART00000124904 |
zgc:152948
|
zgc:152948 |
| chr17_-_7496341 | 0.21 |
ENSDART00000186907
|
si:ch211-278p9.3
|
si:ch211-278p9.3 |
| chr10_-_35257458 | 0.20 |
ENSDART00000143890
ENSDART00000139107 ENSDART00000082445 |
prr11
|
proline rich 11 |
| chr4_+_28997595 | 0.20 |
ENSDART00000133357
|
si:dkey-13e3.1
|
si:dkey-13e3.1 |
| chr12_+_9250107 | 0.20 |
ENSDART00000152249
|
pimr46
|
Pim proto-oncogene, serine/threonine kinase, related 46 |
| chr7_+_4922104 | 0.20 |
ENSDART00000135154
|
si:dkey-28d5.11
|
si:dkey-28d5.11 |
| chr1_+_44127292 | 0.20 |
ENSDART00000160542
|
cabp2a
|
calcium binding protein 2a |
| chr8_+_53080515 | 0.20 |
ENSDART00000143009
|
wdr46
|
WD repeat domain 46 |
| chr13_+_24756486 | 0.20 |
ENSDART00000137074
|
col17a1b
|
collagen, type XVII, alpha 1b |
| chr3_+_23029934 | 0.20 |
ENSDART00000110343
|
nags
|
N-acetylglutamate synthase |
| chr24_+_19415124 | 0.19 |
ENSDART00000186931
|
sulf1
|
sulfatase 1 |
| chr2_+_19578079 | 0.19 |
ENSDART00000144413
|
pimr50
|
Pim proto-oncogene, serine/threonine kinase, related 50 |
| chr20_-_9658405 | 0.19 |
ENSDART00000023809
|
nid2b
|
nidogen 2b (osteonidogen) |
| chr25_-_21816269 | 0.19 |
ENSDART00000152014
|
zgc:158222
|
zgc:158222 |
| chr1_-_9629705 | 0.19 |
ENSDART00000166601
ENSDART00000164079 |
ugt5b5
|
UDP glucuronosyltransferase 5 family, polypeptide B5 |
| chr9_+_48761455 | 0.19 |
ENSDART00000139631
|
abcb11a
|
ATP-binding cassette, sub-family B (MDR/TAP), member 11a |
| chr1_+_40158146 | 0.19 |
ENSDART00000145694
|
si:ch211-113e8.9
|
si:ch211-113e8.9 |
| chr8_-_53044300 | 0.18 |
ENSDART00000191653
|
nr6a1a
|
nuclear receptor subfamily 6, group A, member 1a |
| chr2_-_19520690 | 0.18 |
ENSDART00000133559
|
pimr52
|
Pim proto-oncogene, serine/threonine kinase, related 52 |
| chr2_-_17393216 | 0.18 |
ENSDART00000123137
|
st3gal3b
|
ST3 beta-galactoside alpha-2,3-sialyltransferase 3b |
| chr5_-_24238733 | 0.18 |
ENSDART00000138170
|
plscr3a
|
phospholipid scramblase 3a |
| chr14_+_29941266 | 0.18 |
ENSDART00000112757
|
fam149a
|
family with sequence similarity 149 member A |
| chr12_+_45238292 | 0.18 |
ENSDART00000057983
|
mrpl38
|
mitochondrial ribosomal protein L38 |
| chr3_-_42981739 | 0.18 |
ENSDART00000167844
|
mafk
|
v-maf avian musculoaponeurotic fibrosarcoma oncogene homolog K |
| chr17_-_7473890 | 0.18 |
ENSDART00000192910
|
si:ch211-278p9.2
|
si:ch211-278p9.2 |
| chr23_-_23179417 | 0.18 |
ENSDART00000122945
ENSDART00000091662 |
noc2l
|
NOC2-like nucleolar associated transcriptional repressor |
| chr13_+_7442023 | 0.18 |
ENSDART00000080975
|
tnfaip2b
|
tumor necrosis factor, alpha-induced protein 2b |
| chr4_+_28998230 | 0.18 |
ENSDART00000183828
|
si:dkey-13e3.1
|
si:dkey-13e3.1 |
| chr12_-_17712393 | 0.18 |
ENSDART00000143534
ENSDART00000010144 |
pvalb2
|
parvalbumin 2 |
| chr4_-_72513569 | 0.18 |
ENSDART00000174130
|
CR788316.4
|
|
| chr18_+_33521609 | 0.18 |
ENSDART00000147592
|
v2rh7
|
vomeronasal 2 receptor, h7 |
| chr7_-_2285499 | 0.18 |
ENSDART00000182211
|
si:dkey-187j14.6
|
si:dkey-187j14.6 |
| chr16_+_32736588 | 0.17 |
ENSDART00000075191
ENSDART00000168358 |
zgc:172323
|
zgc:172323 |
| chr13_+_6219551 | 0.17 |
ENSDART00000025910
|
apmap
|
adipocyte plasma membrane associated protein |
| chr12_+_30653047 | 0.17 |
ENSDART00000148562
|
thbs2b
|
thrombospondin 2b |
| chr25_-_13188678 | 0.17 |
ENSDART00000125754
|
si:ch211-147m6.1
|
si:ch211-147m6.1 |
| chr9_+_28598577 | 0.17 |
ENSDART00000142623
ENSDART00000135947 |
si:ch73-7i4.1
|
si:ch73-7i4.1 |
| chr22_+_17261801 | 0.17 |
ENSDART00000192978
ENSDART00000193187 ENSDART00000179953 ENSDART00000134798 |
tdrd5
|
tudor domain containing 5 |
| chr17_+_389218 | 0.17 |
ENSDART00000162898
|
si:rp71-62i8.1
|
si:rp71-62i8.1 |
| chr20_+_98179 | 0.17 |
ENSDART00000022725
|
si:ch1073-155h21.1
|
si:ch1073-155h21.1 |
| chr20_-_38801981 | 0.17 |
ENSDART00000125333
|
cad
|
carbamoyl-phosphate synthetase 2, aspartate transcarbamylase, and dihydroorotase |
| chr18_+_15616167 | 0.17 |
ENSDART00000080454
|
slc17a8
|
solute carrier family 17 (vesicular glutamate transporter), member 8 |
| chr12_+_34896956 | 0.17 |
ENSDART00000055415
|
prph2a
|
peripherin 2a (retinal degeneration, slow) |
| chr24_+_39105051 | 0.17 |
ENSDART00000115297
|
mss51
|
MSS51 mitochondrial translational activator |
| chr13_+_40686133 | 0.17 |
ENSDART00000146112
|
hps1
|
Hermansky-Pudlak syndrome 1 |
| chr7_+_4648398 | 0.17 |
ENSDART00000148099
|
si:dkey-83f18.3
|
si:dkey-83f18.3 |
| chr3_+_23731109 | 0.16 |
ENSDART00000131410
|
hoxb3a
|
homeobox B3a |
| chr9_-_22355391 | 0.16 |
ENSDART00000009115
|
crygm3
|
crystallin, gamma M3 |
| chr10_+_31248036 | 0.16 |
ENSDART00000193574
|
robo4
|
roundabout, axon guidance receptor, homolog 4 (Drosophila) |
| chr23_-_1660708 | 0.16 |
ENSDART00000175138
|
CU693481.1
|
|
| chr7_-_2285314 | 0.16 |
ENSDART00000153614
|
si:dkey-187j14.6
|
si:dkey-187j14.6 |
| chr13_-_15082024 | 0.16 |
ENSDART00000157482
|
sfxn5a
|
sideroflexin 5a |
| chr11_+_30091155 | 0.16 |
ENSDART00000158691
|
si:ch211-161f7.2
|
si:ch211-161f7.2 |
| chr2_-_17235891 | 0.16 |
ENSDART00000144251
|
artnb
|
artemin b |
| chr6_-_39649504 | 0.16 |
ENSDART00000179960
ENSDART00000190951 |
larp4ab
|
La ribonucleoprotein domain family, member 4Ab |
| chr22_+_5532003 | 0.16 |
ENSDART00000106174
|
si:ch73-256j6.2
|
si:ch73-256j6.2 |
| chr11_-_34166960 | 0.16 |
ENSDART00000181571
|
atp13a3
|
ATPase 13A3 |
| chr21_+_10739846 | 0.16 |
ENSDART00000084011
|
cplx4a
|
complexin 4a |
| chr7_+_9008372 | 0.16 |
ENSDART00000144897
|
AL935128.1
|
|
| chr20_+_42881104 | 0.16 |
ENSDART00000131338
|
pimr110
|
Pim proto-oncogene, serine/threonine kinase, related 110 |
| chr2_-_19576640 | 0.16 |
ENSDART00000141021
|
pimr51
|
Pim proto-oncogene, serine/threonine kinase, related 51 |
| chr18_-_40708537 | 0.16 |
ENSDART00000077577
|
si:ch211-132b12.8
|
si:ch211-132b12.8 |
| chr2_+_9990491 | 0.16 |
ENSDART00000011906
|
slc35a3b
|
solute carrier family 35 (UDP-N-acetylglucosamine (UDP-GlcNAc) transporter), member A3b |
| chr23_-_19140781 | 0.16 |
ENSDART00000143580
|
si:ch73-381f5.2
|
si:ch73-381f5.2 |
| chr16_+_32152612 | 0.16 |
ENSDART00000008880
|
gprc6a
|
G protein-coupled receptor, class C, group 6, member A |
| chr11_-_38533505 | 0.16 |
ENSDART00000113894
|
slc45a3
|
solute carrier family 45, member 3 |
| chr24_+_26329018 | 0.16 |
ENSDART00000145752
|
mynn
|
myoneurin |
| chr11_+_31324335 | 0.15 |
ENSDART00000088093
|
sipa1l2
|
signal-induced proliferation-associated 1 like 2 |
| chr22_+_31207799 | 0.15 |
ENSDART00000133267
|
grip2b
|
glutamate receptor interacting protein 2b |
| chr7_+_4682458 | 0.15 |
ENSDART00000139271
|
si:ch211-225k7.2
|
si:ch211-225k7.2 |
| chr21_-_44081540 | 0.15 |
ENSDART00000130833
|
FO704810.1
|
|
| chr18_-_5527050 | 0.15 |
ENSDART00000145400
ENSDART00000132498 ENSDART00000146209 |
zgc:153317
|
zgc:153317 |
| chr18_-_25279323 | 0.15 |
ENSDART00000191547
|
plin1
|
perilipin 1 |
| chr5_+_72194444 | 0.15 |
ENSDART00000165436
|
ddx54
|
DEAD (Asp-Glu-Ala-Asp) box polypeptide 54 |
| chr7_+_20512419 | 0.15 |
ENSDART00000173907
|
si:dkey-19b23.14
|
si:dkey-19b23.14 |
| chr11_-_6877973 | 0.15 |
ENSDART00000160271
|
ddx49
|
DEAD (Asp-Glu-Ala-Asp) box polypeptide 49 |
| chr6_-_52788213 | 0.15 |
ENSDART00000179880
|
rbpjl
|
recombination signal binding protein for immunoglobulin kappa J region-like |
| chr11_+_42474694 | 0.15 |
ENSDART00000056048
ENSDART00000184710 |
si:ch1073-165f9.2
|
si:ch1073-165f9.2 |
| chr15_-_2493771 | 0.15 |
ENSDART00000184906
|
neu4
|
sialidase 4 |
| chr23_+_32039386 | 0.15 |
ENSDART00000133801
|
mylk2
|
myosin light chain kinase 2 |
| chr23_+_4709607 | 0.15 |
ENSDART00000166503
ENSDART00000158752 ENSDART00000163860 ENSDART00000172739 |
raf1a
raf1a
|
Raf-1 proto-oncogene, serine/threonine kinase a Raf-1 proto-oncogene, serine/threonine kinase a |
| chr2_-_31833347 | 0.14 |
ENSDART00000109460
|
nol7
|
nucleolar protein 7 |
| chr17_+_12285285 | 0.14 |
ENSDART00000154336
|
pimr174
|
Pim proto-oncogene, serine/threonine kinase, related 174 |
| chr13_+_733027 | 0.14 |
ENSDART00000149547
|
nrxn1b
|
neurexin 1b |
| chr14_-_26482096 | 0.14 |
ENSDART00000187280
|
smad5
|
SMAD family member 5 |
Network of associatons between targets according to the STRING database.
First level regulatory network of emx2+emx3
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 0.9 | GO:0005985 | sucrose metabolic process(GO:0005985) sucrose biosynthetic process(GO:0005986) disaccharide biosynthetic process(GO:0046351) |
| 0.2 | 0.9 | GO:0006572 | tyrosine catabolic process(GO:0006572) |
| 0.2 | 0.9 | GO:0060967 | negative regulation of posttranscriptional gene silencing(GO:0060149) negative regulation of gene silencing by miRNA(GO:0060965) negative regulation of gene silencing by RNA(GO:0060967) |
| 0.1 | 0.3 | GO:0097053 | L-kynurenine metabolic process(GO:0097052) L-kynurenine catabolic process(GO:0097053) |
| 0.1 | 0.4 | GO:0006526 | arginine biosynthetic process(GO:0006526) |
| 0.1 | 0.5 | GO:0006700 | C21-steroid hormone biosynthetic process(GO:0006700) |
| 0.1 | 0.4 | GO:0072512 | ferric iron transport(GO:0015682) transferrin transport(GO:0033572) trivalent inorganic cation transport(GO:0072512) |
| 0.1 | 0.3 | GO:0010265 | SCF complex assembly(GO:0010265) |
| 0.1 | 0.6 | GO:0033353 | S-adenosylmethionine cycle(GO:0033353) |
| 0.1 | 0.3 | GO:0031120 | snRNA pseudouridine synthesis(GO:0031120) |
| 0.1 | 0.3 | GO:0014856 | skeletal muscle cell proliferation(GO:0014856) regulation of skeletal muscle cell proliferation(GO:0014857) |
| 0.1 | 0.3 | GO:0030238 | male sex determination(GO:0030238) |
| 0.1 | 0.2 | GO:0060879 | peripheral nervous system myelin formation(GO:0032290) semicircular canal fusion(GO:0060879) |
| 0.1 | 0.3 | GO:0090646 | mitochondrial tRNA processing(GO:0090646) |
| 0.1 | 0.2 | GO:0021563 | glossopharyngeal nerve development(GO:0021563) |
| 0.1 | 0.2 | GO:0001954 | positive regulation of cell-matrix adhesion(GO:0001954) |
| 0.0 | 0.0 | GO:0046100 | hypoxanthine metabolic process(GO:0046100) |
| 0.0 | 0.2 | GO:0036498 | IRE1-mediated unfolded protein response(GO:0036498) |
| 0.0 | 0.2 | GO:0015722 | canalicular bile acid transport(GO:0015722) bile acid secretion(GO:0032782) |
| 0.0 | 0.1 | GO:0070314 | G1 to G0 transition(GO:0070314) |
| 0.0 | 0.2 | GO:0010692 | regulation of alkaline phosphatase activity(GO:0010692) negative regulation of alkaline phosphatase activity(GO:0010693) |
| 0.0 | 0.2 | GO:1903232 | melanosome assembly(GO:1903232) |
| 0.0 | 0.3 | GO:0045604 | regulation of epidermal cell differentiation(GO:0045604) |
| 0.0 | 0.4 | GO:0044837 | assembly of actomyosin apparatus involved in cytokinesis(GO:0000912) actomyosin contractile ring assembly(GO:0000915) actomyosin contractile ring organization(GO:0044837) |
| 0.0 | 0.1 | GO:1902455 | negative regulation of stem cell population maintenance(GO:1902455) |
| 0.0 | 0.3 | GO:0018120 | peptidyl-arginine ADP-ribosylation(GO:0018120) |
| 0.0 | 0.4 | GO:0006991 | response to sterol depletion(GO:0006991) SREBP signaling pathway(GO:0032933) cellular response to sterol depletion(GO:0071501) |
| 0.0 | 0.2 | GO:0014707 | branchiomeric skeletal muscle development(GO:0014707) |
| 0.0 | 0.1 | GO:0015882 | L-ascorbic acid transport(GO:0015882) L-ascorbic acid metabolic process(GO:0019852) |
| 0.0 | 0.3 | GO:0071260 | cellular response to mechanical stimulus(GO:0071260) |
| 0.0 | 0.1 | GO:0046333 | octopamine biosynthetic process(GO:0006589) dopamine catabolic process(GO:0042420) norepinephrine biosynthetic process(GO:0042421) octopamine metabolic process(GO:0046333) |
| 0.0 | 1.1 | GO:0071222 | cellular response to molecule of bacterial origin(GO:0071219) cellular response to lipopolysaccharide(GO:0071222) |
| 0.0 | 0.1 | GO:1901187 | regulation of ephrin receptor signaling pathway(GO:1901187) |
| 0.0 | 0.2 | GO:0007608 | sensory perception of smell(GO:0007608) |
| 0.0 | 0.1 | GO:0021531 | spinal cord radial glial cell differentiation(GO:0021531) |
| 0.0 | 0.1 | GO:0036228 | protein targeting to nuclear inner membrane(GO:0036228) protein localization to nuclear envelope(GO:0090435) |
| 0.0 | 0.2 | GO:0021910 | smoothened signaling pathway involved in ventral spinal cord patterning(GO:0021910) regulation of myotome development(GO:2000290) |
| 0.0 | 0.4 | GO:0006000 | fructose metabolic process(GO:0006000) |
| 0.0 | 0.1 | GO:0038107 | nodal signaling pathway involved in determination of left/right asymmetry(GO:0038107) regulation of nodal signaling pathway involved in determination of left/right asymmetry(GO:1900145) |
| 0.0 | 0.3 | GO:0045740 | positive regulation of DNA replication(GO:0045740) |
| 0.0 | 0.1 | GO:0018343 | protein farnesylation(GO:0018343) |
| 0.0 | 0.1 | GO:0006750 | glutathione biosynthetic process(GO:0006750) |
| 0.0 | 0.2 | GO:0098700 | neurotransmitter loading into synaptic vesicle(GO:0098700) |
| 0.0 | 0.2 | GO:0006707 | cholesterol catabolic process(GO:0006707) sterol catabolic process(GO:0016127) |
| 0.0 | 0.2 | GO:0051898 | negative regulation of protein kinase B signaling(GO:0051898) |
| 0.0 | 0.1 | GO:0046487 | glyoxylate metabolic process(GO:0046487) |
| 0.0 | 0.2 | GO:0010890 | positive regulation of sequestering of triglyceride(GO:0010890) |
| 0.0 | 0.3 | GO:0000463 | maturation of LSU-rRNA from tricistronic rRNA transcript (SSU-rRNA, 5.8S rRNA, LSU-rRNA)(GO:0000463) |
| 0.0 | 0.2 | GO:0042572 | retinol metabolic process(GO:0042572) |
| 0.0 | 0.2 | GO:0035065 | regulation of histone acetylation(GO:0035065) |
| 0.0 | 0.2 | GO:0051601 | exocyst localization(GO:0051601) |
| 0.0 | 0.2 | GO:0043046 | DNA methylation involved in gamete generation(GO:0043046) |
| 0.0 | 0.2 | GO:0032264 | IMP salvage(GO:0032264) |
| 0.0 | 0.2 | GO:0021520 | spinal cord motor neuron cell fate specification(GO:0021520) |
| 0.0 | 0.0 | GO:0032755 | positive regulation of interleukin-6 production(GO:0032755) |
| 0.0 | 0.2 | GO:0006842 | tricarboxylic acid transport(GO:0006842) citrate transport(GO:0015746) |
| 0.0 | 0.1 | GO:0010867 | regulation of triglyceride biosynthetic process(GO:0010866) positive regulation of triglyceride biosynthetic process(GO:0010867) |
| 0.0 | 0.2 | GO:0006265 | DNA topological change(GO:0006265) |
| 0.0 | 0.2 | GO:0000186 | activation of MAPKK activity(GO:0000186) |
| 0.0 | 0.2 | GO:0099639 | neurotransmitter receptor transport, endosome to postsynaptic membrane(GO:0098887) endosome to plasma membrane protein transport(GO:0099638) neurotransmitter receptor transport, endosome to plasma membrane(GO:0099639) |
| 0.0 | 0.1 | GO:2000275 | regulation of oxidative phosphorylation uncoupler activity(GO:2000275) |
| 0.0 | 0.1 | GO:0019375 | galactosylceramide biosynthetic process(GO:0006682) galactolipid biosynthetic process(GO:0019375) |
| 0.0 | 0.1 | GO:0030216 | keratinocyte differentiation(GO:0030216) |
| 0.0 | 0.3 | GO:0000027 | ribosomal large subunit assembly(GO:0000027) |
| 0.0 | 0.4 | GO:0007568 | aging(GO:0007568) |
| 0.0 | 0.3 | GO:0031297 | replication fork processing(GO:0031297) |
| 0.0 | 0.7 | GO:0003009 | skeletal muscle contraction(GO:0003009) |
| 0.0 | 0.3 | GO:0042541 | hemoglobin biosynthetic process(GO:0042541) |
| 0.0 | 0.1 | GO:0009313 | oligosaccharide catabolic process(GO:0009313) |
| 0.0 | 0.1 | GO:0031179 | peptide modification(GO:0031179) |
| 0.0 | 0.3 | GO:0070831 | basement membrane assembly(GO:0070831) |
| 0.0 | 0.3 | GO:0030497 | fatty acid elongation(GO:0030497) |
| 0.0 | 0.1 | GO:0019441 | tryptophan catabolic process to kynurenine(GO:0019441) |
| 0.0 | 0.0 | GO:0034729 | histone H3-K79 methylation(GO:0034729) regulation of transcription regulatory region DNA binding(GO:2000677) |
| 0.0 | 0.1 | GO:2000223 | regulation of BMP signaling pathway involved in heart jogging(GO:2000223) |
| 0.0 | 0.1 | GO:0072584 | optomotor response(GO:0071632) caveolin-mediated endocytosis(GO:0072584) negative regulation of clathrin-mediated endocytosis(GO:1900186) regulation of caveolin-mediated endocytosis(GO:2001286) negative regulation of caveolin-mediated endocytosis(GO:2001287) |
| 0.0 | 0.5 | GO:0050727 | regulation of inflammatory response(GO:0050727) |
| 0.0 | 0.0 | GO:0070084 | protein initiator methionine removal(GO:0070084) |
| 0.0 | 0.2 | GO:0015781 | pyrimidine nucleotide-sugar transport(GO:0015781) pyrimidine nucleotide-sugar transmembrane transport(GO:0090481) |
| 0.0 | 0.2 | GO:0015858 | nucleoside transport(GO:0015858) nucleoside transmembrane transport(GO:1901642) |
| 0.0 | 0.1 | GO:0006268 | DNA unwinding involved in DNA replication(GO:0006268) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.4 | GO:0043540 | 6-phosphofructo-2-kinase/fructose-2,6-biphosphatase complex(GO:0043540) |
| 0.1 | 0.2 | GO:1990604 | IRE1-TRAF2-ASK1 complex(GO:1990604) |
| 0.1 | 0.2 | GO:0097124 | cyclin A2-CDK2 complex(GO:0097124) |
| 0.1 | 0.3 | GO:0031429 | box H/ACA snoRNP complex(GO:0031429) box H/ACA RNP complex(GO:0072588) |
| 0.1 | 0.2 | GO:0030689 | Noc complex(GO:0030689) |
| 0.1 | 0.2 | GO:0033391 | chromatoid body(GO:0033391) |
| 0.0 | 0.2 | GO:0031085 | BLOC-3 complex(GO:0031085) |
| 0.0 | 0.3 | GO:0009295 | nucleoid(GO:0009295) mitochondrial nucleoid(GO:0042645) |
| 0.0 | 0.3 | GO:0005662 | DNA replication factor A complex(GO:0005662) |
| 0.0 | 0.1 | GO:0034457 | Mpp10 complex(GO:0034457) |
| 0.0 | 0.1 | GO:0005965 | protein farnesyltransferase complex(GO:0005965) |
| 0.0 | 0.2 | GO:0061617 | MICOS complex(GO:0061617) |
| 0.0 | 0.1 | GO:0044611 | nuclear pore inner ring(GO:0044611) |
| 0.0 | 0.1 | GO:0071595 | Nem1-Spo7 phosphatase complex(GO:0071595) |
| 0.0 | 0.2 | GO:0044545 | NSL complex(GO:0044545) |
| 0.0 | 0.2 | GO:0000940 | condensed chromosome outer kinetochore(GO:0000940) |
| 0.0 | 0.7 | GO:0005861 | troponin complex(GO:0005861) |
| 0.0 | 0.3 | GO:0030687 | preribosome, large subunit precursor(GO:0030687) |
| 0.0 | 0.2 | GO:0031430 | M band(GO:0031430) |
| 0.0 | 0.2 | GO:0098563 | integral component of synaptic vesicle membrane(GO:0030285) intrinsic component of synaptic vesicle membrane(GO:0098563) |
| 0.0 | 0.2 | GO:0005751 | mitochondrial respiratory chain complex IV(GO:0005751) |
| 0.0 | 0.3 | GO:0032040 | small-subunit processome(GO:0032040) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 0.9 | GO:0003868 | 4-hydroxyphenylpyruvate dioxygenase activity(GO:0003868) |
| 0.3 | 0.9 | GO:0042132 | fructose 1,6-bisphosphate 1-phosphatase activity(GO:0042132) |
| 0.2 | 0.6 | GO:0004013 | adenosylhomocysteinase activity(GO:0004013) trialkylsulfonium hydrolase activity(GO:0016802) |
| 0.2 | 0.5 | GO:0008386 | cholesterol monooxygenase (side-chain-cleaving) activity(GO:0008386) oxidoreductase activity, acting on paired donors, with incorporation or reduction of molecular oxygen, reduced iron-sulfur protein as one donor, and incorporation of one atom of oxygen(GO:0016713) |
| 0.1 | 0.4 | GO:0034618 | arginine binding(GO:0034618) |
| 0.1 | 0.4 | GO:0004998 | transferrin receptor activity(GO:0004998) |
| 0.1 | 0.9 | GO:0008494 | translation activator activity(GO:0008494) |
| 0.1 | 1.0 | GO:0045236 | CXCR chemokine receptor binding(GO:0045236) |
| 0.1 | 0.3 | GO:0052905 | tRNA (guanine(9)-N(1))-methyltransferase activity(GO:0052905) |
| 0.1 | 0.3 | GO:0018812 | 3-hydroxyacyl-CoA dehydratase activity(GO:0018812) 3-hydroxy-behenoyl-CoA dehydratase activity(GO:0102344) 3-hydroxy-lignoceroyl-CoA dehydratase activity(GO:0102345) |
| 0.1 | 0.5 | GO:0008118 | N-acetyllactosaminide alpha-2,3-sialyltransferase activity(GO:0008118) |
| 0.1 | 0.3 | GO:0016822 | hydrolase activity, acting on acid carbon-carbon bonds(GO:0016822) hydrolase activity, acting on acid carbon-carbon bonds, in ketonic substances(GO:0016823) |
| 0.1 | 0.2 | GO:0033149 | FFAT motif binding(GO:0033149) |
| 0.1 | 0.2 | GO:0016743 | carboxyl- or carbamoyltransferase activity(GO:0016743) |
| 0.1 | 0.3 | GO:0005412 | glucose:sodium symporter activity(GO:0005412) |
| 0.0 | 0.2 | GO:0015432 | canalicular bile acid transmembrane transporter activity(GO:0015126) bile acid-exporting ATPase activity(GO:0015432) |
| 0.0 | 0.3 | GO:0050262 | ribosylnicotinamide kinase activity(GO:0050262) |
| 0.0 | 0.2 | GO:0033781 | cholesterol 24-hydroxylase activity(GO:0033781) |
| 0.0 | 0.3 | GO:0052650 | NADP-retinol dehydrogenase activity(GO:0052650) |
| 0.0 | 0.3 | GO:0008048 | calcium sensitive guanylate cyclase activator activity(GO:0008048) cyclase activator activity(GO:0010853) guanylate cyclase activator activity(GO:0030250) |
| 0.0 | 0.3 | GO:0003956 | NAD(P)+-protein-arginine ADP-ribosyltransferase activity(GO:0003956) |
| 0.0 | 0.1 | GO:0004377 | GDP-Man:Man3GlcNAc2-PP-Dol alpha-1,2-mannosyltransferase activity(GO:0004377) |
| 0.0 | 0.8 | GO:0047617 | acyl-CoA hydrolase activity(GO:0047617) |
| 0.0 | 0.1 | GO:0016165 | linoleate 13S-lipoxygenase activity(GO:0016165) |
| 0.0 | 0.1 | GO:0004500 | dopamine beta-monooxygenase activity(GO:0004500) |
| 0.0 | 0.6 | GO:0004065 | arylsulfatase activity(GO:0004065) |
| 0.0 | 0.2 | GO:0015137 | citrate transmembrane transporter activity(GO:0015137) tricarboxylic acid transmembrane transporter activity(GO:0015142) |
| 0.0 | 0.4 | GO:0003873 | 6-phosphofructo-2-kinase activity(GO:0003873) |
| 0.0 | 0.1 | GO:0004061 | arylformamidase activity(GO:0004061) |
| 0.0 | 0.2 | GO:0047623 | AMP deaminase activity(GO:0003876) adenosine-phosphate deaminase activity(GO:0047623) |
| 0.0 | 0.1 | GO:0004660 | protein farnesyltransferase activity(GO:0004660) |
| 0.0 | 0.2 | GO:0008422 | beta-glucosidase activity(GO:0008422) |
| 0.0 | 0.1 | GO:0004619 | bisphosphoglycerate mutase activity(GO:0004082) phosphoglycerate mutase activity(GO:0004619) |
| 0.0 | 0.1 | GO:0042834 | peptidoglycan binding(GO:0042834) |
| 0.0 | 0.2 | GO:0005351 | sugar:proton symporter activity(GO:0005351) cation:sugar symporter activity(GO:0005402) sucrose:proton symporter activity(GO:0008506) sucrose transmembrane transporter activity(GO:0008515) disaccharide transmembrane transporter activity(GO:0015154) oligosaccharide transmembrane transporter activity(GO:0015157) |
| 0.0 | 0.2 | GO:0016840 | carbon-nitrogen lyase activity(GO:0016840) |
| 0.0 | 0.1 | GO:0008179 | adenylate cyclase binding(GO:0008179) |
| 0.0 | 0.3 | GO:0009982 | pseudouridine synthase activity(GO:0009982) |
| 0.0 | 0.1 | GO:0070644 | vitamin D response element binding(GO:0070644) |
| 0.0 | 0.1 | GO:0004968 | gonadotropin-releasing hormone receptor activity(GO:0004968) |
| 0.0 | 0.1 | GO:0009378 | four-way junction helicase activity(GO:0009378) |
| 0.0 | 0.2 | GO:0005459 | UDP-galactose transmembrane transporter activity(GO:0005459) |
| 0.0 | 0.2 | GO:0017128 | phospholipid scramblase activity(GO:0017128) |
| 0.0 | 0.0 | GO:0016971 | flavin-linked sulfhydryl oxidase activity(GO:0016971) |
| 0.0 | 0.1 | GO:0048019 | receptor antagonist activity(GO:0048019) |
| 0.0 | 0.0 | GO:0052717 | tRNA-specific adenosine-34 deaminase activity(GO:0052717) |
| 0.0 | 0.1 | GO:0052795 | exo-alpha-(2->3)-sialidase activity(GO:0052794) exo-alpha-(2->6)-sialidase activity(GO:0052795) exo-alpha-(2->8)-sialidase activity(GO:0052796) |
| 0.0 | 0.1 | GO:0050218 | propionate-CoA ligase activity(GO:0050218) |
| 0.0 | 0.2 | GO:0005326 | neurotransmitter transporter activity(GO:0005326) |
| 0.0 | 0.2 | GO:0031729 | CCR1 chemokine receptor binding(GO:0031726) CCR4 chemokine receptor binding(GO:0031729) |
| 0.0 | 0.2 | GO:0002039 | p53 binding(GO:0002039) |
| 0.0 | 0.2 | GO:0005247 | voltage-gated chloride channel activity(GO:0005247) |
| 0.0 | 0.1 | GO:0004687 | myosin light chain kinase activity(GO:0004687) |
| 0.0 | 0.0 | GO:0031151 | histone methyltransferase activity (H3-K79 specific)(GO:0031151) |
| 0.0 | 0.1 | GO:0000048 | peptidyltransferase activity(GO:0000048) glutathione hydrolase activity(GO:0036374) |
| 0.0 | 0.1 | GO:0050694 | galactosylceramide sulfotransferase activity(GO:0001733) galactose 3-O-sulfotransferase activity(GO:0050694) |
| 0.0 | 0.5 | GO:0005212 | structural constituent of eye lens(GO:0005212) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.3 | SA FAS SIGNALING | The TNF-type receptor Fas induces apoptosis on ligand binding. |
| 0.0 | 0.2 | SA REG CASCADE OF CYCLIN EXPR | Expression of cyclins regulates progression through the cell cycle by activating cyclin-dependent kinases. |
| 0.0 | 0.1 | SA MMP CYTOKINE CONNECTION | Cytokines can induce activation of matrix metalloproteinases, which degrade extracellular matrix. |
| 0.0 | 0.5 | PID RHOA REG PATHWAY | Regulation of RhoA activity |
| 0.0 | 0.7 | PID MYC ACTIV PATHWAY | Validated targets of C-MYC transcriptional activation |
| 0.0 | 0.1 | PID ALK2 PATHWAY | ALK2 signaling events |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.5 | REACTOME ENDOGENOUS STEROLS | Genes involved in Endogenous sterols |
| 0.0 | 0.4 | REACTOME THE ACTIVATION OF ARYLSULFATASES | Genes involved in The activation of arylsulfatases |
| 0.0 | 0.4 | REACTOME TRYPTOPHAN CATABOLISM | Genes involved in Tryptophan catabolism |
| 0.0 | 0.7 | REACTOME PHASE II CONJUGATION | Genes involved in Phase II conjugation |
| 0.0 | 0.2 | REACTOME CLASS C 3 METABOTROPIC GLUTAMATE PHEROMONE RECEPTORS | Genes involved in Class C/3 (Metabotropic glutamate/pheromone receptors) |
| 0.0 | 0.2 | REACTOME SIGNALING BY NODAL | Genes involved in Signaling by NODAL |
| 0.0 | 0.4 | REACTOME GLUCONEOGENESIS | Genes involved in Gluconeogenesis |
| 0.0 | 0.2 | REACTOME FGFR LIGAND BINDING AND ACTIVATION | Genes involved in FGFR ligand binding and activation |
| 0.0 | 0.2 | REACTOME RNA POL III CHAIN ELONGATION | Genes involved in RNA Polymerase III Chain Elongation |
| 0.0 | 0.5 | REACTOME AMINO ACID AND OLIGOPEPTIDE SLC TRANSPORTERS | Genes involved in Amino acid and oligopeptide SLC transporters |
| 0.0 | 0.1 | REACTOME SIGNALING BY BMP | Genes involved in Signaling by BMP |