Project
PRJNA207719: Tissue specific transcriptome profiling
Navigation
Downloads
Results for en1b_en2a+en2b_gbx1_emx1
Z-value: 1.14
Transcription factors associated with en1b_en2a+en2b_gbx1_emx1
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
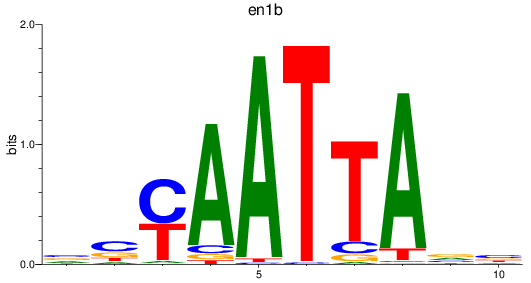
en1b
|
ENSDARG00000098730 | engrailed homeobox 1b |
|
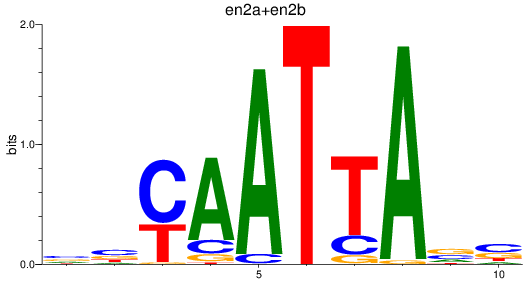
en2a
|
ENSDARG00000026599 | engrailed homeobox 2a |
|
en2b
|
ENSDARG00000038868 | engrailed homeobox 2b |
|
en2a
|
ENSDARG00000115233 | engrailed homeobox 2a |
|
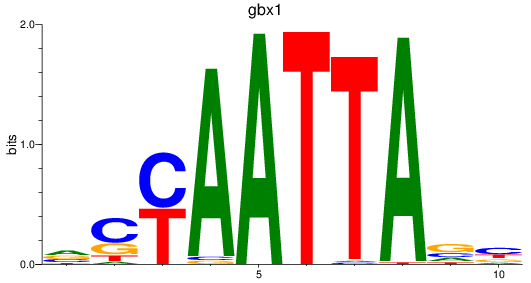
gbx1
|
ENSDARG00000071418 | gastrulation brain homeobox 1 |
|
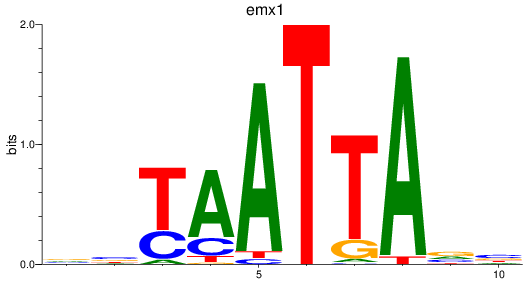
emx1
|
ENSDARG00000039569 | empty spiracles homeobox 1 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| en2a | dr11_v1_chr7_-_40993456_40993456 | 0.99 | 5.9e-04 | Click! |
| en2b | dr11_v1_chr2_+_29976419_29976419 | 0.99 | 1.3e-03 | Click! |
| en1b | dr11_v1_chr1_+_7679328_7679328 | 0.97 | 6.3e-03 | Click! |
| emx1 | dr11_v1_chr13_+_15004398_15004398 | 0.95 | 1.3e-02 | Click! |
| gbx1 | dr11_v1_chr24_+_34113424_34113424 | 0.88 | 5.2e-02 | Click! |
Activity profile of en1b_en2a+en2b_gbx1_emx1 motif
Sorted Z-values of en1b_en2a+en2b_gbx1_emx1 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr5_+_64732036 | 2.10 |
ENSDART00000073950
|
olfm1a
|
olfactomedin 1a |
| chr5_+_64732270 | 2.04 |
ENSDART00000134241
|
olfm1a
|
olfactomedin 1a |
| chr6_+_39232245 | 1.79 |
ENSDART00000187351
|
b4galnt1b
|
beta-1,4-N-acetyl-galactosaminyl transferase 1b |
| chr11_-_42554290 | 1.66 |
ENSDART00000130573
|
atp6ap1la
|
ATPase H+ transporting accessory protein 1 like a |
| chr23_+_40460333 | 1.65 |
ENSDART00000184658
|
soga3b
|
SOGA family member 3b |
| chr19_+_5543072 | 1.57 |
ENSDART00000082080
|
jupb
|
junction plakoglobin b |
| chr23_+_44741500 | 1.53 |
ENSDART00000166421
|
atp1b2a
|
ATPase Na+/K+ transporting subunit beta 2a |
| chr7_+_30787903 | 1.48 |
ENSDART00000174000
|
apba2b
|
amyloid beta (A4) precursor protein-binding, family A, member 2b |
| chr2_+_50608099 | 1.47 |
ENSDART00000185805
ENSDART00000111135 |
neurod6b
|
neuronal differentiation 6b |
| chr7_+_13418812 | 1.39 |
ENSDART00000191905
ENSDART00000091567 |
dagla
|
diacylglycerol lipase, alpha |
| chr22_+_4488454 | 1.27 |
ENSDART00000170620
|
ctxn1
|
cortexin 1 |
| chr21_+_28958471 | 1.24 |
ENSDART00000144331
ENSDART00000005929 |
ppp3ca
|
protein phosphatase 3, catalytic subunit, alpha isozyme |
| chr23_+_26946744 | 1.19 |
ENSDART00000115141
|
cacnb3b
|
calcium channel, voltage-dependent, beta 3b |
| chr16_+_5774977 | 1.19 |
ENSDART00000134202
|
ccka
|
cholecystokinin a |
| chr22_+_17828267 | 1.18 |
ENSDART00000136016
|
hapln4
|
hyaluronan and proteoglycan link protein 4 |
| chr15_+_44366556 | 1.14 |
ENSDART00000133449
|
GUCY1A2
|
guanylate cyclase 1 soluble subunit alpha 2 |
| chr24_-_41320037 | 1.12 |
ENSDART00000129058
|
rheb
|
Ras homolog, mTORC1 binding |
| chr19_-_5805923 | 1.11 |
ENSDART00000134340
|
si:ch211-264f5.8
|
si:ch211-264f5.8 |
| chr24_+_16547035 | 1.09 |
ENSDART00000164319
|
sema5a
|
sema domain, seven thrombospondin repeats (type 1 and type 1-like), transmembrane domain (TM) and short cytoplasmic domain, (semaphorin) 5A |
| chr8_-_50888806 | 1.09 |
ENSDART00000053750
|
acsl2
|
acyl-CoA synthetase long chain family member 2 |
| chr17_-_200316 | 1.08 |
ENSDART00000190561
|
CABZ01083778.1
|
|
| chr24_-_31843173 | 1.08 |
ENSDART00000185782
|
steap2
|
STEAP family member 2, metalloreductase |
| chr15_-_9272328 | 1.06 |
ENSDART00000172114
|
calm2a
|
calmodulin 2a (phosphorylase kinase, delta) |
| chr6_-_6487876 | 1.05 |
ENSDART00000137642
|
cep170ab
|
centrosomal protein 170Ab |
| chr11_+_38280454 | 1.03 |
ENSDART00000171496
|
CDK18
|
si:dkey-166c18.1 |
| chr12_+_24342303 | 1.02 |
ENSDART00000111239
|
nrxn1a
|
neurexin 1a |
| chr16_+_47207691 | 1.01 |
ENSDART00000062507
|
ica1
|
islet cell autoantigen 1 |
| chr18_+_11506561 | 1.01 |
ENSDART00000121647
|
PRMT8
|
protein arginine methyltransferase 8 |
| chr1_-_50859053 | 0.97 |
ENSDART00000132779
ENSDART00000137648 |
si:dkeyp-123h10.2
|
si:dkeyp-123h10.2 |
| chr19_+_14921000 | 0.97 |
ENSDART00000144052
|
oprd1a
|
opioid receptor, delta 1a |
| chr7_+_23515966 | 0.96 |
ENSDART00000186893
ENSDART00000186189 |
zgc:109889
|
zgc:109889 |
| chr5_-_23280098 | 0.92 |
ENSDART00000126540
ENSDART00000051533 |
plp1b
|
proteolipid protein 1b |
| chr1_-_47071979 | 0.91 |
ENSDART00000160817
|
itsn1
|
intersectin 1 (SH3 domain protein) |
| chr7_-_72261721 | 0.91 |
ENSDART00000172229
|
RASGRP2
|
RAS guanyl releasing protein 2 |
| chr1_+_16397063 | 0.91 |
ENSDART00000159794
|
micu3a
|
mitochondrial calcium uptake family, member 3a |
| chr9_-_20372977 | 0.90 |
ENSDART00000113418
|
igsf3
|
immunoglobulin superfamily, member 3 |
| chr4_+_9400012 | 0.90 |
ENSDART00000191960
|
tmtc1
|
transmembrane and tetratricopeptide repeat containing 1 |
| chr25_+_21324588 | 0.87 |
ENSDART00000151842
|
lrrn3a
|
leucine rich repeat neuronal 3a |
| chr9_+_38372216 | 0.86 |
ENSDART00000141895
|
plcd4b
|
phospholipase C, delta 4b |
| chr25_-_32869794 | 0.86 |
ENSDART00000162784
|
tmem266
|
transmembrane protein 266 |
| chr11_+_25129013 | 0.84 |
ENSDART00000132879
|
ndrg3a
|
ndrg family member 3a |
| chr2_-_56385373 | 0.82 |
ENSDART00000169101
|
cers4b
|
ceramide synthase 4b |
| chr20_-_29864390 | 0.81 |
ENSDART00000161834
ENSDART00000132278 |
rnf144ab
|
ring finger protein 144ab |
| chr4_+_11384891 | 0.80 |
ENSDART00000092381
ENSDART00000186577 ENSDART00000191054 ENSDART00000191584 |
pcloa
|
piccolo presynaptic cytomatrix protein a |
| chr10_+_35952532 | 0.80 |
ENSDART00000184730
|
rtn4rl1a
|
reticulon 4 receptor-like 1a |
| chr7_+_32722227 | 0.80 |
ENSDART00000126565
|
si:ch211-150g13.3
|
si:ch211-150g13.3 |
| chr23_-_333457 | 0.77 |
ENSDART00000114486
|
uhrf1bp1
|
UHRF1 binding protein 1 |
| chr19_-_5865766 | 0.77 |
ENSDART00000191007
|
LO018585.1
|
|
| chr10_-_44508249 | 0.77 |
ENSDART00000160018
|
DUSP26
|
dual specificity phosphatase 26 |
| chr23_+_28582865 | 0.77 |
ENSDART00000020296
|
l1cama
|
L1 cell adhesion molecule, paralog a |
| chr8_+_53423408 | 0.76 |
ENSDART00000164792
|
cacna1db
|
calcium channel, voltage-dependent, L type, alpha 1D subunit, b |
| chr22_-_11137268 | 0.76 |
ENSDART00000178882
|
atp6ap2
|
ATPase H+ transporting accessory protein 2 |
| chr2_-_31936966 | 0.76 |
ENSDART00000169484
ENSDART00000192492 ENSDART00000027689 |
amph
|
amphiphysin |
| chr25_-_5119162 | 0.74 |
ENSDART00000153961
|
shisal1b
|
shisa like 1b |
| chr1_-_46981134 | 0.73 |
ENSDART00000130607
|
pknox1.2
|
pbx/knotted 1 homeobox 1.2 |
| chr4_+_21129752 | 0.73 |
ENSDART00000169764
|
syt1a
|
synaptotagmin Ia |
| chr8_+_7756893 | 0.73 |
ENSDART00000191894
|
fgd1
|
FYVE, RhoGEF and PH domain containing 1 |
| chr21_-_39177564 | 0.73 |
ENSDART00000065143
|
unc119b
|
unc-119 homolog b (C. elegans) |
| chr6_+_21001264 | 0.72 |
ENSDART00000044519
ENSDART00000151278 |
cx44.2
|
connexin 44.2 |
| chr9_+_17983463 | 0.71 |
ENSDART00000182150
|
akap11
|
A kinase (PRKA) anchor protein 11 |
| chr18_+_9615147 | 0.71 |
ENSDART00000160284
|
pclob
|
piccolo presynaptic cytomatrix protein b |
| chr2_+_1714640 | 0.67 |
ENSDART00000086761
ENSDART00000111613 |
adgrl2b.1
adgrl2b.1
|
adhesion G protein-coupled receptor L2b, tandem duplicate 1 adhesion G protein-coupled receptor L2b, tandem duplicate 1 |
| chr20_-_46362606 | 0.67 |
ENSDART00000153087
|
bmf2
|
BCL2 modifying factor 2 |
| chr11_+_28476298 | 0.67 |
ENSDART00000122319
|
lrrc38b
|
leucine rich repeat containing 38b |
| chr17_+_24722646 | 0.67 |
ENSDART00000138356
|
mtfr1l
|
mitochondrial fission regulator 1-like |
| chr12_+_31744217 | 0.67 |
ENSDART00000190361
|
RNF157
|
si:dkey-49c17.3 |
| chr15_+_1796313 | 0.66 |
ENSDART00000126253
|
fam124b
|
family with sequence similarity 124B |
| chr14_+_45406299 | 0.66 |
ENSDART00000173142
ENSDART00000112377 |
map1lc3cl
|
microtubule-associated protein 1 light chain 3 gamma, like |
| chr10_+_15255198 | 0.66 |
ENSDART00000139047
ENSDART00000172107 ENSDART00000183413 ENSDART00000185314 |
vldlr
|
very low density lipoprotein receptor |
| chr19_-_20113696 | 0.65 |
ENSDART00000188813
|
npy
|
neuropeptide Y |
| chr10_+_17371356 | 0.65 |
ENSDART00000122663
|
sppl3
|
signal peptide peptidase 3 |
| chr17_+_21902058 | 0.65 |
ENSDART00000142178
|
plekha1a
|
pleckstrin homology domain containing, family A (phosphoinositide binding specific) member 1a |
| chr22_-_11136625 | 0.65 |
ENSDART00000016873
ENSDART00000125561 |
atp6ap2
|
ATPase H+ transporting accessory protein 2 |
| chr22_-_881725 | 0.64 |
ENSDART00000035514
|
cept1b
|
choline/ethanolamine phosphotransferase 1b |
| chr5_+_59392183 | 0.63 |
ENSDART00000082983
ENSDART00000180882 |
clip2
|
CAP-GLY domain containing linker protein 2 |
| chr22_+_17205608 | 0.62 |
ENSDART00000181267
|
rab3b
|
RAB3B, member RAS oncogene family |
| chr3_+_34919810 | 0.62 |
ENSDART00000055264
|
ca10b
|
carbonic anhydrase Xb |
| chr2_-_14390627 | 0.62 |
ENSDART00000172367
|
sgip1b
|
SH3-domain GRB2-like (endophilin) interacting protein 1b |
| chr6_-_12275836 | 0.62 |
ENSDART00000189980
|
pkp4
|
plakophilin 4 |
| chr5_+_30624183 | 0.62 |
ENSDART00000141444
|
abcg4a
|
ATP-binding cassette, sub-family G (WHITE), member 4a |
| chr15_-_11683529 | 0.62 |
ENSDART00000161445
|
fkrp
|
fukutin related protein |
| chr12_+_2446837 | 0.61 |
ENSDART00000112032
|
ARHGAP22
|
si:dkey-191m6.4 |
| chr17_-_12336987 | 0.61 |
ENSDART00000172001
|
snap25b
|
synaptosomal-associated protein, 25b |
| chr19_-_30524952 | 0.60 |
ENSDART00000103506
|
hpcal4
|
hippocalcin like 4 |
| chr15_+_15856178 | 0.59 |
ENSDART00000080338
|
dusp14
|
dual specificity phosphatase 14 |
| chr1_+_49266886 | 0.59 |
ENSDART00000137179
|
caly
|
calcyon neuron-specific vesicular protein |
| chr14_+_8940326 | 0.59 |
ENSDART00000159920
|
rps6kal
|
ribosomal protein S6 kinase a, like |
| chr16_-_35329803 | 0.59 |
ENSDART00000161729
ENSDART00000157700 ENSDART00000184584 ENSDART00000174713 ENSDART00000162518 |
ptprub
|
protein tyrosine phosphatase, receptor type, U, b |
| chr11_-_472547 | 0.57 |
ENSDART00000005923
|
zgc:77375
|
zgc:77375 |
| chr22_+_5176693 | 0.56 |
ENSDART00000160927
|
cers1
|
ceramide synthase 1 |
| chr10_+_15777064 | 0.56 |
ENSDART00000114483
|
apba1b
|
amyloid beta (A4) precursor protein-binding, family A, member 1b |
| chr12_+_47698356 | 0.55 |
ENSDART00000112010
|
lzts2b
|
leucine zipper, putative tumor suppressor 2b |
| chr3_+_13637383 | 0.54 |
ENSDART00000166000
|
si:ch211-194b1.1
|
si:ch211-194b1.1 |
| chr1_+_52929185 | 0.54 |
ENSDART00000147683
|
inpp4b
|
inositol polyphosphate-4-phosphatase type II B |
| chr13_+_38430466 | 0.53 |
ENSDART00000132691
|
adgrb3
|
adhesion G protein-coupled receptor B3 |
| chr21_+_1381276 | 0.53 |
ENSDART00000192907
|
TCF4
|
transcription factor 4 |
| chr18_-_8857137 | 0.53 |
ENSDART00000126331
|
prrt4
|
proline-rich transmembrane protein 4 |
| chr12_-_18393408 | 0.52 |
ENSDART00000159674
|
tom1l2
|
target of myb1 like 2 membrane trafficking protein |
| chr2_-_11258547 | 0.52 |
ENSDART00000165803
ENSDART00000193817 |
slc44a5a
|
solute carrier family 44, member 5a |
| chr16_+_12286148 | 0.52 |
ENSDART00000130736
|
si:dkey-26c10.5
|
si:dkey-26c10.5 |
| chr8_+_49778486 | 0.52 |
ENSDART00000131732
|
ntrk2a
|
neurotrophic tyrosine kinase, receptor, type 2a |
| chr7_-_25895189 | 0.52 |
ENSDART00000173599
ENSDART00000079235 ENSDART00000173786 ENSDART00000173602 ENSDART00000079245 ENSDART00000187568 ENSDART00000173505 |
cd99l2
|
CD99 molecule-like 2 |
| chr16_+_46111849 | 0.52 |
ENSDART00000172232
|
sv2a
|
synaptic vesicle glycoprotein 2A |
| chr19_-_31402429 | 0.52 |
ENSDART00000137292
|
tmem106bb
|
transmembrane protein 106Bb |
| chr5_-_21970881 | 0.51 |
ENSDART00000182907
|
arhgef9a
|
Cdc42 guanine nucleotide exchange factor (GEF) 9a |
| chr23_+_4646194 | 0.51 |
ENSDART00000092344
|
LO017700.1
|
|
| chr2_+_20332044 | 0.50 |
ENSDART00000112131
|
plppr4a
|
phospholipid phosphatase related 4a |
| chr17_+_31185276 | 0.50 |
ENSDART00000062887
|
disp2
|
dispatched homolog 2 (Drosophila) |
| chr1_-_38756870 | 0.50 |
ENSDART00000130324
ENSDART00000148404 |
gpm6ab
|
glycoprotein M6Ab |
| chr11_+_45323455 | 0.50 |
ENSDART00000189249
|
mafgb
|
v-maf avian musculoaponeurotic fibrosarcoma oncogene homolog Gb |
| chr17_+_12813316 | 0.50 |
ENSDART00000155629
|
ralgapa1
|
Ral GTPase activating protein, alpha subunit 1 (catalytic) |
| chr17_-_21418340 | 0.49 |
ENSDART00000007021
|
atp6v1ba
|
ATPase, H+ transporting, lysosomal, V1 subunit B, member a |
| chr14_+_23717165 | 0.49 |
ENSDART00000006373
|
ndfip1
|
Nedd4 family interacting protein 1 |
| chr2_-_11512819 | 0.49 |
ENSDART00000142013
|
penka
|
proenkephalin a |
| chr2_-_32558795 | 0.49 |
ENSDART00000140026
|
smarcd3a
|
SWI/SNF related, matrix associated, actin dependent regulator of chromatin, subfamily d, member 3a |
| chr16_-_29437373 | 0.49 |
ENSDART00000148405
|
si:ch211-113g11.6
|
si:ch211-113g11.6 |
| chr20_-_31808779 | 0.49 |
ENSDART00000133788
|
stxbp5a
|
syntaxin binding protein 5a (tomosyn) |
| chr4_+_12612145 | 0.48 |
ENSDART00000181201
|
lmo3
|
LIM domain only 3 |
| chr15_-_33925851 | 0.48 |
ENSDART00000187807
ENSDART00000187780 |
mag
|
myelin associated glycoprotein |
| chr19_-_30510259 | 0.48 |
ENSDART00000135128
ENSDART00000186169 ENSDART00000182974 ENSDART00000187797 |
bag6l
|
BCL2 associated athanogene 6, like |
| chr8_-_1838315 | 0.46 |
ENSDART00000114476
ENSDART00000140077 |
pi4kab
|
phosphatidylinositol 4-kinase, catalytic, alpha b |
| chr22_+_5176255 | 0.46 |
ENSDART00000092647
|
cers1
|
ceramide synthase 1 |
| chr15_-_22074315 | 0.46 |
ENSDART00000149830
|
drd2a
|
dopamine receptor D2a |
| chr9_+_52492639 | 0.46 |
ENSDART00000078939
|
march4
|
membrane-associated ring finger (C3HC4) 4 |
| chr5_-_12219572 | 0.46 |
ENSDART00000167834
|
nos1
|
nitric oxide synthase 1 (neuronal) |
| chr3_-_60827402 | 0.45 |
ENSDART00000053494
|
anks4b
|
ankyrin repeat and sterile alpha motif domain containing 4B |
| chr3_+_45364849 | 0.45 |
ENSDART00000153974
|
ube2ia
|
ubiquitin-conjugating enzyme E2Ia |
| chr25_+_7784582 | 0.45 |
ENSDART00000155016
|
dgkzb
|
diacylglycerol kinase, zeta b |
| chr16_-_29458806 | 0.44 |
ENSDART00000047931
|
lingo4b
|
leucine rich repeat and Ig domain containing 4b |
| chr8_+_18624658 | 0.44 |
ENSDART00000089141
|
fsd1
|
fibronectin type III and SPRY domain containing 1 |
| chr3_-_61099004 | 0.43 |
ENSDART00000112043
|
cacng4b
|
calcium channel, voltage-dependent, gamma subunit 4b |
| chr21_-_42100471 | 0.43 |
ENSDART00000166148
|
gabra1
|
gamma-aminobutyric acid (GABA) A receptor, alpha 1 |
| chr8_-_19649617 | 0.43 |
ENSDART00000189033
|
fam78bb
|
family with sequence similarity 78, member B b |
| chr15_-_35252522 | 0.43 |
ENSDART00000144153
ENSDART00000059195 |
mff
|
mitochondrial fission factor |
| chr20_+_19238382 | 0.42 |
ENSDART00000136757
|
fndc4a
|
fibronectin type III domain containing 4a |
| chr3_+_32443395 | 0.42 |
ENSDART00000188447
|
prr12b
|
proline rich 12b |
| chr24_-_1341543 | 0.42 |
ENSDART00000169341
|
nrp1a
|
neuropilin 1a |
| chr10_+_3049636 | 0.42 |
ENSDART00000081794
ENSDART00000183167 ENSDART00000191634 ENSDART00000183514 |
rasgrf2a
|
Ras protein-specific guanine nucleotide-releasing factor 2a |
| chr3_+_33341640 | 0.42 |
ENSDART00000186352
|
pyya
|
peptide YYa |
| chr16_+_35661331 | 0.41 |
ENSDART00000161725
|
map7d1a
|
MAP7 domain containing 1a |
| chr20_-_9436521 | 0.41 |
ENSDART00000133000
|
zgc:101840
|
zgc:101840 |
| chr7_-_24699985 | 0.41 |
ENSDART00000052802
|
calb2b
|
calbindin 2b |
| chr20_+_27020201 | 0.41 |
ENSDART00000126919
ENSDART00000016014 |
chga
|
chromogranin A |
| chr18_-_1185772 | 0.41 |
ENSDART00000143245
|
nptnb
|
neuroplastin b |
| chr1_+_7517454 | 0.40 |
ENSDART00000016139
|
lancl1
|
LanC antibiotic synthetase component C-like 1 (bacterial) |
| chr10_+_42690374 | 0.40 |
ENSDART00000123496
|
rhobtb2b
|
Rho-related BTB domain containing 2b |
| chr3_+_56366395 | 0.40 |
ENSDART00000154367
|
cacng5b
|
calcium channel, voltage-dependent, gamma subunit 5b |
| chr20_+_34933183 | 0.40 |
ENSDART00000062738
|
snap25a
|
synaptosomal-associated protein, 25a |
| chr20_+_26349002 | 0.40 |
ENSDART00000152842
|
syne1a
|
spectrin repeat containing, nuclear envelope 1a |
| chr3_+_13842554 | 0.40 |
ENSDART00000162317
ENSDART00000158068 |
ilf3b
|
interleukin enhancer binding factor 3b |
| chr13_+_15838151 | 0.39 |
ENSDART00000008987
|
klc1a
|
kinesin light chain 1a |
| chr18_+_1154189 | 0.39 |
ENSDART00000135090
|
si:ch1073-75f15.2
|
si:ch1073-75f15.2 |
| chr6_-_40352215 | 0.39 |
ENSDART00000103992
|
ttll3
|
tubulin tyrosine ligase-like family, member 3 |
| chr10_+_42423318 | 0.39 |
ENSDART00000134282
|
npy8ar
|
neuropeptide Y receptor Y8a |
| chr16_+_20161805 | 0.38 |
ENSDART00000192146
|
c16h2orf66
|
chromosome 16 C2orf66 homolog |
| chr11_+_30057762 | 0.38 |
ENSDART00000164139
|
nhsb
|
Nance-Horan syndrome b (congenital cataracts and dental anomalies) |
| chr8_+_29986265 | 0.38 |
ENSDART00000148258
|
ptch1
|
patched 1 |
| chr4_+_4849789 | 0.38 |
ENSDART00000130818
ENSDART00000127751 |
ptprz1b
|
protein tyrosine phosphatase, receptor-type, Z polypeptide 1b |
| chr10_+_15255012 | 0.37 |
ENSDART00000023766
|
vldlr
|
very low density lipoprotein receptor |
| chr14_-_34044369 | 0.37 |
ENSDART00000149396
ENSDART00000123607 ENSDART00000190746 |
cyfip2
|
cytoplasmic FMR1 interacting protein 2 |
| chr2_+_366429 | 0.37 |
ENSDART00000111746
|
si:dkey-33c14.6
|
si:dkey-33c14.6 |
| chr25_-_19224298 | 0.37 |
ENSDART00000149917
|
acanb
|
aggrecan b |
| chr23_-_35790235 | 0.37 |
ENSDART00000142369
ENSDART00000141141 ENSDART00000011004 |
mfsd5
|
major facilitator superfamily domain containing 5 |
| chr18_+_38908903 | 0.37 |
ENSDART00000159834
|
myo5aa
|
myosin VAa |
| chr8_+_52503340 | 0.36 |
ENSDART00000168838
|
si:ch1073-392o20.1
|
si:ch1073-392o20.1 |
| chr3_-_23406964 | 0.36 |
ENSDART00000114723
|
rapgefl1
|
Rap guanine nucleotide exchange factor (GEF)-like 1 |
| chr4_+_12612723 | 0.36 |
ENSDART00000133767
|
lmo3
|
LIM domain only 3 |
| chr23_-_14990865 | 0.36 |
ENSDART00000147799
|
ndrg3b
|
ndrg family member 3b |
| chr17_-_28198099 | 0.36 |
ENSDART00000156143
|
htr1d
|
5-hydroxytryptamine (serotonin) receptor 1D, G protein-coupled |
| chr24_-_5786759 | 0.36 |
ENSDART00000152069
|
chst2b
|
carbohydrate (N-acetylglucosamine-6-O) sulfotransferase 2b |
| chr24_-_11057305 | 0.36 |
ENSDART00000186494
|
asap1b
|
ArfGAP with SH3 domain, ankyrin repeat and PH domain 1b |
| chr20_+_52442870 | 0.36 |
ENSDART00000163100
|
arhgap39
|
Rho GTPase activating protein 39 |
| chr24_+_10310577 | 0.36 |
ENSDART00000141718
|
otulina
|
OTU deubiquitinase with linear linkage specificity a |
| chr5_-_62925282 | 0.36 |
ENSDART00000188048
|
specc1
|
sperm antigen with calponin homology and coiled-coil domains 1 |
| chr1_-_44701313 | 0.35 |
ENSDART00000193926
|
si:dkey-28b4.8
|
si:dkey-28b4.8 |
| chr25_+_16116740 | 0.35 |
ENSDART00000139778
|
far1
|
fatty acyl CoA reductase 1 |
| chr20_-_5052786 | 0.35 |
ENSDART00000138818
ENSDART00000181655 ENSDART00000164274 |
arid1b
|
AT rich interactive domain 1B (SWI1-like) |
| chr3_-_61065055 | 0.35 |
ENSDART00000074199
|
cacng1b
|
calcium channel, voltage-dependent, gamma subunit 1b |
| chr8_+_49778756 | 0.35 |
ENSDART00000083790
|
ntrk2a
|
neurotrophic tyrosine kinase, receptor, type 2a |
| chr20_+_27087539 | 0.35 |
ENSDART00000062094
|
tmem251
|
transmembrane protein 251 |
| chr8_-_49908978 | 0.35 |
ENSDART00000172642
|
agtpbp1
|
ATP/GTP binding protein 1 |
| chr18_+_10884996 | 0.34 |
ENSDART00000147613
|
mical3a
|
microtubule associated monooxygenase, calponin and LIM domain containing 3a |
| chr3_-_46811611 | 0.34 |
ENSDART00000134092
|
elavl3
|
ELAV like neuron-specific RNA binding protein 3 |
| chr1_+_33668236 | 0.34 |
ENSDART00000122316
ENSDART00000102184 |
arl13b
|
ADP-ribosylation factor-like 13b |
| chr8_+_22930627 | 0.34 |
ENSDART00000187860
|
sypa
|
synaptophysin a |
| chr20_+_35382482 | 0.34 |
ENSDART00000135284
|
vsnl1a
|
visinin-like 1a |
| chr20_-_53321499 | 0.33 |
ENSDART00000179894
ENSDART00000127427 ENSDART00000084952 |
wasf1
|
WAS protein family, member 1 |
| chr20_+_53181017 | 0.33 |
ENSDART00000189692
ENSDART00000177109 |
FIG4
|
FIG4 phosphoinositide 5-phosphatase |
| chr25_+_18889332 | 0.33 |
ENSDART00000017043
|
cax2
|
cation/H+ exchanger protein 2 |
| chr20_-_47188966 | 0.33 |
ENSDART00000152965
|
si:dkeyp-104f11.9
|
si:dkeyp-104f11.9 |
| chr20_-_45060241 | 0.33 |
ENSDART00000185227
|
klhl29
|
kelch-like family member 29 |
| chr2_-_31302615 | 0.33 |
ENSDART00000034784
ENSDART00000060812 |
adcyap1b
|
adenylate cyclase activating polypeptide 1b |
| chr20_-_32110882 | 0.33 |
ENSDART00000030324
|
grm1a
|
glutamate receptor, metabotropic 1a |
| chr1_-_23157583 | 0.33 |
ENSDART00000144208
|
adgrl3.1
|
adhesion G protein-coupled receptor L3.1 |
| chr18_+_24921587 | 0.32 |
ENSDART00000191345
|
rgma
|
repulsive guidance molecule family member a |
| chr18_-_2433011 | 0.32 |
ENSDART00000181922
ENSDART00000193276 |
CR769778.1
|
|
| chr17_+_23298928 | 0.32 |
ENSDART00000153652
|
zgc:165461
|
zgc:165461 |
| chr17_-_21162821 | 0.32 |
ENSDART00000157283
|
abhd12
|
abhydrolase domain containing 12 |
| chr14_-_30587814 | 0.32 |
ENSDART00000144912
ENSDART00000149714 |
tmem265
|
transmembrane protein 265 |
| chr19_-_20430892 | 0.32 |
ENSDART00000111409
|
tbc1d5
|
TBC1 domain family, member 5 |
Network of associatons between targets according to the STRING database.
First level regulatory network of en1b_en2a+en2b_gbx1_emx1
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.5 | 4.1 | GO:0061075 | positive regulation of neural retina development(GO:0061075) positive regulation of retina development in camera-type eye(GO:1902868) |
| 0.3 | 1.4 | GO:0071926 | endocannabinoid signaling pathway(GO:0071926) retrograde trans-synaptic signaling by lipid(GO:0098920) retrograde trans-synaptic signaling by endocannabinoid(GO:0098921) |
| 0.3 | 1.6 | GO:0002159 | desmosome assembly(GO:0002159) |
| 0.3 | 0.9 | GO:0099538 | synaptic signaling via neuropeptide(GO:0099538) trans-synaptic signaling by neuropeptide(GO:0099540) trans-synaptic signaling by neuropeptide, modulating synaptic transmission(GO:0099551) cellular response to brain-derived neurotrophic factor stimulus(GO:1990416) |
| 0.3 | 1.1 | GO:0015677 | copper ion import(GO:0015677) |
| 0.2 | 1.0 | GO:0038003 | opioid receptor signaling pathway(GO:0038003) |
| 0.2 | 0.7 | GO:0036306 | embryonic heart tube elongation(GO:0036306) |
| 0.2 | 0.7 | GO:0045938 | positive regulation of circadian sleep/wake cycle, sleep(GO:0045938) |
| 0.2 | 0.8 | GO:0016322 | neuron remodeling(GO:0016322) |
| 0.2 | 0.5 | GO:0098917 | retrograde trans-synaptic signaling(GO:0098917) |
| 0.2 | 1.5 | GO:1904071 | presynaptic active zone assembly(GO:1904071) |
| 0.1 | 0.4 | GO:0006089 | lactate metabolic process(GO:0006089) |
| 0.1 | 0.4 | GO:0021519 | optic cup formation involved in camera-type eye development(GO:0003408) spinal cord association neuron specification(GO:0021519) |
| 0.1 | 1.8 | GO:0001574 | ganglioside biosynthetic process(GO:0001574) |
| 0.1 | 0.4 | GO:0002792 | negative regulation of peptide secretion(GO:0002792) negative regulation of peptide hormone secretion(GO:0090278) |
| 0.1 | 0.6 | GO:0060049 | regulation of protein glycosylation(GO:0060049) |
| 0.1 | 0.5 | GO:0060074 | synapse maturation(GO:0060074) |
| 0.1 | 0.6 | GO:0042772 | DNA damage response, signal transduction by p53 class mediator resulting in transcription of p21 class mediator(GO:0006978) DNA damage response, signal transduction resulting in transcription(GO:0042772) |
| 0.1 | 0.1 | GO:0001659 | temperature homeostasis(GO:0001659) |
| 0.1 | 0.3 | GO:0097500 | receptor localization to nonmotile primary cilium(GO:0097500) protein localization to ciliary membrane(GO:1903441) |
| 0.1 | 0.4 | GO:1900271 | regulation of presynaptic cytosolic calcium ion concentration(GO:0099509) regulation of long-term synaptic potentiation(GO:1900271) |
| 0.1 | 0.5 | GO:0060055 | angiogenesis involved in wound healing(GO:0060055) |
| 0.1 | 0.3 | GO:1900620 | acetylcholine biosynthetic process(GO:0008292) acetate ester biosynthetic process(GO:1900620) |
| 0.1 | 0.7 | GO:2001286 | caveolin-mediated endocytosis(GO:0072584) negative regulation of clathrin-mediated endocytosis(GO:1900186) regulation of caveolin-mediated endocytosis(GO:2001286) negative regulation of caveolin-mediated endocytosis(GO:2001287) |
| 0.1 | 0.8 | GO:0006465 | signal peptide processing(GO:0006465) |
| 0.1 | 0.3 | GO:0035610 | protein side chain deglutamylation(GO:0035610) |
| 0.1 | 1.3 | GO:0051127 | positive regulation of actin nucleation(GO:0051127) positive regulation of Arp2/3 complex-mediated actin nucleation(GO:2000601) |
| 0.1 | 0.4 | GO:0070208 | protein heterotrimerization(GO:0070208) |
| 0.1 | 0.3 | GO:0045579 | positive regulation of B cell differentiation(GO:0045579) |
| 0.1 | 1.2 | GO:0033173 | calcineurin-NFAT signaling cascade(GO:0033173) |
| 0.1 | 1.2 | GO:0045162 | clustering of voltage-gated sodium channels(GO:0045162) |
| 0.1 | 0.2 | GO:0007414 | axonal defasciculation(GO:0007414) |
| 0.1 | 1.5 | GO:0071436 | cellular potassium ion homeostasis(GO:0030007) sodium ion export from cell(GO:0036376) sodium ion export(GO:0071436) |
| 0.1 | 0.2 | GO:0090219 | negative regulation of phosphatidylinositol 3-kinase activity(GO:0043553) negative regulation of lipid kinase activity(GO:0090219) |
| 0.1 | 0.9 | GO:0006646 | phosphatidylethanolamine biosynthetic process(GO:0006646) |
| 0.1 | 0.5 | GO:0045604 | regulation of epidermal cell differentiation(GO:0045604) |
| 0.1 | 0.4 | GO:1902514 | calcium ion transmembrane transport via high voltage-gated calcium channel(GO:0061577) regulation of calcium ion transmembrane transport via high voltage-gated calcium channel(GO:1902514) |
| 0.1 | 0.5 | GO:0051013 | microtubule severing(GO:0051013) |
| 0.1 | 0.2 | GO:0097378 | interneuron axon guidance(GO:0097376) spinal cord interneuron axon guidance(GO:0097377) dorsal spinal cord interneuron axon guidance(GO:0097378) |
| 0.1 | 0.3 | GO:0046166 | methylglyoxal biosynthetic process(GO:0019242) glyceraldehyde-3-phosphate biosynthetic process(GO:0046166) |
| 0.1 | 1.2 | GO:0036444 | calcium ion transmembrane import into mitochondrion(GO:0036444) |
| 0.1 | 0.3 | GO:0031293 | membrane protein intracellular domain proteolysis(GO:0031293) |
| 0.1 | 0.3 | GO:0046462 | phosphatidylserine catabolic process(GO:0006660) monoacylglycerol metabolic process(GO:0046462) monoacylglycerol catabolic process(GO:0052651) |
| 0.1 | 0.6 | GO:0071816 | tail-anchored membrane protein insertion into ER membrane(GO:0071816) |
| 0.1 | 0.3 | GO:1901387 | positive regulation of voltage-gated calcium channel activity(GO:1901387) |
| 0.1 | 1.2 | GO:0050796 | regulation of insulin secretion(GO:0050796) |
| 0.1 | 0.2 | GO:0021742 | abducens nucleus development(GO:0021742) |
| 0.1 | 0.2 | GO:0039015 | spinal cord anterior/posterior patterning(GO:0021512) cell proliferation involved in pronephros development(GO:0039015) cell proliferation involved in kidney development(GO:0072111) |
| 0.1 | 0.4 | GO:1990108 | protein linear deubiquitination(GO:1990108) |
| 0.1 | 0.4 | GO:0090243 | fibroblast growth factor receptor signaling pathway involved in somitogenesis(GO:0090243) |
| 0.1 | 0.3 | GO:0035627 | ceramide transport(GO:0035627) |
| 0.1 | 0.2 | GO:0043152 | induction of bacterial agglutination(GO:0043152) |
| 0.1 | 0.2 | GO:0046069 | cGMP catabolic process(GO:0046069) |
| 0.1 | 0.5 | GO:0031498 | nucleosome disassembly(GO:0006337) chromatin disassembly(GO:0031498) protein-DNA complex disassembly(GO:0032986) |
| 0.1 | 0.8 | GO:0051481 | adenylate cyclase-inhibiting dopamine receptor signaling pathway(GO:0007195) negative regulation of cytosolic calcium ion concentration(GO:0051481) negative regulation of synaptic transmission, glutamatergic(GO:0051967) |
| 0.1 | 0.3 | GO:0015744 | thiosulfate transport(GO:0015709) oxaloacetate transport(GO:0015729) malate transport(GO:0015743) succinate transport(GO:0015744) succinate transmembrane transport(GO:0071422) malate transmembrane transport(GO:0071423) |
| 0.1 | 0.2 | GO:2000434 | regulation of protein neddylation(GO:2000434) |
| 0.0 | 0.2 | GO:0071543 | diphosphoinositol polyphosphate metabolic process(GO:0071543) diadenosine pentaphosphate metabolic process(GO:1901906) diadenosine pentaphosphate catabolic process(GO:1901907) diadenosine hexaphosphate metabolic process(GO:1901908) diadenosine hexaphosphate catabolic process(GO:1901909) adenosine 5'-(hexahydrogen pentaphosphate) metabolic process(GO:1901910) adenosine 5'-(hexahydrogen pentaphosphate) catabolic process(GO:1901911) |
| 0.0 | 0.8 | GO:0098943 | neurotransmitter receptor transport, postsynaptic endosome to lysosome(GO:0098943) |
| 0.0 | 0.2 | GO:1903251 | multi-ciliated epithelial cell differentiation(GO:1903251) |
| 0.0 | 0.9 | GO:0022010 | central nervous system myelination(GO:0022010) axon ensheathment in central nervous system(GO:0032291) |
| 0.0 | 1.1 | GO:0035338 | long-chain fatty-acyl-CoA biosynthetic process(GO:0035338) |
| 0.0 | 0.2 | GO:0034454 | microtubule anchoring at centrosome(GO:0034454) |
| 0.0 | 0.5 | GO:0001957 | intramembranous ossification(GO:0001957) direct ossification(GO:0036072) |
| 0.0 | 1.4 | GO:0048268 | clathrin coat assembly(GO:0048268) |
| 0.0 | 0.0 | GO:0035767 | endothelial cell chemotaxis(GO:0035767) |
| 0.0 | 0.6 | GO:0035336 | long-chain fatty-acyl-CoA metabolic process(GO:0035336) |
| 0.0 | 1.1 | GO:0035622 | intrahepatic bile duct development(GO:0035622) |
| 0.0 | 0.4 | GO:0050795 | regulation of behavior(GO:0050795) |
| 0.0 | 0.4 | GO:0006044 | N-acetylglucosamine metabolic process(GO:0006044) |
| 0.0 | 0.2 | GO:0007217 | tachykinin receptor signaling pathway(GO:0007217) |
| 0.0 | 0.7 | GO:0048791 | calcium ion-regulated exocytosis of neurotransmitter(GO:0048791) |
| 0.0 | 0.3 | GO:0033278 | cell proliferation in midbrain(GO:0033278) |
| 0.0 | 0.3 | GO:0031954 | positive regulation of protein autophosphorylation(GO:0031954) |
| 0.0 | 0.2 | GO:0048755 | branching morphogenesis of a nerve(GO:0048755) |
| 0.0 | 0.1 | GO:0051574 | positive regulation of histone H3-K9 methylation(GO:0051574) |
| 0.0 | 0.1 | GO:0014814 | regulation of synaptic growth at neuromuscular junction(GO:0008582) axon regeneration at neuromuscular junction(GO:0014814) positive regulation of synaptic growth at neuromuscular junction(GO:0045887) |
| 0.0 | 1.2 | GO:0046847 | filopodium assembly(GO:0046847) |
| 0.0 | 1.8 | GO:0046513 | ceramide biosynthetic process(GO:0046513) |
| 0.0 | 0.3 | GO:0006543 | glutamate biosynthetic process(GO:0006537) glutamine catabolic process(GO:0006543) |
| 0.0 | 0.2 | GO:2000582 | positive regulation of microtubule motor activity(GO:2000576) regulation of ATP-dependent microtubule motor activity, plus-end-directed(GO:2000580) positive regulation of ATP-dependent microtubule motor activity, plus-end-directed(GO:2000582) |
| 0.0 | 0.6 | GO:0032467 | positive regulation of cytokinesis(GO:0032467) |
| 0.0 | 0.5 | GO:0032418 | lysosome localization(GO:0032418) |
| 0.0 | 0.9 | GO:0032008 | positive regulation of TOR signaling(GO:0032008) |
| 0.0 | 0.2 | GO:0090104 | pancreatic D cell differentiation(GO:0003311) pancreatic epsilon cell differentiation(GO:0090104) |
| 0.0 | 0.2 | GO:0018214 | peptidyl-glutamic acid carboxylation(GO:0017187) protein carboxylation(GO:0018214) |
| 0.0 | 2.2 | GO:1902600 | hydrogen ion transmembrane transport(GO:1902600) |
| 0.0 | 0.7 | GO:0007205 | protein kinase C-activating G-protein coupled receptor signaling pathway(GO:0007205) |
| 0.0 | 0.1 | GO:1903003 | positive regulation of protein deubiquitination(GO:1903003) |
| 0.0 | 0.1 | GO:0021960 | anterior commissure morphogenesis(GO:0021960) |
| 0.0 | 0.3 | GO:0006054 | N-acetylneuraminate metabolic process(GO:0006054) |
| 0.0 | 0.3 | GO:0071300 | cellular response to retinoic acid(GO:0071300) |
| 0.0 | 0.4 | GO:0060325 | face morphogenesis(GO:0060325) |
| 0.0 | 0.2 | GO:0000395 | mRNA 5'-splice site recognition(GO:0000395) |
| 0.0 | 0.1 | GO:0000012 | single strand break repair(GO:0000012) |
| 0.0 | 0.1 | GO:0046951 | ketone body biosynthetic process(GO:0046951) |
| 0.0 | 0.1 | GO:0042832 | response to protozoan(GO:0001562) defense response to protozoan(GO:0042832) |
| 0.0 | 0.5 | GO:0050708 | regulation of protein secretion(GO:0050708) |
| 0.0 | 0.1 | GO:0045669 | positive regulation of osteoblast differentiation(GO:0045669) |
| 0.0 | 0.4 | GO:0016082 | synaptic vesicle priming(GO:0016082) |
| 0.0 | 0.1 | GO:0035124 | embryonic caudal fin morphogenesis(GO:0035124) |
| 0.0 | 0.2 | GO:0031116 | positive regulation of microtubule polymerization(GO:0031116) |
| 0.0 | 0.6 | GO:0045773 | positive regulation of axon extension(GO:0045773) |
| 0.0 | 0.2 | GO:0050820 | positive regulation of blood coagulation(GO:0030194) positive regulation of coagulation(GO:0050820) positive regulation of hemostasis(GO:1900048) |
| 0.0 | 0.3 | GO:0045842 | positive regulation of mitotic metaphase/anaphase transition(GO:0045842) positive regulation of mitotic sister chromatid separation(GO:1901970) positive regulation of metaphase/anaphase transition of cell cycle(GO:1902101) |
| 0.0 | 0.1 | GO:0031061 | negative regulation of histone methylation(GO:0031061) |
| 0.0 | 0.5 | GO:1990035 | calcium ion import across plasma membrane(GO:0098703) calcium ion import into cell(GO:1990035) |
| 0.0 | 1.4 | GO:1902668 | negative regulation of axon extension involved in axon guidance(GO:0048843) negative regulation of axon guidance(GO:1902668) |
| 0.0 | 0.2 | GO:0006283 | transcription-coupled nucleotide-excision repair(GO:0006283) |
| 0.0 | 0.3 | GO:0001964 | startle response(GO:0001964) |
| 0.0 | 0.1 | GO:0090148 | membrane fission(GO:0090148) |
| 0.0 | 0.1 | GO:0017006 | protein-heme linkage(GO:0017003) protein-tetrapyrrole linkage(GO:0017006) cytochrome c-heme linkage(GO:0018063) |
| 0.0 | 0.2 | GO:0071467 | cellular response to pH(GO:0071467) |
| 0.0 | 0.3 | GO:0035493 | SNARE complex assembly(GO:0035493) |
| 0.0 | 0.7 | GO:1903322 | positive regulation of protein modification by small protein conjugation or removal(GO:1903322) |
| 0.0 | 0.4 | GO:0010507 | negative regulation of autophagy(GO:0010507) |
| 0.0 | 0.9 | GO:1901800 | positive regulation of proteasomal ubiquitin-dependent protein catabolic process(GO:0032436) positive regulation of proteasomal protein catabolic process(GO:1901800) |
| 0.0 | 0.2 | GO:0043328 | protein targeting to vacuole involved in ubiquitin-dependent protein catabolic process via the multivesicular body sorting pathway(GO:0043328) |
| 0.0 | 0.3 | GO:0048923 | posterior lateral line neuromast hair cell differentiation(GO:0048923) |
| 0.0 | 0.2 | GO:2000650 | negative regulation of sodium ion transmembrane transport(GO:1902306) negative regulation of sodium ion transmembrane transporter activity(GO:2000650) |
| 0.0 | 0.1 | GO:0035994 | response to muscle stretch(GO:0035994) |
| 0.0 | 0.1 | GO:0097476 | spinal cord motor neuron migration(GO:0097476) lateral motor column neuron migration(GO:0097477) |
| 0.0 | 1.5 | GO:0035249 | synaptic transmission, glutamatergic(GO:0035249) |
| 0.0 | 0.5 | GO:0007631 | feeding behavior(GO:0007631) |
| 0.0 | 0.1 | GO:1900182 | positive regulation of protein localization to nucleus(GO:1900182) regulation of protein localization to Cajal body(GO:1904869) positive regulation of protein localization to Cajal body(GO:1904871) |
| 0.0 | 0.2 | GO:1902038 | positive regulation of hematopoietic stem cell differentiation(GO:1902038) |
| 0.0 | 0.7 | GO:0036269 | swimming behavior(GO:0036269) |
| 0.0 | 0.2 | GO:0098900 | regulation of action potential(GO:0098900) |
| 0.0 | 0.2 | GO:0003428 | growth plate cartilage morphogenesis(GO:0003422) chondrocyte intercalation involved in growth plate cartilage morphogenesis(GO:0003428) |
| 0.0 | 0.0 | GO:0042364 | water-soluble vitamin biosynthetic process(GO:0042364) |
| 0.0 | 0.2 | GO:0038007 | netrin-activated signaling pathway(GO:0038007) |
| 0.0 | 0.1 | GO:0072592 | oxygen metabolic process(GO:0072592) regulation of oxygen metabolic process(GO:2000374) positive regulation of oxygen metabolic process(GO:2000376) |
| 0.0 | 1.6 | GO:0010506 | regulation of autophagy(GO:0010506) |
| 0.0 | 0.1 | GO:0097032 | respiratory chain complex II assembly(GO:0034552) mitochondrial respiratory chain complex II assembly(GO:0034553) mitochondrial respiratory chain complex II biogenesis(GO:0097032) |
| 0.0 | 0.6 | GO:0006904 | vesicle docking involved in exocytosis(GO:0006904) |
| 0.0 | 0.1 | GO:1901825 | tetraterpenoid metabolic process(GO:0016108) tetraterpenoid biosynthetic process(GO:0016109) carotenoid metabolic process(GO:0016116) carotenoid biosynthetic process(GO:0016117) xanthophyll metabolic process(GO:0016122) xanthophyll biosynthetic process(GO:0016123) zeaxanthin metabolic process(GO:1901825) zeaxanthin biosynthetic process(GO:1901827) |
| 0.0 | 0.1 | GO:0070973 | protein localization to endoplasmic reticulum exit site(GO:0070973) |
| 0.0 | 0.1 | GO:0006703 | estrogen biosynthetic process(GO:0006703) |
| 0.0 | 0.2 | GO:0030073 | insulin secretion(GO:0030073) |
| 0.0 | 0.1 | GO:0006895 | Golgi to endosome transport(GO:0006895) |
| 0.0 | 0.2 | GO:1903828 | negative regulation of cellular protein localization(GO:1903828) |
| 0.0 | 0.1 | GO:0044528 | regulation of mitochondrial mRNA stability(GO:0044528) |
| 0.0 | 0.8 | GO:0048488 | synaptic vesicle endocytosis(GO:0048488) |
| 0.0 | 0.2 | GO:0021702 | cerebellar Purkinje cell layer formation(GO:0021694) cerebellar Purkinje cell differentiation(GO:0021702) |
| 0.0 | 0.3 | GO:0048026 | positive regulation of mRNA splicing, via spliceosome(GO:0048026) |
| 0.0 | 0.1 | GO:0021877 | forebrain neuron fate commitment(GO:0021877) |
| 0.0 | 0.1 | GO:0061088 | sequestering of zinc ion(GO:0032119) regulation of sequestering of zinc ion(GO:0061088) |
| 0.0 | 0.1 | GO:0071276 | cellular response to cadmium ion(GO:0071276) |
| 0.0 | 0.0 | GO:0030043 | actin filament fragmentation(GO:0030043) |
| 0.0 | 0.2 | GO:0045471 | response to ethanol(GO:0045471) |
| 0.0 | 0.5 | GO:0006506 | GPI anchor biosynthetic process(GO:0006506) |
| 0.0 | 0.0 | GO:0051660 | establishment of centrosome localization(GO:0051660) |
| 0.0 | 0.1 | GO:0006370 | 7-methylguanosine mRNA capping(GO:0006370) |
| 0.0 | 0.6 | GO:0046474 | glycerophospholipid biosynthetic process(GO:0046474) |
| 0.0 | 0.0 | GO:0061182 | negative regulation of chondrocyte differentiation(GO:0032331) negative regulation of cartilage development(GO:0061037) negative regulation of chondrocyte development(GO:0061182) regulation of bone mineralization involved in bone maturation(GO:1900157) negative regulation of bone mineralization involved in bone maturation(GO:1900158) negative regulation of bone development(GO:1903011) |
| 0.0 | 1.2 | GO:0007218 | neuropeptide signaling pathway(GO:0007218) |
| 0.0 | 0.6 | GO:0048384 | retinoic acid receptor signaling pathway(GO:0048384) |
| 0.0 | 0.2 | GO:0034453 | microtubule anchoring(GO:0034453) |
| 0.0 | 0.1 | GO:0048312 | intracellular distribution of mitochondria(GO:0048312) |
| 0.0 | 1.1 | GO:0019722 | calcium-mediated signaling(GO:0019722) |
| 0.0 | 0.1 | GO:0045670 | regulation of osteoclast differentiation(GO:0045670) |
| 0.0 | 0.5 | GO:0007586 | digestion(GO:0007586) |
| 0.0 | 0.0 | GO:0060898 | eye field cell fate commitment involved in camera-type eye formation(GO:0060898) |
| 0.0 | 0.1 | GO:0036372 | opsin transport(GO:0036372) |
| 0.0 | 0.1 | GO:0060114 | vestibular receptor cell differentiation(GO:0060114) vestibular receptor cell development(GO:0060118) |
| 0.0 | 0.2 | GO:0099633 | protein localization to postsynaptic specialization membrane(GO:0099633) neurotransmitter receptor localization to postsynaptic specialization membrane(GO:0099645) |
| 0.0 | 0.0 | GO:0006145 | purine nucleobase catabolic process(GO:0006145) |
| 0.0 | 0.3 | GO:0007379 | somite specification(GO:0001757) segment specification(GO:0007379) |
| 0.0 | 0.9 | GO:0048015 | phosphatidylinositol-mediated signaling(GO:0048015) |
| 0.0 | 0.2 | GO:0072583 | clathrin-mediated endocytosis(GO:0072583) |
| 0.0 | 0.0 | GO:0033540 | fatty acid beta-oxidation using acyl-CoA oxidase(GO:0033540) |
| 0.0 | 1.9 | GO:0070588 | calcium ion transmembrane transport(GO:0070588) |
| 0.0 | 0.3 | GO:0035305 | negative regulation of dephosphorylation(GO:0035305) negative regulation of protein dephosphorylation(GO:0035308) |
| 0.0 | 0.7 | GO:0046839 | phospholipid dephosphorylation(GO:0046839) |
| 0.0 | 0.1 | GO:0035721 | intraciliary retrograde transport(GO:0035721) |
| 0.0 | 0.1 | GO:0010043 | response to zinc ion(GO:0010043) |
| 0.0 | 0.1 | GO:2000096 | positive regulation of Wnt signaling pathway, planar cell polarity pathway(GO:2000096) |
| 0.0 | 0.2 | GO:0042759 | long-chain fatty acid biosynthetic process(GO:0042759) |
| 0.0 | 0.4 | GO:0018231 | peptidyl-L-cysteine S-palmitoylation(GO:0018230) peptidyl-S-diacylglycerol-L-cysteine biosynthetic process from peptidyl-cysteine(GO:0018231) |
| 0.0 | 0.5 | GO:0071391 | cellular response to estrogen stimulus(GO:0071391) |
| 0.0 | 0.1 | GO:0001881 | receptor recycling(GO:0001881) |
| 0.0 | 0.2 | GO:0015936 | coenzyme A metabolic process(GO:0015936) |
| 0.0 | 0.1 | GO:0021576 | hindbrain formation(GO:0021576) |
| 0.0 | 0.1 | GO:0050908 | detection of light stimulus involved in visual perception(GO:0050908) detection of light stimulus involved in sensory perception(GO:0050962) |
| 0.0 | 0.1 | GO:0055017 | cardiac muscle tissue growth(GO:0055017) cardiac muscle cell proliferation(GO:0060038) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 1.7 | GO:0033181 | plasma membrane proton-transporting V-type ATPase complex(GO:0033181) |
| 0.2 | 1.3 | GO:0070032 | synaptobrevin 2-SNAP-25-syntaxin-1a-complexin I complex(GO:0070032) |
| 0.2 | 1.2 | GO:0005955 | calcineurin complex(GO:0005955) |
| 0.2 | 0.8 | GO:0071556 | integral component of cytoplasmic side of endoplasmic reticulum membrane(GO:0071458) integral component of lumenal side of endoplasmic reticulum membrane(GO:0071556) lumenal side of endoplasmic reticulum membrane(GO:0098553) |
| 0.2 | 1.5 | GO:0005890 | sodium:potassium-exchanging ATPase complex(GO:0005890) cation-transporting ATPase complex(GO:0090533) |
| 0.2 | 1.5 | GO:0098982 | GABA-ergic synapse(GO:0098982) |
| 0.1 | 0.7 | GO:0042584 | chromaffin granule membrane(GO:0042584) |
| 0.1 | 0.8 | GO:0043083 | synaptic cleft(GO:0043083) |
| 0.1 | 1.3 | GO:0044295 | axonal growth cone(GO:0044295) |
| 0.1 | 1.7 | GO:0031209 | SCAR complex(GO:0031209) |
| 0.1 | 1.2 | GO:1990246 | uniplex complex(GO:1990246) |
| 0.1 | 0.6 | GO:0071818 | BAT3 complex(GO:0071818) |
| 0.1 | 1.4 | GO:0032591 | dendritic spine membrane(GO:0032591) |
| 0.1 | 0.4 | GO:0042583 | chromaffin granule(GO:0042583) |
| 0.1 | 1.0 | GO:0034361 | very-low-density lipoprotein particle(GO:0034361) |
| 0.1 | 0.7 | GO:0045171 | intercellular bridge(GO:0045171) |
| 0.1 | 0.4 | GO:0000506 | glycosylphosphatidylinositol-N-acetylglucosaminyltransferase (GPI-GnT) complex(GO:0000506) |
| 0.1 | 1.2 | GO:0030057 | desmosome(GO:0030057) |
| 0.1 | 0.2 | GO:0097189 | apoptotic body(GO:0097189) |
| 0.1 | 1.0 | GO:0030667 | secretory granule membrane(GO:0030667) |
| 0.1 | 0.4 | GO:1990454 | L-type voltage-gated calcium channel complex(GO:1990454) |
| 0.0 | 0.2 | GO:1990513 | CLOCK-BMAL transcription complex(GO:1990513) |
| 0.0 | 0.9 | GO:0043209 | myelin sheath(GO:0043209) |
| 0.0 | 0.4 | GO:0035060 | brahma complex(GO:0035060) |
| 0.0 | 0.2 | GO:0005854 | nascent polypeptide-associated complex(GO:0005854) |
| 0.0 | 0.5 | GO:0033180 | proton-transporting V-type ATPase, V1 domain(GO:0033180) |
| 0.0 | 0.2 | GO:0016533 | cyclin-dependent protein kinase 5 holoenzyme complex(GO:0016533) |
| 0.0 | 1.0 | GO:0035371 | microtubule plus-end(GO:0035371) |
| 0.0 | 0.3 | GO:0071797 | LUBAC complex(GO:0071797) |
| 0.0 | 2.4 | GO:0005891 | voltage-gated calcium channel complex(GO:0005891) |
| 0.0 | 0.3 | GO:0016586 | RSC complex(GO:0016586) |
| 0.0 | 2.0 | GO:0005905 | clathrin-coated pit(GO:0005905) |
| 0.0 | 0.4 | GO:0071564 | npBAF complex(GO:0071564) |
| 0.0 | 0.3 | GO:0030670 | phagocytic vesicle membrane(GO:0030670) |
| 0.0 | 0.1 | GO:0005828 | kinetochore microtubule(GO:0005828) |
| 0.0 | 0.7 | GO:0005881 | cytoplasmic microtubule(GO:0005881) |
| 0.0 | 0.5 | GO:0005640 | nuclear outer membrane(GO:0005640) |
| 0.0 | 0.4 | GO:0005868 | cytoplasmic dynein complex(GO:0005868) |
| 0.0 | 0.9 | GO:0043679 | axon terminus(GO:0043679) |
| 0.0 | 0.5 | GO:0043204 | perikaryon(GO:0043204) |
| 0.0 | 0.3 | GO:0005964 | phosphorylase kinase complex(GO:0005964) |
| 0.0 | 3.9 | GO:0008021 | synaptic vesicle(GO:0008021) |
| 0.0 | 0.2 | GO:0044232 | organelle membrane contact site(GO:0044232) |
| 0.0 | 0.7 | GO:0098978 | glutamatergic synapse(GO:0098978) |
| 0.0 | 2.7 | GO:0030424 | axon(GO:0030424) |
| 0.0 | 0.5 | GO:0043025 | neuronal cell body(GO:0043025) |
| 0.0 | 0.1 | GO:0070724 | BMP receptor complex(GO:0070724) |
| 0.0 | 0.8 | GO:0008076 | voltage-gated potassium channel complex(GO:0008076) potassium channel complex(GO:0034705) |
| 0.0 | 0.2 | GO:0044224 | juxtaparanode region of axon(GO:0044224) |
| 0.0 | 0.7 | GO:1902710 | GABA receptor complex(GO:1902710) GABA-A receptor complex(GO:1902711) |
| 0.0 | 0.6 | GO:0032281 | AMPA glutamate receptor complex(GO:0032281) |
| 0.0 | 0.3 | GO:0046658 | anchored component of plasma membrane(GO:0046658) |
| 0.0 | 0.2 | GO:0032021 | NELF complex(GO:0032021) |
| 0.0 | 0.1 | GO:0005879 | axonemal microtubule(GO:0005879) |
| 0.0 | 0.8 | GO:0005778 | peroxisomal membrane(GO:0005778) microbody membrane(GO:0031903) |
| 0.0 | 0.1 | GO:0035517 | PR-DUB complex(GO:0035517) |
| 0.0 | 1.2 | GO:0030173 | integral component of Golgi membrane(GO:0030173) intrinsic component of Golgi membrane(GO:0031228) |
| 0.0 | 9.2 | GO:0045202 | synapse(GO:0045202) |
| 0.0 | 0.1 | GO:0032426 | stereocilia coupling link(GO:0002139) stereocilia ankle link(GO:0002141) stereocilia ankle link complex(GO:0002142) stereocilium tip(GO:0032426) |
| 0.0 | 0.1 | GO:0030991 | intraciliary transport particle A(GO:0030991) |
| 0.0 | 0.2 | GO:0002116 | semaphorin receptor complex(GO:0002116) |
| 0.0 | 0.2 | GO:0000813 | ESCRT I complex(GO:0000813) |
| 0.0 | 0.2 | GO:0060170 | ciliary membrane(GO:0060170) |
| 0.0 | 0.3 | GO:0005680 | anaphase-promoting complex(GO:0005680) |
| 0.0 | 0.1 | GO:0005943 | phosphatidylinositol 3-kinase complex, class IA(GO:0005943) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 1.0 | GO:0038046 | enkephalin receptor activity(GO:0038046) |
| 0.3 | 0.9 | GO:0048403 | brain-derived neurotrophic factor binding(GO:0048403) brain-derived neurotrophic factor-activated receptor activity(GO:0060175) |
| 0.3 | 1.1 | GO:0052851 | cupric reductase activity(GO:0008823) ferric-chelate reductase (NADPH) activity(GO:0052851) |
| 0.3 | 1.0 | GO:0030228 | lipoprotein particle receptor activity(GO:0030228) |
| 0.2 | 1.1 | GO:0031841 | neuropeptide Y receptor binding(GO:0031841) type 2 neuropeptide Y receptor binding(GO:0031843) |
| 0.2 | 1.2 | GO:0033192 | calcium-dependent protein serine/threonine phosphatase activity(GO:0004723) calmodulin-dependent protein phosphatase activity(GO:0033192) |
| 0.2 | 0.9 | GO:0004307 | diacylglycerol cholinephosphotransferase activity(GO:0004142) ethanolaminephosphotransferase activity(GO:0004307) |
| 0.2 | 1.6 | GO:0045294 | alpha-catenin binding(GO:0045294) |
| 0.2 | 0.7 | GO:0061656 | SUMO conjugating enzyme activity(GO:0061656) |
| 0.2 | 0.5 | GO:0070738 | protein-glycine ligase activity(GO:0070735) tubulin-glycine ligase activity(GO:0070738) |
| 0.2 | 0.8 | GO:0008046 | axon guidance receptor activity(GO:0008046) |
| 0.1 | 1.3 | GO:0071933 | Arp2/3 complex binding(GO:0071933) |
| 0.1 | 0.5 | GO:0016521 | pituitary adenylate cyclase activating polypeptide activity(GO:0016521) |
| 0.1 | 1.8 | GO:0050291 | sphingosine N-acyltransferase activity(GO:0050291) |
| 0.1 | 1.2 | GO:0004465 | lipoprotein lipase activity(GO:0004465) |
| 0.1 | 0.5 | GO:0050699 | WW domain binding(GO:0050699) |
| 0.1 | 1.1 | GO:0047676 | arachidonate-CoA ligase activity(GO:0047676) |
| 0.1 | 0.3 | GO:0035620 | ceramide transporter activity(GO:0035620) ceramide 1-phosphate binding(GO:1902387) ceramide 1-phosphate transporter activity(GO:1902388) |
| 0.1 | 0.8 | GO:0042500 | aspartic endopeptidase activity, intramembrane cleaving(GO:0042500) |
| 0.1 | 0.4 | GO:0097108 | hedgehog family protein binding(GO:0097108) |
| 0.1 | 2.0 | GO:0001540 | beta-amyloid binding(GO:0001540) |
| 0.1 | 1.5 | GO:0098882 | structural constituent of presynaptic active zone(GO:0098882) |
| 0.1 | 0.6 | GO:0080019 | fatty-acyl-CoA reductase (alcohol-forming) activity(GO:0080019) |
| 0.1 | 1.1 | GO:0004169 | dolichyl-phosphate-mannose-protein mannosyltransferase activity(GO:0004169) |
| 0.1 | 0.5 | GO:0016316 | phosphatidylinositol-3,4-bisphosphate 4-phosphatase activity(GO:0016316) |
| 0.1 | 1.4 | GO:0001671 | ATPase activator activity(GO:0001671) |
| 0.1 | 0.3 | GO:0008929 | triose-phosphate isomerase activity(GO:0004807) methylglyoxal synthase activity(GO:0008929) |
| 0.1 | 0.5 | GO:0004430 | 1-phosphatidylinositol 4-kinase activity(GO:0004430) |
| 0.1 | 0.5 | GO:0004517 | nitric-oxide synthase activity(GO:0004517) |
| 0.1 | 0.5 | GO:0004457 | lactate dehydrogenase activity(GO:0004457) |
| 0.1 | 0.9 | GO:0019911 | structural constituent of myelin sheath(GO:0019911) |
| 0.1 | 0.3 | GO:0043295 | glutathione binding(GO:0043295) |
| 0.1 | 0.3 | GO:0004689 | phosphorylase kinase activity(GO:0004689) |
| 0.1 | 0.4 | GO:0017176 | phosphatidylinositol N-acetylglucosaminyltransferase activity(GO:0017176) |
| 0.1 | 2.1 | GO:0017075 | syntaxin-1 binding(GO:0017075) |
| 0.1 | 0.8 | GO:0001591 | dopamine neurotransmitter receptor activity, coupled via Gi/Go(GO:0001591) |
| 0.1 | 0.3 | GO:0015140 | thiosulfate transmembrane transporter activity(GO:0015117) oxaloacetate transmembrane transporter activity(GO:0015131) malate transmembrane transporter activity(GO:0015140) |
| 0.1 | 0.7 | GO:0032051 | clathrin light chain binding(GO:0032051) |
| 0.0 | 0.2 | GO:0000298 | endopolyphosphatase activity(GO:0000298) diphosphoinositol-polyphosphate diphosphatase activity(GO:0008486) bis(5'-adenosyl)-hexaphosphatase activity(GO:0034431) bis(5'-adenosyl)-pentaphosphatase activity(GO:0034432) |
| 0.0 | 0.6 | GO:0004711 | ribosomal protein S6 kinase activity(GO:0004711) |
| 0.0 | 0.2 | GO:0008488 | gamma-glutamyl carboxylase activity(GO:0008488) |
| 0.0 | 1.6 | GO:0099529 | neurotransmitter receptor activity involved in regulation of postsynaptic membrane potential(GO:0099529) transmitter-gated ion channel activity involved in regulation of postsynaptic membrane potential(GO:1904315) |
| 0.0 | 0.4 | GO:0001517 | N-acetylglucosamine 6-O-sulfotransferase activity(GO:0001517) |
| 0.0 | 0.3 | GO:0099530 | G-protein coupled receptor activity involved in regulation of postsynaptic membrane potential(GO:0099530) |
| 0.0 | 0.1 | GO:0031716 | calcitonin receptor binding(GO:0031716) |
| 0.0 | 0.5 | GO:0005021 | vascular endothelial growth factor-activated receptor activity(GO:0005021) |
| 0.0 | 1.8 | GO:0008376 | acetylgalactosaminyltransferase activity(GO:0008376) |
| 0.0 | 1.3 | GO:0005184 | neuropeptide hormone activity(GO:0005184) |
| 0.0 | 0.2 | GO:0051139 | calcium:proton antiporter activity(GO:0015369) metal ion:proton antiporter activity(GO:0051139) |
| 0.0 | 1.3 | GO:0051010 | microtubule plus-end binding(GO:0051010) |
| 0.0 | 0.3 | GO:0005161 | platelet-derived growth factor receptor binding(GO:0005161) |
| 0.0 | 0.5 | GO:0005432 | calcium:sodium antiporter activity(GO:0005432) |
| 0.0 | 1.4 | GO:0005540 | hyaluronic acid binding(GO:0005540) |
| 0.0 | 0.1 | GO:1990226 | histone methyltransferase binding(GO:1990226) |
| 0.0 | 3.1 | GO:0005245 | voltage-gated calcium channel activity(GO:0005245) |
| 0.0 | 0.2 | GO:1990380 | Lys48-specific deubiquitinase activity(GO:1990380) |
| 0.0 | 0.5 | GO:0031628 | opioid receptor binding(GO:0031628) |
| 0.0 | 0.1 | GO:0019777 | Atg12 transferase activity(GO:0019777) |
| 0.0 | 0.2 | GO:0070888 | E-box binding(GO:0070888) |
| 0.0 | 0.1 | GO:0008440 | inositol-1,4,5-trisphosphate 3-kinase activity(GO:0008440) |
| 0.0 | 0.1 | GO:0080132 | fatty acid alpha-hydroxylase activity(GO:0080132) |
| 0.0 | 0.5 | GO:0070628 | proteasome binding(GO:0070628) |
| 0.0 | 0.1 | GO:0047035 | testosterone dehydrogenase (NAD+) activity(GO:0047035) |
| 0.0 | 0.2 | GO:0044736 | acid-sensing ion channel activity(GO:0044736) |
| 0.0 | 0.7 | GO:0004143 | diacylglycerol kinase activity(GO:0004143) |
| 0.0 | 1.2 | GO:0031624 | ubiquitin conjugating enzyme binding(GO:0031624) |
| 0.0 | 0.3 | GO:0004359 | glutaminase activity(GO:0004359) |
| 0.0 | 1.1 | GO:0019003 | GDP binding(GO:0019003) |
| 0.0 | 0.3 | GO:0004865 | protein serine/threonine phosphatase inhibitor activity(GO:0004865) |
| 0.0 | 0.6 | GO:0017017 | MAP kinase tyrosine/serine/threonine phosphatase activity(GO:0017017) MAP kinase phosphatase activity(GO:0033549) |
| 0.0 | 0.8 | GO:0004089 | carbonate dehydratase activity(GO:0004089) |
| 0.0 | 0.5 | GO:0000340 | RNA 7-methylguanosine cap binding(GO:0000340) |
| 0.0 | 0.5 | GO:0045159 | myosin II binding(GO:0045159) |
| 0.0 | 0.1 | GO:0004419 | hydroxymethylglutaryl-CoA lyase activity(GO:0004419) |
| 0.0 | 0.3 | GO:0070567 | cytidylyltransferase activity(GO:0070567) |
| 0.0 | 0.5 | GO:0033691 | sialic acid binding(GO:0033691) |
| 0.0 | 1.6 | GO:0030215 | semaphorin receptor binding(GO:0030215) |
| 0.0 | 0.6 | GO:0005246 | calcium channel regulator activity(GO:0005246) |
| 0.0 | 0.6 | GO:0051018 | protein kinase A binding(GO:0051018) |
| 0.0 | 0.1 | GO:0004061 | arylformamidase activity(GO:0004061) |
| 0.0 | 0.1 | GO:0022889 | serine transmembrane transporter activity(GO:0022889) |
| 0.0 | 0.3 | GO:0047372 | acylglycerol lipase activity(GO:0047372) |
| 0.0 | 0.4 | GO:0005388 | calcium-transporting ATPase activity(GO:0005388) |
| 0.0 | 0.1 | GO:0042975 | peroxisome proliferator activated receptor binding(GO:0042975) |
| 0.0 | 1.2 | GO:0044325 | ion channel binding(GO:0044325) |
| 0.0 | 0.1 | GO:0004408 | holocytochrome-c synthase activity(GO:0004408) |
| 0.0 | 0.2 | GO:0015057 | thrombin receptor activity(GO:0015057) |
| 0.0 | 0.7 | GO:0030165 | PDZ domain binding(GO:0030165) |
| 0.0 | 0.4 | GO:0004983 | neuropeptide Y receptor activity(GO:0004983) |
| 0.0 | 0.1 | GO:0016415 | octanoyltransferase activity(GO:0016415) |
| 0.0 | 0.2 | GO:0019871 | sodium channel inhibitor activity(GO:0019871) |
| 0.0 | 0.9 | GO:0001786 | phosphatidylserine binding(GO:0001786) |
| 0.0 | 0.3 | GO:0004724 | magnesium-dependent protein serine/threonine phosphatase activity(GO:0004724) |
| 0.0 | 0.2 | GO:0016286 | small conductance calcium-activated potassium channel activity(GO:0016286) |
| 0.0 | 0.2 | GO:0005042 | netrin receptor activity(GO:0005042) |
| 0.0 | 0.5 | GO:0042577 | lipid phosphatase activity(GO:0042577) |
| 0.0 | 0.1 | GO:0016422 | mRNA (2'-O-methyladenosine-N6-)-methyltransferase activity(GO:0016422) |
| 0.0 | 0.1 | GO:0032422 | purine-rich negative regulatory element binding(GO:0032422) |
| 0.0 | 0.4 | GO:0005154 | epidermal growth factor receptor binding(GO:0005154) |
| 0.0 | 0.1 | GO:0052885 | retinol isomerase activity(GO:0050251) all-trans-retinyl-ester hydrolase, 11-cis retinol forming activity(GO:0052885) |
| 0.0 | 0.1 | GO:0050220 | prostaglandin-E synthase activity(GO:0050220) |
| 0.0 | 0.3 | GO:0015026 | coreceptor activity(GO:0015026) |
| 0.0 | 0.1 | GO:0016863 | intramolecular oxidoreductase activity, transposing C=C bonds(GO:0016863) |
| 0.0 | 0.2 | GO:0061575 | cyclin-dependent protein serine/threonine kinase activator activity(GO:0061575) |
| 0.0 | 0.2 | GO:0035925 | AU-rich element binding(GO:0017091) mRNA 3'-UTR AU-rich region binding(GO:0035925) |
| 0.0 | 0.2 | GO:0004971 | AMPA glutamate receptor activity(GO:0004971) |
| 0.0 | 0.2 | GO:0016413 | O-acetyltransferase activity(GO:0016413) |
| 0.0 | 0.2 | GO:0004190 | aspartic-type endopeptidase activity(GO:0004190) aspartic-type peptidase activity(GO:0070001) |
| 0.0 | 0.2 | GO:0098748 | clathrin adaptor activity(GO:0035615) endocytic adaptor activity(GO:0098748) |
| 0.0 | 0.3 | GO:0043176 | amine binding(GO:0043176) serotonin binding(GO:0051378) |
| 0.0 | 0.5 | GO:0004435 | phosphatidylinositol phospholipase C activity(GO:0004435) phospholipase C activity(GO:0004629) |
| 0.0 | 0.0 | GO:0003960 | NADPH:quinone reductase activity(GO:0003960) |
| 0.0 | 0.2 | GO:0004890 | GABA-A receptor activity(GO:0004890) |
| 0.0 | 0.2 | GO:0003785 | actin monomer binding(GO:0003785) |
| 0.0 | 0.1 | GO:0035804 | structural constituent of egg coat(GO:0035804) |
| 0.0 | 0.1 | GO:0046934 | phosphatidylinositol-4,5-bisphosphate 3-kinase activity(GO:0046934) phosphatidylinositol-3,4-bisphosphate 5-kinase activity(GO:0052812) |
| 0.0 | 0.2 | GO:0017154 | semaphorin receptor activity(GO:0017154) |
| 0.0 | 0.0 | GO:0031005 | filamin binding(GO:0031005) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 1.4 | PID WNT SIGNALING PATHWAY | Wnt signaling network |
| 0.1 | 0.7 | PID TCR CALCIUM PATHWAY | Calcium signaling in the CD4+ TCR pathway |
| 0.0 | 1.0 | PID REELIN PATHWAY | Reelin signaling pathway |
| 0.0 | 1.6 | PID CDC42 REG PATHWAY | Regulation of CDC42 activity |
| 0.0 | 1.0 | PID NCADHERIN PATHWAY | N-cadherin signaling events |
| 0.0 | 0.9 | PID RAS PATHWAY | Regulation of Ras family activation |
| 0.0 | 0.9 | NABA PROTEOGLYCANS | Genes encoding proteoglycans |
| 0.0 | 0.4 | PID NFKAPPAB ATYPICAL PATHWAY | Atypical NF-kappaB pathway |
| 0.0 | 0.4 | PID HEDGEHOG 2PATHWAY | Signaling events mediated by the Hedgehog family |
| 0.0 | 0.8 | PID ERBB1 INTERNALIZATION PATHWAY | Internalization of ErbB1 |
| 0.0 | 1.0 | PID MTOR 4PATHWAY | mTOR signaling pathway |
| 0.0 | 0.8 | PID RAC1 PATHWAY | RAC1 signaling pathway |
| 0.0 | 0.2 | PID INTEGRIN5 PATHWAY | Beta5 beta6 beta7 and beta8 integrin cell surface interactions |
| 0.0 | 0.2 | PID LPA4 PATHWAY | LPA4-mediated signaling events |
| 0.0 | 0.2 | PID ARF 3PATHWAY | Arf1 pathway |
| 0.0 | 0.2 | PID P38 MKK3 6PATHWAY | p38 MAPK signaling pathway |
| 0.0 | 0.4 | PID NOTCH PATHWAY | Notch signaling pathway |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 1.1 | REACTOME REGULATION OF RHEB GTPASE ACTIVITY BY AMPK | Genes involved in Regulation of Rheb GTPase activity by AMPK |
| 0.1 | 0.9 | REACTOME RAP1 SIGNALLING | Genes involved in Rap1 signalling |
| 0.1 | 1.5 | REACTOME NITRIC OXIDE STIMULATES GUANYLATE CYCLASE | Genes involved in Nitric oxide stimulates guanylate cyclase |
| 0.1 | 1.2 | REACTOME DARPP 32 EVENTS | Genes involved in DARPP-32 events |
| 0.1 | 1.7 | REACTOME EFFECTS OF PIP2 HYDROLYSIS | Genes involved in Effects of PIP2 hydrolysis |
| 0.1 | 0.4 | REACTOME SEROTONIN RECEPTORS | Genes involved in Serotonin receptors |
| 0.1 | 1.0 | REACTOME OTHER SEMAPHORIN INTERACTIONS | Genes involved in Other semaphorin interactions |
| 0.1 | 0.9 | REACTOME SYNTHESIS OF PIPS AT THE EARLY ENDOSOME MEMBRANE | Genes involved in Synthesis of PIPs at the early endosome membrane |
| 0.0 | 1.9 | REACTOME NRAGE SIGNALS DEATH THROUGH JNK | Genes involved in NRAGE signals death through JNK |
| 0.0 | 0.4 | REACTOME GABA A RECEPTOR ACTIVATION | Genes involved in GABA A receptor activation |
| 0.0 | 0.5 | REACTOME SYNTHESIS OF GLYCOSYLPHOSPHATIDYLINOSITOL GPI | Genes involved in Synthesis of glycosylphosphatidylinositol (GPI) |
| 0.0 | 0.2 | REACTOME PEPTIDE HORMONE BIOSYNTHESIS | Genes involved in Peptide hormone biosynthesis |
| 0.0 | 0.2 | REACTOME PKA MEDIATED PHOSPHORYLATION OF CREB | Genes involved in PKA-mediated phosphorylation of CREB |
| 0.0 | 0.2 | REACTOME RETROGRADE NEUROTROPHIN SIGNALLING | Genes involved in Retrograde neurotrophin signalling |
| 0.0 | 0.2 | REACTOME GAMMA CARBOXYLATION TRANSPORT AND AMINO TERMINAL CLEAVAGE OF PROTEINS | Genes involved in Gamma-carboxylation, transport, and amino-terminal cleavage of proteins |
| 0.0 | 0.4 | REACTOME PEROXISOMAL LIPID METABOLISM | Genes involved in Peroxisomal lipid metabolism |
| 0.0 | 0.5 | REACTOME P75 NTR RECEPTOR MEDIATED SIGNALLING | Genes involved in p75 NTR receptor-mediated signalling |
| 0.0 | 0.4 | REACTOME ACTIVATED NOTCH1 TRANSMITS SIGNAL TO THE NUCLEUS | Genes involved in Activated NOTCH1 Transmits Signal to the Nucleus |
| 0.0 | 0.2 | REACTOME MEMBRANE BINDING AND TARGETTING OF GAG PROTEINS | Genes involved in Membrane binding and targetting of GAG proteins |
| 0.0 | 0.2 | REACTOME SEMA4D INDUCED CELL MIGRATION AND GROWTH CONE COLLAPSE | Genes involved in Sema4D induced cell migration and growth-cone collapse |
| 0.0 | 0.3 | REACTOME CONVERSION FROM APC C CDC20 TO APC C CDH1 IN LATE ANAPHASE | Genes involved in Conversion from APC/C:Cdc20 to APC/C:Cdh1 in late anaphase |
| 0.0 | 0.3 | REACTOME REGULATION OF GENE EXPRESSION IN BETA CELLS | Genes involved in Regulation of gene expression in beta cells |
| 0.0 | 0.3 | REACTOME REGULATION OF SIGNALING BY CBL | Genes involved in Regulation of signaling by CBL |
| 0.0 | 0.3 | REACTOME GLUCONEOGENESIS | Genes involved in Gluconeogenesis |
| 0.0 | 0.2 | REACTOME GLUTATHIONE CONJUGATION | Genes involved in Glutathione conjugation |
| 0.0 | 0.8 | REACTOME PEPTIDE LIGAND BINDING RECEPTORS | Genes involved in Peptide ligand-binding receptors |