Project
PRJNA207719: Tissue specific transcriptome profiling
Navigation
Downloads
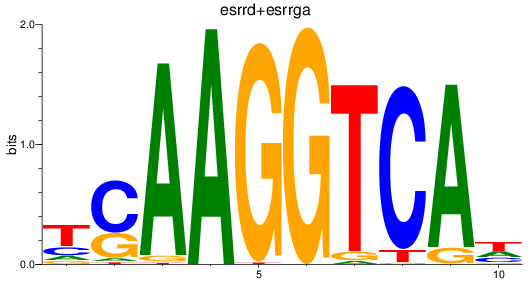
Results for esrrd+esrrga_esrrb_esrra+esrrgb
Z-value: 1.16
Transcription factors associated with esrrd+esrrga_esrrb_esrra+esrrgb
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
esrrga
|
ENSDARG00000004861 | estrogen-related receptor gamma a |
|
esrrd
|
ENSDARG00000015064 | estrogen-related receptor delta |
|
esrrb
|
ENSDARG00000100847 | estrogen-related receptor beta |
|
esrrgb
|
ENSDARG00000011696 | estrogen-related receptor gamma b |
|
esrra
|
ENSDARG00000069266 | estrogen-related receptor alpha |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| esrra | dr11_v1_chr21_+_26620867_26620867 | 0.93 | 2.4e-02 | Click! |
| esrrb | dr11_v1_chr17_-_29877336_29877336 | 0.84 | 7.6e-02 | Click! |
| esrrgb | dr11_v1_chr20_-_46817223_46817223 | 0.58 | 3.1e-01 | Click! |
| esrrd | dr11_v1_chr18_-_48992363_48992363 | 0.08 | 8.9e-01 | Click! |
Activity profile of esrrd+esrrga_esrrb_esrra+esrrgb motif
Sorted Z-values of esrrd+esrrga_esrrb_esrra+esrrgb motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr4_+_2620751 | 1.39 |
ENSDART00000013924
|
gpr22a
|
G protein-coupled receptor 22a |
| chr18_-_226800 | 1.11 |
ENSDART00000165180
|
tarsl2
|
threonyl-tRNA synthetase-like 2 |
| chr2_-_24289641 | 0.98 |
ENSDART00000128784
ENSDART00000123565 ENSDART00000141922 ENSDART00000184550 ENSDART00000191469 |
myh7l
|
myosin heavy chain 7-like |
| chr19_-_32487469 | 0.92 |
ENSDART00000050130
|
gmpr
|
guanosine monophosphate reductase |
| chr14_-_33454595 | 0.91 |
ENSDART00000109615
ENSDART00000173267 ENSDART00000185737 ENSDART00000190989 |
tmem255a
|
transmembrane protein 255A |
| chr3_+_39568290 | 0.88 |
ENSDART00000020741
|
aldoaa
|
aldolase a, fructose-bisphosphate, a |
| chr7_+_67494107 | 0.84 |
ENSDART00000185653
|
cpne7
|
copine VII |
| chr11_+_30513656 | 0.83 |
ENSDART00000008594
|
tmem178
|
transmembrane protein 178 |
| chr10_+_45128375 | 0.83 |
ENSDART00000164805
|
camk2b2
|
calcium/calmodulin-dependent protein kinase (CaM kinase) II beta 2 |
| chr21_-_32487061 | 0.83 |
ENSDART00000114359
ENSDART00000131591 ENSDART00000131477 |
si:dkeyp-72g9.4
|
si:dkeyp-72g9.4 |
| chr11_+_25111846 | 0.77 |
ENSDART00000128705
ENSDART00000190058 |
ndrg3a
|
ndrg family member 3a |
| chr3_-_60142530 | 0.76 |
ENSDART00000153247
|
si:ch211-120g10.1
|
si:ch211-120g10.1 |
| chr14_+_1014109 | 0.76 |
ENSDART00000157945
|
f8
|
coagulation factor VIII, procoagulant component |
| chr3_-_21280373 | 0.72 |
ENSDART00000003939
|
syngr1a
|
synaptogyrin 1a |
| chr5_-_10768258 | 0.66 |
ENSDART00000157043
|
rtn4r
|
reticulon 4 receptor |
| chr10_+_10677697 | 0.66 |
ENSDART00000188705
|
fam163b
|
family with sequence similarity 163, member B |
| chr9_-_29844596 | 0.66 |
ENSDART00000138574
|
il1rapl1a
|
interleukin 1 receptor accessory protein-like 1a |
| chr6_-_15653494 | 0.64 |
ENSDART00000038133
|
trim63a
|
tripartite motif containing 63a |
| chr17_-_12385308 | 0.64 |
ENSDART00000080927
|
snap25b
|
synaptosomal-associated protein, 25b |
| chr7_+_38716048 | 0.63 |
ENSDART00000024590
|
syt13
|
synaptotagmin XIII |
| chr17_-_33289304 | 0.63 |
ENSDART00000135118
ENSDART00000040346 |
efr3ba
|
EFR3 homolog Ba (S. cerevisiae) |
| chr5_-_26566435 | 0.62 |
ENSDART00000146070
|
arvcfb
|
ARVCF, delta catenin family member b |
| chr1_-_23110740 | 0.62 |
ENSDART00000171848
ENSDART00000086797 ENSDART00000189344 ENSDART00000190858 |
adgrl3.1
|
adhesion G protein-coupled receptor L3.1 |
| chr5_-_57820873 | 0.60 |
ENSDART00000089961
|
sik2a
|
salt-inducible kinase 2a |
| chr1_+_45080897 | 0.59 |
ENSDART00000129819
|
si:ch211-151p13.8
|
si:ch211-151p13.8 |
| chr22_-_21150845 | 0.59 |
ENSDART00000027345
|
tmem59l
|
transmembrane protein 59-like |
| chr6_+_48618512 | 0.59 |
ENSDART00000111190
|
FAM19A3
|
si:dkey-238f9.1 |
| chr21_+_30563115 | 0.58 |
ENSDART00000028566
|
si:ch211-200p22.4
|
si:ch211-200p22.4 |
| chr11_-_7320211 | 0.57 |
ENSDART00000091664
|
apc2
|
adenomatosis polyposis coli 2 |
| chr3_-_46817838 | 0.55 |
ENSDART00000028610
|
elavl3
|
ELAV like neuron-specific RNA binding protein 3 |
| chr3_+_43460696 | 0.55 |
ENSDART00000164581
|
galr2b
|
galanin receptor 2b |
| chr6_+_28124393 | 0.55 |
ENSDART00000089195
|
gpr17
|
G protein-coupled receptor 17 |
| chr14_-_9199968 | 0.53 |
ENSDART00000146113
|
arhgef9b
|
Cdc42 guanine nucleotide exchange factor (GEF) 9b |
| chr11_-_25213651 | 0.53 |
ENSDART00000097316
ENSDART00000152186 |
myh7ba
|
myosin, heavy chain 7B, cardiac muscle, beta a |
| chr3_-_46818001 | 0.53 |
ENSDART00000166505
|
elavl3
|
ELAV like neuron-specific RNA binding protein 3 |
| chr21_+_19008168 | 0.51 |
ENSDART00000136196
ENSDART00000128381 ENSDART00000176624 |
nefla
|
neurofilament, light polypeptide a |
| chr21_-_12119711 | 0.51 |
ENSDART00000131538
|
celf4
|
CUGBP, Elav-like family member 4 |
| chr7_+_30875273 | 0.51 |
ENSDART00000173693
|
apba2b
|
amyloid beta (A4) precursor protein-binding, family A, member 2b |
| chr7_+_25913225 | 0.51 |
ENSDART00000129924
|
hmgb3a
|
high mobility group box 3a |
| chr10_+_34623183 | 0.50 |
ENSDART00000114630
|
nbeaa
|
neurobeachin a |
| chr14_-_29826659 | 0.50 |
ENSDART00000138413
|
sorbs2b
|
sorbin and SH3 domain containing 2b |
| chr15_+_45994123 | 0.50 |
ENSDART00000124704
|
lrfn1
|
leucine rich repeat and fibronectin type III domain containing 1 |
| chr11_+_28476298 | 0.48 |
ENSDART00000122319
|
lrrc38b
|
leucine rich repeat containing 38b |
| chr19_-_9712530 | 0.48 |
ENSDART00000134816
|
slc2a3a
|
solute carrier family 2 (facilitated glucose transporter), member 3a |
| chr11_-_43200994 | 0.48 |
ENSDART00000164700
|
sptbn1
|
spectrin, beta, non-erythrocytic 1 |
| chr12_+_45200744 | 0.48 |
ENSDART00000098932
|
wbp2
|
WW domain binding protein 2 |
| chr4_+_14360372 | 0.47 |
ENSDART00000007103
|
nuak1a
|
NUAK family, SNF1-like kinase, 1a |
| chr7_-_58130703 | 0.46 |
ENSDART00000172082
|
ank2b
|
ankyrin 2b, neuronal |
| chr19_-_5103313 | 0.45 |
ENSDART00000037007
|
tpi1a
|
triosephosphate isomerase 1a |
| chr6_-_4214297 | 0.45 |
ENSDART00000191433
|
trak2
|
trafficking protein, kinesin binding 2 |
| chr11_-_29650930 | 0.44 |
ENSDART00000166969
|
chd5
|
chromodomain helicase DNA binding protein 5 |
| chr8_+_24854600 | 0.43 |
ENSDART00000156570
|
slc6a17
|
solute carrier family 6 (neutral amino acid transporter), member 17 |
| chr23_-_19953089 | 0.40 |
ENSDART00000153828
|
atp2b3b
|
ATPase plasma membrane Ca2+ transporting 3b |
| chr10_+_8437930 | 0.40 |
ENSDART00000074553
|
pptc7b
|
PTC7 protein phosphatase homolog b |
| chr7_+_568819 | 0.40 |
ENSDART00000173716
|
nrxn2b
|
neurexin 2b |
| chr5_+_1278092 | 0.38 |
ENSDART00000147972
ENSDART00000159783 |
dnm1a
|
dynamin 1a |
| chr2_+_34767171 | 0.38 |
ENSDART00000145451
|
astn1
|
astrotactin 1 |
| chr23_-_10017245 | 0.38 |
ENSDART00000131669
|
plxnb1a
|
plexin b1a |
| chr7_-_48805181 | 0.38 |
ENSDART00000015884
|
mfge8a
|
milk fat globule-EGF factor 8 protein a |
| chr11_-_10770053 | 0.38 |
ENSDART00000179213
|
slc4a10a
|
solute carrier family 4, sodium bicarbonate transporter, member 10a |
| chr10_+_21737745 | 0.38 |
ENSDART00000170498
ENSDART00000167997 |
pcdh1g18
|
protocadherin 1 gamma 18 |
| chr10_+_22782522 | 0.37 |
ENSDART00000079498
ENSDART00000145558 |
si:ch211-237l4.6
|
si:ch211-237l4.6 |
| chr5_-_57879138 | 0.37 |
ENSDART00000145959
|
sik2a
|
salt-inducible kinase 2a |
| chr25_-_29316193 | 0.36 |
ENSDART00000088249
|
hcn4l
|
hyperpolarization activated cyclic nucleotide-gated potassium channel 4l |
| chr19_+_30662529 | 0.36 |
ENSDART00000175662
|
fam49al
|
family with sequence similarity 49, member A-like |
| chr4_-_1360495 | 0.36 |
ENSDART00000164623
|
ptn
|
pleiotrophin |
| chr5_+_66355153 | 0.36 |
ENSDART00000082745
|
si:ch211-261c8.5
|
si:ch211-261c8.5 |
| chr19_-_7291733 | 0.36 |
ENSDART00000015559
|
sdha
|
succinate dehydrogenase complex, subunit A, flavoprotein (Fp) |
| chr17_+_23554932 | 0.35 |
ENSDART00000135814
|
pank1a
|
pantothenate kinase 1a |
| chr5_-_35301800 | 0.35 |
ENSDART00000085142
|
map1b
|
microtubule-associated protein 1B |
| chr2_-_24462277 | 0.35 |
ENSDART00000033922
|
kcnn1a
|
potassium intermediate/small conductance calcium-activated channel, subfamily N, member 1a |
| chr5_-_24044724 | 0.35 |
ENSDART00000023784
|
pdha1a
|
pyruvate dehydrogenase E1 alpha 1 subunit a |
| chr10_+_1638876 | 0.35 |
ENSDART00000184484
ENSDART00000060946 ENSDART00000181251 |
sgsm1b
|
small G protein signaling modulator 1b |
| chr8_+_26879358 | 0.34 |
ENSDART00000132485
|
rimkla
|
ribosomal modification protein rimK-like family member A |
| chr4_+_17280868 | 0.34 |
ENSDART00000145349
|
bcat1
|
branched chain amino-acid transaminase 1, cytosolic |
| chr12_-_26415499 | 0.34 |
ENSDART00000185779
|
synpo2lb
|
synaptopodin 2-like b |
| chr21_-_25416391 | 0.34 |
ENSDART00000114081
|
sms
|
spermine synthase |
| chr1_-_54718863 | 0.34 |
ENSDART00000122601
|
pgam1b
|
phosphoglycerate mutase 1b |
| chr10_+_35952532 | 0.33 |
ENSDART00000184730
|
rtn4rl1a
|
reticulon 4 receptor-like 1a |
| chr5_-_24044877 | 0.33 |
ENSDART00000161131
|
pdha1a
|
pyruvate dehydrogenase E1 alpha 1 subunit a |
| chr21_-_18648861 | 0.33 |
ENSDART00000112113
|
si:dkey-112m2.1
|
si:dkey-112m2.1 |
| chr2_+_55982940 | 0.32 |
ENSDART00000097753
ENSDART00000097751 |
nmrk2
|
nicotinamide riboside kinase 2 |
| chr16_-_33059246 | 0.32 |
ENSDART00000171718
ENSDART00000168305 ENSDART00000166401 |
snap91
|
synaptosomal-associated protein 91 |
| chr17_-_8692722 | 0.32 |
ENSDART00000148931
ENSDART00000192891 |
ctbp2a
|
C-terminal binding protein 2a |
| chr24_-_21471389 | 0.32 |
ENSDART00000109848
|
atp8a2
|
ATPase phospholipid transporting 8A2 |
| chr16_+_50741154 | 0.32 |
ENSDART00000101627
|
IGLON5
|
zgc:110372 |
| chr5_-_63109232 | 0.32 |
ENSDART00000115128
|
usp2b
|
ubiquitin specific peptidase 2b |
| chr16_-_17197546 | 0.32 |
ENSDART00000139939
ENSDART00000135146 ENSDART00000063800 ENSDART00000163606 |
gapdh
|
glyceraldehyde-3-phosphate dehydrogenase |
| chr21_+_26620867 | 0.32 |
ENSDART00000176008
|
esrra
|
estrogen-related receptor alpha |
| chr2_-_19109304 | 0.31 |
ENSDART00000168028
|
si:dkey-225f23.5
|
si:dkey-225f23.5 |
| chr2_-_9646857 | 0.31 |
ENSDART00000056901
|
zgc:153615
|
zgc:153615 |
| chr23_+_18722915 | 0.31 |
ENSDART00000025057
|
myh7bb
|
myosin, heavy chain 7B, cardiac muscle, beta b |
| chr10_+_22775253 | 0.31 |
ENSDART00000190141
|
tmem88a
|
transmembrane protein 88 a |
| chr6_-_11523987 | 0.31 |
ENSDART00000189363
|
gulp1b
|
GULP, engulfment adaptor PTB domain containing 1b |
| chr1_+_16573982 | 0.30 |
ENSDART00000166317
|
mtus1b
|
microtubule associated tumor suppressor 1b |
| chr24_-_20641000 | 0.30 |
ENSDART00000166135
|
zbtb47b
|
zinc finger and BTB domain containing 47b |
| chr17_-_45254585 | 0.30 |
ENSDART00000185507
ENSDART00000172080 |
ttbk2a
|
tau tubulin kinase 2a |
| chr8_-_14052349 | 0.30 |
ENSDART00000135811
|
atp2b3a
|
ATPase plasma membrane Ca2+ transporting 3a |
| chr11_+_23704410 | 0.30 |
ENSDART00000112655
|
nfasca
|
neurofascin homolog (chicken) a |
| chr12_+_3912544 | 0.29 |
ENSDART00000013465
|
tbx6
|
T-box 6 |
| chr5_-_35252761 | 0.29 |
ENSDART00000051278
|
tnpo1
|
transportin 1 |
| chr21_+_39432248 | 0.29 |
ENSDART00000179938
|
pafah1b1b
|
platelet-activating factor acetylhydrolase 1b, regulatory subunit 1b |
| chr23_-_14918276 | 0.29 |
ENSDART00000179831
|
ndrg3b
|
ndrg family member 3b |
| chr22_+_9472814 | 0.29 |
ENSDART00000112125
ENSDART00000138850 |
cacna2d2b
|
calcium channel, voltage-dependent, alpha 2/delta subunit 2b |
| chr20_+_218886 | 0.28 |
ENSDART00000002661
|
lama4
|
laminin, alpha 4 |
| chr6_-_29143463 | 0.28 |
ENSDART00000087440
|
fam69ab
|
family with sequence similarity 69, member Ab |
| chr4_-_11163112 | 0.28 |
ENSDART00000188854
|
prmt8b
|
protein arginine methyltransferase 8b |
| chr4_-_11165611 | 0.28 |
ENSDART00000059489
|
prmt8b
|
protein arginine methyltransferase 8b |
| chr22_-_11137268 | 0.28 |
ENSDART00000178882
|
atp6ap2
|
ATPase H+ transporting accessory protein 2 |
| chr10_+_1849874 | 0.28 |
ENSDART00000158897
ENSDART00000149956 |
apc
|
adenomatous polyposis coli |
| chr4_-_20235904 | 0.28 |
ENSDART00000146621
ENSDART00000193655 |
stk38l
|
serine/threonine kinase 38 like |
| chr22_-_17574511 | 0.28 |
ENSDART00000181496
|
pip5k1ca
|
phosphatidylinositol-4-phosphate 5-kinase, type I, gamma a |
| chr25_+_25124684 | 0.28 |
ENSDART00000167542
|
ldha
|
lactate dehydrogenase A4 |
| chr19_-_5103141 | 0.27 |
ENSDART00000150952
|
tpi1a
|
triosephosphate isomerase 1a |
| chr18_-_26510545 | 0.27 |
ENSDART00000135133
|
FRMD5
|
si:ch211-69m14.1 |
| chr1_-_1627487 | 0.27 |
ENSDART00000166094
|
clic6
|
chloride intracellular channel 6 |
| chr9_-_42861080 | 0.27 |
ENSDART00000193688
|
ttn.1
|
titin, tandem duplicate 1 |
| chr8_-_22639794 | 0.27 |
ENSDART00000188029
|
iqsec2a
|
IQ motif and Sec7 domain 2a |
| chr19_+_712127 | 0.27 |
ENSDART00000093281
ENSDART00000180002 ENSDART00000146050 |
fhod3a
|
formin homology 2 domain containing 3a |
| chr23_+_18722715 | 0.27 |
ENSDART00000137438
|
myh7bb
|
myosin, heavy chain 7B, cardiac muscle, beta b |
| chr5_-_42544522 | 0.26 |
ENSDART00000157968
|
BX510949.1
|
|
| chr10_+_17235370 | 0.26 |
ENSDART00000038780
|
sppl3
|
signal peptide peptidase 3 |
| chr23_-_20002459 | 0.26 |
ENSDART00000163396
|
b4galt3
|
UDP-Gal:betaGlcNAc beta 1,4- galactosyltransferase, polypeptide 3 |
| chr3_-_26524934 | 0.26 |
ENSDART00000087118
|
xylt1
|
xylosyltransferase I |
| chr22_+_33362552 | 0.26 |
ENSDART00000101580
|
nicn1
|
nicolin 1 |
| chr13_-_42724645 | 0.26 |
ENSDART00000046066
|
capn1a
|
calpain 1, (mu/I) large subunit a |
| chr5_+_59397739 | 0.26 |
ENSDART00000148659
|
clip2
|
CAP-GLY domain containing linker protein 2 |
| chr4_-_11165961 | 0.26 |
ENSDART00000193393
|
prmt8b
|
protein arginine methyltransferase 8b |
| chr5_-_30475011 | 0.25 |
ENSDART00000187501
|
phldb1a
|
pleckstrin homology-like domain, family B, member 1a |
| chr8_+_31248917 | 0.25 |
ENSDART00000112170
|
unm_hu7912
|
un-named hu7912 |
| chr20_-_27857676 | 0.25 |
ENSDART00000192934
|
syndig1l
|
synapse differentiation inducing 1-like |
| chr17_-_21278582 | 0.25 |
ENSDART00000157518
|
hspa12a
|
heat shock protein 12A |
| chr2_+_6926100 | 0.25 |
ENSDART00000153289
|
nos1apb
|
nitric oxide synthase 1 (neuronal) adaptor protein b |
| chr6_+_22337081 | 0.25 |
ENSDART00000128047
ENSDART00000138930 |
uqcrc1
|
ubiquinol-cytochrome c reductase core protein 1 |
| chr21_+_13383413 | 0.24 |
ENSDART00000151345
|
zgc:113162
|
zgc:113162 |
| chr23_-_24488696 | 0.24 |
ENSDART00000155593
|
tmem82
|
transmembrane protein 82 |
| chr14_-_34026316 | 0.24 |
ENSDART00000186062
|
cyfip2
|
cytoplasmic FMR1 interacting protein 2 |
| chr10_+_44584614 | 0.24 |
ENSDART00000163523
|
sez6l
|
seizure related 6 homolog (mouse)-like |
| chr5_-_46896541 | 0.24 |
ENSDART00000133240
|
edil3a
|
EGF-like repeats and discoidin I-like domains 3a |
| chr5_-_69316142 | 0.24 |
ENSDART00000157238
ENSDART00000144570 |
smtnb
|
smoothelin b |
| chr12_+_28799988 | 0.24 |
ENSDART00000022724
|
pnpo
|
pyridoxamine 5'-phosphate oxidase |
| chr5_-_37871526 | 0.24 |
ENSDART00000136450
|
arhgap35b
|
Rho GTPase activating protein 35b |
| chr3_-_45250924 | 0.23 |
ENSDART00000109017
|
usp31
|
ubiquitin specific peptidase 31 |
| chr9_+_7909490 | 0.23 |
ENSDART00000133063
ENSDART00000109288 |
myo16
|
myosin XVI |
| chr16_+_21242491 | 0.23 |
ENSDART00000145886
|
osbpl3b
|
oxysterol binding protein-like 3b |
| chr18_+_48608366 | 0.23 |
ENSDART00000151229
|
kcnj5
|
potassium inwardly-rectifying channel, subfamily J, member 5 |
| chr23_-_15284757 | 0.23 |
ENSDART00000139135
|
sulf2b
|
sulfatase 2b |
| chr24_+_28525507 | 0.23 |
ENSDART00000191121
|
arhgap29a
|
Rho GTPase activating protein 29a |
| chr25_+_21832938 | 0.23 |
ENSDART00000148299
|
ckmt1
|
creatine kinase, mitochondrial 1 |
| chr5_+_20693724 | 0.23 |
ENSDART00000141368
|
si:ch211-240b21.2
|
si:ch211-240b21.2 |
| chr23_-_28239750 | 0.23 |
ENSDART00000003548
|
znf385a
|
zinc finger protein 385A |
| chr23_-_21515182 | 0.22 |
ENSDART00000142000
|
rnf207b
|
ring finger protein 207b |
| chr17_-_14696529 | 0.22 |
ENSDART00000190205
|
CU694317.1
|
|
| chr16_+_29630965 | 0.22 |
ENSDART00000185820
ENSDART00000192105 ENSDART00000153683 ENSDART00000186713 |
VPS72
|
si:ch211-203d17.1 |
| chr7_-_19638319 | 0.22 |
ENSDART00000163686
|
si:ch211-212k18.4
|
si:ch211-212k18.4 |
| chr2_+_53359234 | 0.22 |
ENSDART00000147581
|
celf5b
|
cugbp, Elav-like family member 5b |
| chr19_-_5805923 | 0.22 |
ENSDART00000134340
|
si:ch211-264f5.8
|
si:ch211-264f5.8 |
| chr22_-_26865361 | 0.22 |
ENSDART00000182504
|
hmox2a
|
heme oxygenase 2a |
| chr6_+_2190214 | 0.22 |
ENSDART00000156716
|
acvr1bb
|
activin A receptor type 1Bb |
| chr1_-_17569793 | 0.22 |
ENSDART00000125125
|
acsl1a
|
acyl-CoA synthetase long chain family member 1a |
| chr24_+_37080771 | 0.22 |
ENSDART00000159942
|
kcnc3b
|
potassium voltage-gated channel, Shaw-related subfamily, member 3b |
| chr20_+_19212962 | 0.22 |
ENSDART00000063706
|
fndc4a
|
fibronectin type III domain containing 4a |
| chr12_+_47162456 | 0.22 |
ENSDART00000186272
ENSDART00000180811 |
RYR2
|
ryanodine receptor 2 |
| chr13_-_37127970 | 0.22 |
ENSDART00000135510
|
syne2b
|
spectrin repeat containing, nuclear envelope 2b |
| chr17_-_20711735 | 0.21 |
ENSDART00000150056
|
ank3b
|
ankyrin 3b |
| chr16_-_30655980 | 0.21 |
ENSDART00000146508
|
ldlrad4b
|
low density lipoprotein receptor class A domain containing 4b |
| chr12_+_47162761 | 0.21 |
ENSDART00000192339
ENSDART00000167726 |
RYR2
|
ryanodine receptor 2 |
| chr4_-_4129187 | 0.21 |
ENSDART00000146889
|
asb15b
|
ankyrin repeat and SOCS box containing 15b |
| chr7_+_44484853 | 0.21 |
ENSDART00000189079
ENSDART00000121826 |
bean1
|
brain expressed, associated with NEDD4, 1 |
| chr25_+_21177326 | 0.21 |
ENSDART00000149142
|
erc1a
|
ELKS/RAB6-interacting/CAST family member 1a |
| chr18_-_39787040 | 0.21 |
ENSDART00000169916
|
dmxl2
|
Dmx-like 2 |
| chr14_+_12142828 | 0.21 |
ENSDART00000012438
|
hspa9
|
heat shock protein 9 |
| chr23_+_16430559 | 0.21 |
ENSDART00000112436
|
ntsr1
|
neurotensin receptor 1 (high affinity) |
| chr12_-_33359052 | 0.21 |
ENSDART00000135943
|
slc16a3
|
solute carrier family 16 (monocarboxylate transporter), member 3 |
| chr3_-_6519691 | 0.21 |
ENSDART00000165273
ENSDART00000179882 ENSDART00000172292 |
GGA3 (1 of many)
|
si:ch73-157i16.3 |
| chr21_+_27513859 | 0.21 |
ENSDART00000065420
|
pacs1a
|
phosphofurin acidic cluster sorting protein 1a |
| chr7_+_48761875 | 0.21 |
ENSDART00000003690
|
acana
|
aggrecan a |
| chr10_+_35953068 | 0.21 |
ENSDART00000015279
|
rtn4rl1a
|
reticulon 4 receptor-like 1a |
| chr25_+_22730490 | 0.20 |
ENSDART00000149455
|
abcc8
|
ATP-binding cassette, sub-family C (CFTR/MRP), member 8 |
| chr19_-_37154436 | 0.20 |
ENSDART00000103155
|
cx39.4
|
connexin 39.4 |
| chr15_-_10341048 | 0.20 |
ENSDART00000171013
|
tenm4
|
teneurin transmembrane protein 4 |
| chr8_-_49908978 | 0.20 |
ENSDART00000172642
|
agtpbp1
|
ATP/GTP binding protein 1 |
| chr18_+_910992 | 0.20 |
ENSDART00000161206
ENSDART00000167229 |
pkma
|
pyruvate kinase M1/2a |
| chr2_+_55982300 | 0.20 |
ENSDART00000183903
|
nmrk2
|
nicotinamide riboside kinase 2 |
| chr22_-_11136625 | 0.20 |
ENSDART00000016873
ENSDART00000125561 |
atp6ap2
|
ATPase H+ transporting accessory protein 2 |
| chr18_+_16744307 | 0.20 |
ENSDART00000179872
ENSDART00000133490 |
lyve1b
|
lymphatic vessel endothelial hyaluronic receptor 1b |
| chr9_+_24159280 | 0.20 |
ENSDART00000184624
ENSDART00000178422 |
hecw2a
|
HECT, C2 and WW domain containing E3 ubiquitin protein ligase 2a |
| chr16_-_12914288 | 0.20 |
ENSDART00000184221
|
cacng8b
|
calcium channel, voltage-dependent, gamma subunit 8b |
| chr11_-_18791834 | 0.20 |
ENSDART00000156431
|
pfkfb2b
|
6-phosphofructo-2-kinase/fructose-2,6-biphosphatase 2b |
| chr24_-_1021318 | 0.20 |
ENSDART00000181403
|
ralaa
|
v-ral simian leukemia viral oncogene homolog Aa (ras related) |
| chr16_+_22618620 | 0.20 |
ENSDART00000185728
ENSDART00000041625 |
chrnb2b
|
cholinergic receptor, nicotinic, beta 2b |
| chr25_+_21176937 | 0.20 |
ENSDART00000163751
ENSDART00000188930 ENSDART00000187155 |
erc1a
|
ELKS/RAB6-interacting/CAST family member 1a |
| chr12_+_15622621 | 0.19 |
ENSDART00000079784
|
plcd3b
|
phospholipase C, delta 3b |
| chr8_-_44015210 | 0.19 |
ENSDART00000186879
ENSDART00000188965 ENSDART00000001313 ENSDART00000188902 ENSDART00000185935 ENSDART00000147869 |
rimbp2
rimbp2
|
RIMS binding protein 2 RIMS binding protein 2 |
| chr5_-_67115872 | 0.19 |
ENSDART00000065262
|
rps6ka4
|
ribosomal protein S6 kinase, polypeptide 4 |
| chr6_+_52235441 | 0.19 |
ENSDART00000056319
|
cox6c
|
cytochrome c oxidase subunit VIc |
| chr22_-_31517300 | 0.19 |
ENSDART00000164799
|
slc6a6b
|
solute carrier family 6 (neurotransmitter transporter), member 6b |
| chr3_+_4997545 | 0.19 |
ENSDART00000181237
|
CABZ01117706.1
|
|
| chr6_-_1514767 | 0.19 |
ENSDART00000067586
|
chchd6b
|
coiled-coil-helix-coiled-coil-helix domain containing 6b |
| chr21_+_15709061 | 0.19 |
ENSDART00000065772
|
ddt
|
D-dopachrome tautomerase |
Network of associatons between targets according to the STRING database.
First level regulatory network of esrrd+esrrga_esrrb_esrra+esrrgb
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 0.8 | GO:0045671 | negative regulation of osteoclast differentiation(GO:0045671) |
| 0.2 | 0.7 | GO:0046166 | methylglyoxal biosynthetic process(GO:0019242) glyceraldehyde-3-phosphate biosynthetic process(GO:0046166) |
| 0.1 | 0.6 | GO:1905067 | negative regulation of canonical Wnt signaling pathway involved in heart development(GO:1905067) |
| 0.1 | 0.4 | GO:0015824 | proline transport(GO:0015824) |
| 0.1 | 0.4 | GO:0042364 | water-soluble vitamin biosynthetic process(GO:0042364) |
| 0.1 | 0.8 | GO:0006086 | acetyl-CoA biosynthetic process from pyruvate(GO:0006086) |
| 0.1 | 0.5 | GO:0006121 | mitochondrial electron transport, succinate to ubiquinone(GO:0006121) |
| 0.1 | 0.5 | GO:0036371 | protein localization to T-tubule(GO:0036371) |
| 0.1 | 0.3 | GO:0021785 | branchiomotor neuron axon guidance(GO:0021785) |
| 0.1 | 0.3 | GO:0009098 | leucine biosynthetic process(GO:0009098) |
| 0.1 | 0.2 | GO:0051196 | regulation of glycolytic process(GO:0006110) regulation of nucleotide catabolic process(GO:0030811) regulation of cofactor metabolic process(GO:0051193) regulation of coenzyme metabolic process(GO:0051196) |
| 0.1 | 0.5 | GO:0098957 | anterograde axonal transport of mitochondrion(GO:0098957) |
| 0.1 | 0.2 | GO:1903779 | regulation of cardiac conduction(GO:1903779) |
| 0.1 | 0.3 | GO:0051012 | microtubule sliding(GO:0051012) |
| 0.1 | 0.2 | GO:1901546 | regulation of cellular pH reduction(GO:0032847) synaptic vesicle lumen acidification(GO:0097401) regulation of synaptic vesicle lumen acidification(GO:1901546) |
| 0.1 | 0.4 | GO:0003151 | outflow tract morphogenesis(GO:0003151) |
| 0.1 | 0.3 | GO:0035332 | positive regulation of hippo signaling(GO:0035332) |
| 0.1 | 1.4 | GO:0030388 | fructose 1,6-bisphosphate metabolic process(GO:0030388) |
| 0.1 | 0.8 | GO:0045162 | clustering of voltage-gated sodium channels(GO:0045162) |
| 0.1 | 0.6 | GO:0031111 | negative regulation of microtubule depolymerization(GO:0007026) negative regulation of microtubule polymerization or depolymerization(GO:0031111) |
| 0.1 | 0.3 | GO:0048714 | positive regulation of glial cell differentiation(GO:0045687) positive regulation of oligodendrocyte differentiation(GO:0048714) |
| 0.1 | 0.2 | GO:0035610 | protein side chain deglutamylation(GO:0035610) |
| 0.0 | 1.4 | GO:0071907 | determination of digestive tract left/right asymmetry(GO:0071907) |
| 0.0 | 0.5 | GO:0099558 | maintenance of presynaptic active zone structure(GO:0048790) maintenance of synapse structure(GO:0099558) |
| 0.0 | 0.2 | GO:0006531 | aspartate metabolic process(GO:0006531) |
| 0.0 | 0.3 | GO:0070073 | clustering of voltage-gated calcium channels(GO:0070073) |
| 0.0 | 0.4 | GO:2000650 | negative regulation of sodium ion transmembrane transporter activity(GO:2000650) |
| 0.0 | 0.7 | GO:0018216 | peptidyl-arginine methylation(GO:0018216) |
| 0.0 | 0.3 | GO:0021523 | somatic motor neuron differentiation(GO:0021523) |
| 0.0 | 0.7 | GO:0045920 | negative regulation of exocytosis(GO:0045920) |
| 0.0 | 0.1 | GO:0006089 | lactate metabolic process(GO:0006089) |
| 0.0 | 0.1 | GO:0033615 | mitochondrial proton-transporting ATP synthase complex assembly(GO:0033615) |
| 0.0 | 1.4 | GO:0042149 | cellular response to glucose starvation(GO:0042149) |
| 0.0 | 0.4 | GO:0031998 | regulation of fatty acid beta-oxidation(GO:0031998) |
| 0.0 | 1.1 | GO:0016185 | synaptic vesicle budding from presynaptic endocytic zone membrane(GO:0016185) |
| 0.0 | 0.1 | GO:0070125 | mitochondrial translational elongation(GO:0070125) |
| 0.0 | 0.2 | GO:0006788 | heme oxidation(GO:0006788) |
| 0.0 | 0.1 | GO:0070358 | actin polymerization-dependent cell motility(GO:0070358) |
| 0.0 | 0.2 | GO:0098529 | neuromuscular junction development, skeletal muscle fiber(GO:0098529) |
| 0.0 | 0.2 | GO:0019375 | galactosylceramide biosynthetic process(GO:0006682) galactolipid biosynthetic process(GO:0019375) |
| 0.0 | 0.2 | GO:0016572 | histone phosphorylation(GO:0016572) |
| 0.0 | 0.4 | GO:0031114 | regulation of microtubule depolymerization(GO:0031114) |
| 0.0 | 0.7 | GO:0048936 | peripheral nervous system neuron axonogenesis(GO:0048936) |
| 0.0 | 1.7 | GO:0071277 | cellular response to calcium ion(GO:0071277) |
| 0.0 | 0.2 | GO:0060394 | negative regulation of pathway-restricted SMAD protein phosphorylation(GO:0060394) |
| 0.0 | 0.1 | GO:1902410 | mitotic cytokinetic process(GO:1902410) mitotic cleavage furrow formation(GO:1903673) |
| 0.0 | 0.8 | GO:0030168 | platelet activation(GO:0030168) |
| 0.0 | 0.1 | GO:1901492 | positive regulation of lymphangiogenesis(GO:1901492) |
| 0.0 | 0.3 | GO:0006122 | mitochondrial electron transport, ubiquinol to cytochrome c(GO:0006122) |
| 0.0 | 0.1 | GO:0048795 | swim bladder morphogenesis(GO:0048795) |
| 0.0 | 0.4 | GO:0006123 | mitochondrial electron transport, cytochrome c to oxygen(GO:0006123) |
| 0.0 | 0.1 | GO:0090069 | regulation of ribosome biogenesis(GO:0090069) |
| 0.0 | 0.4 | GO:0015937 | coenzyme A biosynthetic process(GO:0015937) |
| 0.0 | 0.5 | GO:0048026 | positive regulation of mRNA splicing, via spliceosome(GO:0048026) |
| 0.0 | 0.4 | GO:0007158 | neuron cell-cell adhesion(GO:0007158) |
| 0.0 | 0.1 | GO:0045041 | protein import into mitochondrial intermembrane space(GO:0045041) |
| 0.0 | 0.4 | GO:1902307 | positive regulation of sodium ion transport(GO:0010765) positive regulation of sodium ion transmembrane transport(GO:1902307) regulation of potassium ion import(GO:1903286) positive regulation of potassium ion import(GO:1903288) positive regulation of sodium ion transmembrane transporter activity(GO:2000651) |
| 0.0 | 0.2 | GO:0006465 | signal peptide processing(GO:0006465) |
| 0.0 | 0.4 | GO:0048769 | sarcomerogenesis(GO:0048769) |
| 0.0 | 0.1 | GO:0006844 | acyl carnitine transport(GO:0006844) |
| 0.0 | 0.1 | GO:0045724 | positive regulation of cilium assembly(GO:0045724) |
| 0.0 | 0.1 | GO:0015988 | energy coupled proton transmembrane transport, against electrochemical gradient(GO:0015988) electron transport coupled proton transport(GO:0015990) |
| 0.0 | 0.3 | GO:0035860 | esophagus smooth muscle contraction(GO:0014846) glial cell-derived neurotrophic factor receptor signaling pathway(GO:0035860) |
| 0.0 | 0.1 | GO:0032218 | riboflavin transport(GO:0032218) |
| 0.0 | 0.8 | GO:0035118 | embryonic pectoral fin morphogenesis(GO:0035118) |
| 0.0 | 0.3 | GO:0006596 | polyamine biosynthetic process(GO:0006596) |
| 0.0 | 0.5 | GO:1902285 | semaphorin-plexin signaling pathway involved in neuron projection guidance(GO:1902285) semaphorin-plexin signaling pathway involved in axon guidance(GO:1902287) |
| 0.0 | 1.1 | GO:0071391 | cellular response to estrogen stimulus(GO:0071391) |
| 0.0 | 0.2 | GO:0070572 | positive regulation of axon regeneration(GO:0048680) positive regulation of neuron projection regeneration(GO:0070572) |
| 0.0 | 0.1 | GO:0002082 | regulation of oxidative phosphorylation(GO:0002082) |
| 0.0 | 0.1 | GO:0006740 | NADPH regeneration(GO:0006740) |
| 0.0 | 0.1 | GO:0060074 | synapse maturation(GO:0060074) |
| 0.0 | 0.1 | GO:0045023 | G0 to G1 transition(GO:0045023) regulation of G0 to G1 transition(GO:0070316) negative regulation of G0 to G1 transition(GO:0070317) |
| 0.0 | 0.1 | GO:1903964 | monounsaturated fatty acid metabolic process(GO:1903964) monounsaturated fatty acid biosynthetic process(GO:1903966) |
| 0.0 | 0.6 | GO:0016339 | calcium-dependent cell-cell adhesion via plasma membrane cell adhesion molecules(GO:0016339) |
| 0.0 | 0.1 | GO:1901380 | negative regulation of potassium ion transmembrane transporter activity(GO:1901017) negative regulation of potassium ion transmembrane transport(GO:1901380) negative regulation of delayed rectifier potassium channel activity(GO:1902260) negative regulation of voltage-gated potassium channel activity(GO:1903817) |
| 0.0 | 0.2 | GO:0006797 | polyphosphate metabolic process(GO:0006797) polyphosphate catabolic process(GO:0006798) |
| 0.0 | 0.1 | GO:0060986 | regulation of endocrine process(GO:0044060) positive regulation of circadian sleep/wake cycle, sleep(GO:0045938) endocrine hormone secretion(GO:0060986) |
| 0.0 | 0.2 | GO:0001964 | startle response(GO:0001964) |
| 0.0 | 0.1 | GO:0033212 | iron assimilation(GO:0033212) |
| 0.0 | 0.1 | GO:0032978 | protein insertion into membrane from inner side(GO:0032978) protein insertion into mitochondrial membrane from inner side(GO:0032979) protein insertion into mitochondrial membrane(GO:0051204) |
| 0.0 | 0.4 | GO:0015701 | bicarbonate transport(GO:0015701) |
| 0.0 | 0.6 | GO:0006119 | oxidative phosphorylation(GO:0006119) |
| 0.0 | 0.1 | GO:0050847 | progesterone receptor signaling pathway(GO:0050847) |
| 0.0 | 0.1 | GO:0060976 | coronary vasculature development(GO:0060976) |
| 0.0 | 0.2 | GO:0008089 | anterograde axonal transport(GO:0008089) |
| 0.0 | 0.1 | GO:0003321 | positive regulation of heart rate by epinephrine-norepinephrine(GO:0001996) positive regulation of blood pressure by epinephrine-norepinephrine(GO:0003321) positive regulation of heart rate(GO:0010460) |
| 0.0 | 0.1 | GO:0071684 | hatching(GO:0035188) organism emergence from protective structure(GO:0071684) |
| 0.0 | 0.2 | GO:0060142 | regulation of syncytium formation by plasma membrane fusion(GO:0060142) |
| 0.0 | 0.1 | GO:0032732 | positive regulation of interleukin-1 production(GO:0032732) |
| 0.0 | 0.2 | GO:0071816 | tail-anchored membrane protein insertion into ER membrane(GO:0071816) |
| 0.0 | 0.3 | GO:0016203 | muscle attachment(GO:0016203) |
| 0.0 | 0.6 | GO:0006096 | glycolytic process(GO:0006096) |
| 0.0 | 0.1 | GO:0097198 | histone H3-K36 trimethylation(GO:0097198) |
| 0.0 | 0.4 | GO:0002027 | regulation of heart rate(GO:0002027) |
| 0.0 | 0.4 | GO:0015986 | energy coupled proton transport, down electrochemical gradient(GO:0015985) ATP synthesis coupled proton transport(GO:0015986) |
| 0.0 | 0.0 | GO:0051228 | mitotic spindle disassembly(GO:0051228) spindle disassembly(GO:0051230) |
| 0.0 | 0.2 | GO:0007274 | neuromuscular synaptic transmission(GO:0007274) |
| 0.0 | 0.1 | GO:1903292 | protein localization to Golgi membrane(GO:1903292) |
| 0.0 | 0.2 | GO:0006003 | fructose 2,6-bisphosphate metabolic process(GO:0006003) |
| 0.0 | 0.2 | GO:0042026 | protein refolding(GO:0042026) |
| 0.0 | 0.0 | GO:0070129 | regulation of mitochondrial translation(GO:0070129) positive regulation of mitochondrial translation(GO:0070131) |
| 0.0 | 0.1 | GO:0007197 | adenylate cyclase-inhibiting G-protein coupled acetylcholine receptor signaling pathway(GO:0007197) |
| 0.0 | 0.5 | GO:0035025 | positive regulation of Rho protein signal transduction(GO:0035025) |
| 0.0 | 0.3 | GO:0032233 | positive regulation of actin filament bundle assembly(GO:0032233) |
| 0.0 | 0.1 | GO:0048069 | eye pigmentation(GO:0048069) |
| 0.0 | 0.1 | GO:0035094 | response to nicotine(GO:0035094) |
| 0.0 | 0.0 | GO:0070070 | proton-transporting V-type ATPase complex assembly(GO:0070070) vacuolar proton-transporting V-type ATPase complex assembly(GO:0070072) |
| 0.0 | 0.2 | GO:0099590 | neurotransmitter receptor internalization(GO:0099590) |
| 0.0 | 0.0 | GO:0051660 | establishment of centrosome localization(GO:0051660) |
| 0.0 | 0.2 | GO:0043550 | regulation of lipid kinase activity(GO:0043550) regulation of phosphatidylinositol 3-kinase activity(GO:0043551) |
| 0.0 | 0.3 | GO:0051639 | actin filament network formation(GO:0051639) |
| 0.0 | 0.1 | GO:0006189 | 'de novo' IMP biosynthetic process(GO:0006189) |
| 0.0 | 0.3 | GO:0050650 | chondroitin sulfate proteoglycan biosynthetic process(GO:0050650) |
| 0.0 | 0.1 | GO:0044805 | late nucleophagy(GO:0044805) |
| 0.0 | 0.4 | GO:0018230 | peptidyl-L-cysteine S-palmitoylation(GO:0018230) peptidyl-S-diacylglycerol-L-cysteine biosynthetic process from peptidyl-cysteine(GO:0018231) |
| 0.0 | 0.0 | GO:0007509 | mesoderm migration involved in gastrulation(GO:0007509) |
| 0.0 | 0.2 | GO:0006144 | purine nucleobase metabolic process(GO:0006144) |
| 0.0 | 0.3 | GO:0035622 | intrahepatic bile duct development(GO:0035622) |
| 0.0 | 0.3 | GO:0070536 | protein K63-linked deubiquitination(GO:0070536) |
| 0.0 | 0.4 | GO:0045332 | phospholipid translocation(GO:0045332) |
| 0.0 | 0.3 | GO:0019226 | transmission of nerve impulse(GO:0019226) |
| 0.0 | 0.2 | GO:0097031 | NADH dehydrogenase complex assembly(GO:0010257) mitochondrial respiratory chain complex I assembly(GO:0032981) mitochondrial respiratory chain complex I biogenesis(GO:0097031) |
| 0.0 | 0.1 | GO:0035735 | intraciliary transport involved in cilium morphogenesis(GO:0035735) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 0.8 | GO:0098753 | intrinsic component of the cytoplasmic side of the plasma membrane(GO:0031235) anchored component of the cytoplasmic side of the plasma membrane(GO:0098753) |
| 0.3 | 0.8 | GO:1902560 | GMP reductase complex(GO:1902560) |
| 0.1 | 0.8 | GO:0070032 | synaptobrevin 2-SNAP-25-syntaxin-1a-complexin I complex(GO:0070032) |
| 0.1 | 0.8 | GO:0005967 | mitochondrial pyruvate dehydrogenase complex(GO:0005967) |
| 0.1 | 0.8 | GO:0030877 | beta-catenin destruction complex(GO:0030877) |
| 0.1 | 0.5 | GO:0045257 | mitochondrial respiratory chain complex II, succinate dehydrogenase complex (ubiquinone)(GO:0005749) succinate dehydrogenase complex (ubiquinone)(GO:0045257) respiratory chain complex II(GO:0045273) succinate dehydrogenase complex(GO:0045281) fumarate reductase complex(GO:0045283) |
| 0.1 | 0.5 | GO:0008091 | spectrin(GO:0008091) |
| 0.1 | 0.9 | GO:0098835 | presynaptic endocytic zone(GO:0098833) presynaptic endocytic zone membrane(GO:0098835) extrinsic component of presynaptic membrane(GO:0098888) extrinsic component of presynaptic endocytic zone(GO:0098894) |
| 0.1 | 0.3 | GO:0008247 | 1-alkyl-2-acetylglycerophosphocholine esterase complex(GO:0008247) |
| 0.1 | 0.3 | GO:0098553 | integral component of cytoplasmic side of endoplasmic reticulum membrane(GO:0071458) integral component of lumenal side of endoplasmic reticulum membrane(GO:0071556) lumenal side of endoplasmic reticulum membrane(GO:0098553) |
| 0.1 | 0.2 | GO:0043291 | RAVE complex(GO:0043291) |
| 0.0 | 0.3 | GO:0000221 | vacuolar proton-transporting V-type ATPase, V1 domain(GO:0000221) |
| 0.0 | 0.4 | GO:0005945 | 6-phosphofructokinase complex(GO:0005945) |
| 0.0 | 0.3 | GO:0042583 | chromaffin granule(GO:0042583) |
| 0.0 | 0.2 | GO:0045261 | proton-transporting ATP synthase complex, catalytic core F(1)(GO:0045261) |
| 0.0 | 0.5 | GO:0032591 | dendritic spine membrane(GO:0032591) |
| 0.0 | 0.3 | GO:0045275 | mitochondrial respiratory chain complex III(GO:0005750) respiratory chain complex III(GO:0045275) |
| 0.0 | 0.5 | GO:0030315 | T-tubule(GO:0030315) |
| 0.0 | 0.7 | GO:0031594 | neuromuscular junction(GO:0031594) |
| 0.0 | 0.2 | GO:0071818 | BAT3 complex(GO:0071818) |
| 0.0 | 0.5 | GO:0002116 | semaphorin receptor complex(GO:0002116) |
| 0.0 | 0.1 | GO:0044609 | DBIRD complex(GO:0044609) |
| 0.0 | 0.2 | GO:0061617 | MICOS complex(GO:0061617) |
| 0.0 | 0.3 | GO:0002178 | palmitoyltransferase complex(GO:0002178) |
| 0.0 | 2.1 | GO:0016459 | myosin complex(GO:0016459) |
| 0.0 | 0.2 | GO:0048179 | activin receptor complex(GO:0048179) |
| 0.0 | 0.2 | GO:0070449 | elongin complex(GO:0070449) |
| 0.0 | 0.3 | GO:0043256 | basal lamina(GO:0005605) laminin complex(GO:0043256) |
| 0.0 | 0.2 | GO:0031209 | SCAR complex(GO:0031209) |
| 0.0 | 0.4 | GO:0098831 | cytoskeleton of presynaptic active zone(GO:0048788) presynaptic active zone cytoplasmic component(GO:0098831) |
| 0.0 | 0.0 | GO:0060171 | stereocilium membrane(GO:0060171) |
| 0.0 | 0.2 | GO:0005892 | acetylcholine-gated channel complex(GO:0005892) |
| 0.0 | 0.2 | GO:0098844 | postsynaptic endocytic zone membrane(GO:0098844) |
| 0.0 | 0.8 | GO:0005746 | mitochondrial respiratory chain(GO:0005746) |
| 0.0 | 0.3 | GO:0035371 | microtubule plus-end(GO:0035371) |
| 0.0 | 0.2 | GO:0032809 | neuronal cell body membrane(GO:0032809) cell body membrane(GO:0044298) |
| 0.0 | 0.1 | GO:0045277 | respiratory chain complex IV(GO:0045277) |
| 0.0 | 0.1 | GO:0005744 | mitochondrial inner membrane presequence translocase complex(GO:0005744) |
| 0.0 | 0.2 | GO:0000276 | mitochondrial proton-transporting ATP synthase complex, coupling factor F(o)(GO:0000276) |
| 0.0 | 0.3 | GO:0045180 | basal cortex(GO:0045180) |
| 0.0 | 0.1 | GO:0043034 | costamere(GO:0043034) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 0.8 | GO:0003920 | GMP reductase activity(GO:0003920) oxidoreductase activity, acting on NAD(P)H, nitrogenous group as acceptor(GO:0016657) |
| 0.2 | 0.7 | GO:0050135 | NAD(P)+ nucleosidase activity(GO:0050135) |
| 0.2 | 0.7 | GO:0008929 | triose-phosphate isomerase activity(GO:0004807) methylglyoxal synthase activity(GO:0008929) |
| 0.2 | 0.8 | GO:0004739 | pyruvate dehydrogenase (acetyl-transferring) activity(GO:0004739) |
| 0.1 | 0.4 | GO:0000104 | succinate dehydrogenase activity(GO:0000104) |
| 0.1 | 0.9 | GO:0004332 | fructose-bisphosphate aldolase activity(GO:0004332) |
| 0.1 | 0.8 | GO:0035242 | protein-arginine omega-N asymmetric methyltransferase activity(GO:0035242) |
| 0.1 | 0.3 | GO:0072591 | N-acetyl-L-aspartate-L-glutamate ligase activity(GO:0072590) citrate-L-glutamate ligase activity(GO:0072591) |
| 0.1 | 0.4 | GO:0043891 | glyceraldehyde-3-phosphate dehydrogenase (NAD+) (phosphorylating) activity(GO:0004365) glyceraldehyde-3-phosphate dehydrogenase (NAD(P)+) (phosphorylating) activity(GO:0043891) |
| 0.1 | 0.6 | GO:0004966 | galanin receptor activity(GO:0004966) |
| 0.1 | 0.3 | GO:0052656 | branched-chain-amino-acid transaminase activity(GO:0004084) L-leucine transaminase activity(GO:0052654) L-valine transaminase activity(GO:0052655) L-isoleucine transaminase activity(GO:0052656) |
| 0.1 | 0.5 | GO:0050262 | ribosylnicotinamide kinase activity(GO:0050262) |
| 0.1 | 0.5 | GO:0004095 | carnitine O-palmitoyltransferase activity(GO:0004095) O-palmitoyltransferase activity(GO:0016416) |
| 0.1 | 0.9 | GO:0005545 | 1-phosphatidylinositol binding(GO:0005545) |
| 0.1 | 0.3 | GO:0004082 | bisphosphoglycerate mutase activity(GO:0004082) phosphoglycerate mutase activity(GO:0004619) |
| 0.1 | 0.4 | GO:0004594 | pantothenate kinase activity(GO:0004594) |
| 0.1 | 0.4 | GO:0004457 | lactate dehydrogenase activity(GO:0004457) |
| 0.0 | 0.2 | GO:0004069 | L-aspartate:2-oxoglutarate aminotransferase activity(GO:0004069) |
| 0.0 | 0.4 | GO:0015467 | G-protein activated inward rectifier potassium channel activity(GO:0015467) |
| 0.0 | 0.2 | GO:0070412 | R-SMAD binding(GO:0070412) |
| 0.0 | 0.7 | GO:0048495 | Roundabout binding(GO:0048495) |
| 0.0 | 0.2 | GO:0050998 | nitric-oxide synthase binding(GO:0050998) |
| 0.0 | 0.2 | GO:0008281 | sulfonylurea receptor activity(GO:0008281) |
| 0.0 | 0.2 | GO:0070891 | lipoteichoic acid binding(GO:0070891) |
| 0.0 | 0.4 | GO:0070095 | 6-phosphofructokinase activity(GO:0003872) fructose-6-phosphate binding(GO:0070095) |
| 0.0 | 0.3 | GO:0016286 | small conductance calcium-activated potassium channel activity(GO:0016286) |
| 0.0 | 0.3 | GO:0035252 | UDP-xylosyltransferase activity(GO:0035252) |
| 0.0 | 0.2 | GO:0004392 | heme oxygenase (decyclizing) activity(GO:0004392) |
| 0.0 | 0.2 | GO:0016361 | activin receptor activity, type I(GO:0016361) |
| 0.0 | 0.1 | GO:0030273 | melanin-concentrating hormone receptor activity(GO:0030273) |
| 0.0 | 0.2 | GO:0004743 | pyruvate kinase activity(GO:0004743) |
| 0.0 | 0.3 | GO:0042500 | aspartic endopeptidase activity, intramembrane cleaving(GO:0042500) |
| 0.0 | 0.9 | GO:0030165 | PDZ domain binding(GO:0030165) |
| 0.0 | 0.4 | GO:0008510 | sodium:bicarbonate symporter activity(GO:0008510) |
| 0.0 | 0.1 | GO:0003844 | 1,4-alpha-glucan branching enzyme activity(GO:0003844) |
| 0.0 | 0.1 | GO:0047777 | (3S)-citramalyl-CoA lyase activity(GO:0047777) |
| 0.0 | 0.5 | GO:0008301 | DNA binding, bending(GO:0008301) |
| 0.0 | 0.6 | GO:0001540 | beta-amyloid binding(GO:0001540) |
| 0.0 | 0.3 | GO:0070001 | aspartic-type endopeptidase activity(GO:0004190) aspartic-type peptidase activity(GO:0070001) |
| 0.0 | 0.4 | GO:0019870 | chloride channel inhibitor activity(GO:0019869) potassium channel inhibitor activity(GO:0019870) |
| 0.0 | 0.7 | GO:0016676 | cytochrome-c oxidase activity(GO:0004129) heme-copper terminal oxidase activity(GO:0015002) oxidoreductase activity, acting on a heme group of donors(GO:0016675) oxidoreductase activity, acting on a heme group of donors, oxygen as acceptor(GO:0016676) |
| 0.0 | 0.1 | GO:0015227 | acyl carnitine transmembrane transporter activity(GO:0015227) |
| 0.0 | 0.5 | GO:0050811 | GABA receptor binding(GO:0050811) |
| 0.0 | 0.4 | GO:0098882 | structural constituent of presynaptic active zone(GO:0098882) |
| 0.0 | 0.1 | GO:0032217 | riboflavin transporter activity(GO:0032217) |
| 0.0 | 0.5 | GO:0008137 | NADH dehydrogenase (ubiquinone) activity(GO:0008137) NADH dehydrogenase (quinone) activity(GO:0050136) |
| 0.0 | 0.1 | GO:0008746 | NAD(P)+ transhydrogenase activity(GO:0008746) oxidoreductase activity, acting on NAD(P)H, NAD(P) as acceptor(GO:0016652) |
| 0.0 | 0.1 | GO:0050508 | glucuronosyl-N-acetylglucosaminyl-proteoglycan 4-alpha-N-acetylglucosaminyltransferase activity(GO:0050508) |
| 0.0 | 0.3 | GO:0070840 | dynein complex binding(GO:0070840) |
| 0.0 | 0.4 | GO:0030552 | cAMP binding(GO:0030552) |
| 0.0 | 0.2 | GO:0008273 | calcium, potassium:sodium antiporter activity(GO:0008273) |
| 0.0 | 0.1 | GO:0016215 | stearoyl-CoA 9-desaturase activity(GO:0004768) acyl-CoA desaturase activity(GO:0016215) palmitoyl-CoA 9-desaturase activity(GO:0032896) |
| 0.0 | 0.1 | GO:0016842 | amidine-lyase activity(GO:0016842) |
| 0.0 | 0.8 | GO:0017075 | syntaxin-1 binding(GO:0017075) |
| 0.0 | 0.2 | GO:0047676 | arachidonate-CoA ligase activity(GO:0047676) |
| 0.0 | 0.2 | GO:0004309 | exopolyphosphatase activity(GO:0004309) |
| 0.0 | 0.2 | GO:0015129 | lactate transmembrane transporter activity(GO:0015129) |
| 0.0 | 0.7 | GO:0005540 | hyaluronic acid binding(GO:0005540) |
| 0.0 | 0.1 | GO:0048039 | ubiquinone binding(GO:0048039) |
| 0.0 | 0.9 | GO:0001786 | phosphatidylserine binding(GO:0001786) |
| 0.0 | 0.8 | GO:0008013 | beta-catenin binding(GO:0008013) |
| 0.0 | 0.1 | GO:0015086 | cadmium ion transmembrane transporter activity(GO:0015086) |
| 0.0 | 0.3 | GO:0008449 | N-acetylglucosamine-6-sulfatase activity(GO:0008449) |
| 0.0 | 0.8 | GO:0004683 | calmodulin-dependent protein kinase activity(GO:0004683) |
| 0.0 | 0.1 | GO:0008131 | primary amine oxidase activity(GO:0008131) |
| 0.0 | 0.2 | GO:0005332 | gamma-aminobutyric acid:sodium symporter activity(GO:0005332) |
| 0.0 | 0.5 | GO:0017154 | semaphorin receptor activity(GO:0017154) |
| 0.0 | 0.1 | GO:0015450 | P-P-bond-hydrolysis-driven protein transmembrane transporter activity(GO:0015450) |
| 0.0 | 0.1 | GO:0016922 | ligand-dependent nuclear receptor binding(GO:0016922) |
| 0.0 | 0.1 | GO:0016421 | CoA carboxylase activity(GO:0016421) |
| 0.0 | 0.1 | GO:0004937 | alpha1-adrenergic receptor activity(GO:0004937) |
| 0.0 | 0.1 | GO:0030504 | inorganic diphosphate transmembrane transporter activity(GO:0030504) |
| 0.0 | 0.4 | GO:0051787 | misfolded protein binding(GO:0051787) |
| 0.0 | 0.2 | GO:0003873 | 6-phosphofructo-2-kinase activity(GO:0003873) phosphofructokinase activity(GO:0008443) |
| 0.0 | 0.1 | GO:0004449 | isocitrate dehydrogenase (NAD+) activity(GO:0004449) |
| 0.0 | 0.8 | GO:0044325 | ion channel binding(GO:0044325) |
| 0.0 | 0.1 | GO:0004977 | melanocortin receptor activity(GO:0004977) |
| 0.0 | 0.3 | GO:0010181 | FMN binding(GO:0010181) |
| 0.0 | 0.8 | GO:0005544 | calcium-dependent phospholipid binding(GO:0005544) |
| 0.0 | 0.1 | GO:0003747 | translation release factor activity(GO:0003747) translation termination factor activity(GO:0008079) |
| 0.0 | 0.1 | GO:0016681 | ubiquinol-cytochrome-c reductase activity(GO:0008121) oxidoreductase activity, acting on diphenols and related substances as donors, cytochrome as acceptor(GO:0016681) |
| 0.0 | 0.1 | GO:0035613 | RNA stem-loop binding(GO:0035613) |
| 0.0 | 0.0 | GO:0017081 | chloride channel regulator activity(GO:0017081) |
| 0.0 | 0.1 | GO:0004706 | JUN kinase kinase kinase activity(GO:0004706) |
| 0.0 | 0.3 | GO:0046961 | proton-transporting ATPase activity, rotational mechanism(GO:0046961) |
| 0.0 | 0.2 | GO:0030544 | Hsp70 protein binding(GO:0030544) |
| 0.0 | 0.2 | GO:0003841 | 1-acylglycerol-3-phosphate O-acyltransferase activity(GO:0003841) |
| 0.0 | 0.1 | GO:0004687 | myosin light chain kinase activity(GO:0004687) |
| 0.0 | 0.1 | GO:0016907 | G-protein coupled acetylcholine receptor activity(GO:0016907) |
| 0.0 | 0.2 | GO:0061578 | Lys63-specific deubiquitinase activity(GO:0061578) |
| 0.0 | 0.1 | GO:0005522 | profilin binding(GO:0005522) |
| 0.0 | 0.0 | GO:0071889 | 14-3-3 protein binding(GO:0071889) |
| 0.0 | 0.1 | GO:0031419 | cobalamin binding(GO:0031419) |
| 0.0 | 0.2 | GO:0050291 | sphingosine N-acyltransferase activity(GO:0050291) |
| 0.0 | 0.3 | GO:0051010 | microtubule plus-end binding(GO:0051010) |
| 0.0 | 0.1 | GO:0010997 | anaphase-promoting complex binding(GO:0010997) ubiquitin ligase activator activity(GO:1990757) |
| 0.0 | 0.1 | GO:0034596 | phosphatidylinositol phosphate 4-phosphatase activity(GO:0034596) |
| 0.0 | 0.1 | GO:0080019 | fatty-acyl-CoA reductase (alcohol-forming) activity(GO:0080019) |
| 0.0 | 0.1 | GO:0032977 | membrane insertase activity(GO:0032977) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.4 | PID SYNDECAN 3 PATHWAY | Syndecan-3-mediated signaling events |
| 0.0 | 0.5 | ST MYOCYTE AD PATHWAY | Myocyte Adrenergic Pathway is a specific case of the generalized Adrenergic Pathway. |
| 0.0 | 0.5 | PID WNT SIGNALING PATHWAY | Wnt signaling network |
| 0.0 | 0.4 | PID REELIN PATHWAY | Reelin signaling pathway |
| 0.0 | 0.3 | PID RETINOIC ACID PATHWAY | Retinoic acid receptors-mediated signaling |
| 0.0 | 0.4 | PID INTEGRIN3 PATHWAY | Beta3 integrin cell surface interactions |
| 0.0 | 0.1 | ST GRANULE CELL SURVIVAL PATHWAY | Granule Cell Survival Pathway is a specific case of more general PAC1 Receptor Pathway. |
| 0.0 | 0.2 | PID TRAIL PATHWAY | TRAIL signaling pathway |
| 0.0 | 0.7 | PID P75 NTR PATHWAY | p75(NTR)-mediated signaling |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.8 | REACTOME PURINE SALVAGE | Genes involved in Purine salvage |
| 0.0 | 0.5 | REACTOME EICOSANOID LIGAND BINDING RECEPTORS | Genes involved in Eicosanoid ligand-binding receptors |
| 0.0 | 0.5 | REACTOME METABOLISM OF POLYAMINES | Genes involved in Metabolism of polyamines |
| 0.0 | 0.1 | REACTOME CYTOSOLIC SULFONATION OF SMALL MOLECULES | Genes involved in Cytosolic sulfonation of small molecules |
| 0.0 | 0.5 | REACTOME INTERACTION BETWEEN L1 AND ANKYRINS | Genes involved in Interaction between L1 and Ankyrins |
| 0.0 | 1.9 | REACTOME RESPIRATORY ELECTRON TRANSPORT | Genes involved in Respiratory electron transport |
| 0.0 | 0.6 | REACTOME PYRUVATE METABOLISM | Genes involved in Pyruvate metabolism |
| 0.0 | 0.1 | REACTOME DOPAMINE NEUROTRANSMITTER RELEASE CYCLE | Genes involved in Dopamine Neurotransmitter Release Cycle |
| 0.0 | 0.3 | REACTOME DESTABILIZATION OF MRNA BY TRISTETRAPROLIN TTP | Genes involved in Destabilization of mRNA by Tristetraprolin (TTP) |
| 0.0 | 0.3 | REACTOME INTRINSIC PATHWAY | Genes involved in Intrinsic Pathway |
| 0.0 | 0.3 | REACTOME CTNNB1 PHOSPHORYLATION CASCADE | Genes involved in Beta-catenin phosphorylation cascade |
| 0.0 | 0.4 | REACTOME BRANCHED CHAIN AMINO ACID CATABOLISM | Genes involved in Branched-chain amino acid catabolism |
| 0.0 | 0.5 | REACTOME INWARDLY RECTIFYING K CHANNELS | Genes involved in Inwardly rectifying K+ channels |
| 0.0 | 0.3 | REACTOME YAP1 AND WWTR1 TAZ STIMULATED GENE EXPRESSION | Genes involved in YAP1- and WWTR1 (TAZ)-stimulated gene expression |
| 0.0 | 0.2 | REACTOME IL 7 SIGNALING | Genes involved in Interleukin-7 signaling |
| 0.0 | 0.2 | REACTOME IL 6 SIGNALING | Genes involved in Interleukin-6 signaling |
| 0.0 | 0.1 | REACTOME DCC MEDIATED ATTRACTIVE SIGNALING | Genes involved in DCC mediated attractive signaling |
| 0.0 | 0.3 | REACTOME PYRUVATE METABOLISM AND CITRIC ACID TCA CYCLE | Genes involved in Pyruvate metabolism and Citric Acid (TCA) cycle |
| 0.0 | 0.1 | REACTOME SYNTHESIS SECRETION AND INACTIVATION OF GIP | Genes involved in Synthesis, Secretion, and Inactivation of Glucose-dependent Insulinotropic Polypeptide (GIP) |
| 0.0 | 0.2 | REACTOME TRAFFICKING OF GLUR2 CONTAINING AMPA RECEPTORS | Genes involved in Trafficking of GluR2-containing AMPA receptors |
| 0.0 | 0.5 | REACTOME MITOCHONDRIAL PROTEIN IMPORT | Genes involved in Mitochondrial Protein Import |