Project
PRJNA207719: Tissue specific transcriptome profiling
Navigation
Downloads
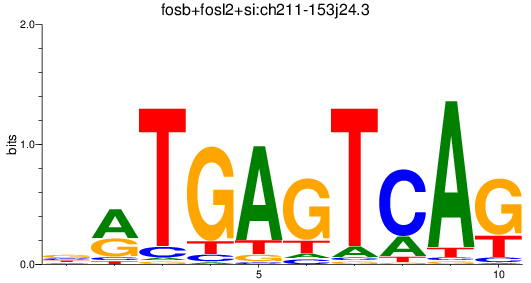
Results for fosb+fosl2+si:ch211-153j24.3
Z-value: 1.29
Transcription factors associated with fosb+fosl2+si:ch211-153j24.3
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
fosl2
|
ENSDARG00000040623 | fos-like antigen 2 |
|
fosb
|
ENSDARG00000055751 | FBJ murine osteosarcoma viral oncogene homolog B |
|
si_ch211-153j24.3
|
ENSDARG00000068428 | si_ch211-153j24.3 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| si:ch211-153j24.3 | dr11_v1_chr20_+_46544052_46544052 | -0.97 | 6.7e-03 | Click! |
| fosb | dr11_v1_chr18_+_36770166_36770166 | -0.89 | 4.2e-02 | Click! |
| fosl2 | dr11_v1_chr17_+_41302660_41302660 | -0.76 | 1.4e-01 | Click! |
Activity profile of fosb+fosl2+si:ch211-153j24.3 motif
Sorted Z-values of fosb+fosl2+si:ch211-153j24.3 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr11_-_18253111 | 1.01 |
ENSDART00000125984
|
mustn1b
|
musculoskeletal, embryonic nuclear protein 1b |
| chr25_-_13408760 | 0.96 |
ENSDART00000154445
|
gins3
|
GINS complex subunit 3 |
| chr19_+_48359259 | 0.91 |
ENSDART00000167353
|
sgo1
|
shugoshin 1 |
| chr25_-_22187397 | 0.82 |
ENSDART00000123211
ENSDART00000139110 |
pkp3a
|
plakophilin 3a |
| chr12_+_25945560 | 0.80 |
ENSDART00000109799
|
mmrn2b
|
multimerin 2b |
| chr13_+_24280380 | 0.75 |
ENSDART00000184115
|
acta1b
|
actin, alpha 1b, skeletal muscle |
| chr21_-_2415808 | 0.72 |
ENSDART00000171179
|
si:ch211-241b2.5
|
si:ch211-241b2.5 |
| chr6_-_35439406 | 0.70 |
ENSDART00000073784
|
rgs5a
|
regulator of G protein signaling 5a |
| chr16_-_22930925 | 0.67 |
ENSDART00000133819
|
si:dkey-246i14.3
|
si:dkey-246i14.3 |
| chr25_+_13191391 | 0.67 |
ENSDART00000109937
|
si:ch211-147m6.2
|
si:ch211-147m6.2 |
| chr16_+_23403602 | 0.64 |
ENSDART00000159848
|
s100w
|
S100 calcium binding protein W |
| chr6_+_51713076 | 0.63 |
ENSDART00000146281
|
ripor3
|
RIPOR family member 3 |
| chr17_-_53359028 | 0.57 |
ENSDART00000185218
|
CABZ01068208.1
|
|
| chr19_+_15441022 | 0.57 |
ENSDART00000098970
ENSDART00000140276 |
lin28a
|
lin-28 homolog A (C. elegans) |
| chr13_-_37122217 | 0.57 |
ENSDART00000133242
|
syne2b
|
spectrin repeat containing, nuclear envelope 2b |
| chr19_+_4061699 | 0.56 |
ENSDART00000158309
ENSDART00000166512 |
btr25
btr26
|
bloodthirsty-related gene family, member 25 bloodthirsty-related gene family, member 26 |
| chr15_+_963292 | 0.56 |
ENSDART00000156586
|
alox5b.2
|
arachidonate 5-lipoxygenase b, tandem duplicate 2 |
| chr14_-_11456724 | 0.54 |
ENSDART00000110424
|
si:ch211-153b23.4
|
si:ch211-153b23.4 |
| chr18_-_5595546 | 0.53 |
ENSDART00000191825
|
cyp1a
|
cytochrome P450, family 1, subfamily A |
| chr5_+_24089334 | 0.52 |
ENSDART00000183748
|
tp53
|
tumor protein p53 |
| chr23_+_4324625 | 0.52 |
ENSDART00000146302
ENSDART00000136792 ENSDART00000135027 ENSDART00000179819 |
sgk2a
|
serum/glucocorticoid regulated kinase 2a |
| chr5_+_9037650 | 0.51 |
ENSDART00000158226
|
si:ch211-155m12.1
|
si:ch211-155m12.1 |
| chr5_-_25582721 | 0.51 |
ENSDART00000123986
|
anxa1a
|
annexin A1a |
| chr22_-_15593824 | 0.51 |
ENSDART00000123125
|
tpm4a
|
tropomyosin 4a |
| chr7_-_32981559 | 0.49 |
ENSDART00000175614
|
pkp3b
|
plakophilin 3b |
| chr4_+_77971104 | 0.49 |
ENSDART00000188609
|
zgc:113921
|
zgc:113921 |
| chr4_+_77735212 | 0.47 |
ENSDART00000160716
|
si:dkey-238k10.1
|
si:dkey-238k10.1 |
| chr13_-_15982707 | 0.46 |
ENSDART00000186911
ENSDART00000181072 |
ikzf1
|
IKAROS family zinc finger 1 (Ikaros) |
| chr25_+_13191615 | 0.46 |
ENSDART00000168849
|
si:ch211-147m6.2
|
si:ch211-147m6.2 |
| chr7_-_44963154 | 0.45 |
ENSDART00000073735
|
rrad
|
Ras-related associated with diabetes |
| chr20_+_26880668 | 0.44 |
ENSDART00000077769
|
serpinb1
|
serpin peptidase inhibitor, clade B (ovalbumin), member 1 |
| chr15_-_38129845 | 0.43 |
ENSDART00000057095
|
si:dkey-24p1.1
|
si:dkey-24p1.1 |
| chr21_+_5636008 | 0.42 |
ENSDART00000158385
|
shroom3
|
shroom family member 3 |
| chr10_-_44560165 | 0.42 |
ENSDART00000181217
ENSDART00000076084 |
npm2b
|
nucleophosmin/nucleoplasmin, 2b |
| chr18_-_45617146 | 0.42 |
ENSDART00000146543
|
wt1b
|
wilms tumor 1b |
| chr18_+_50650512 | 0.42 |
ENSDART00000161022
|
si:dkey-151j17.4
|
si:dkey-151j17.4 |
| chr1_+_14253118 | 0.42 |
ENSDART00000161996
|
cxcl8a
|
chemokine (C-X-C motif) ligand 8a |
| chr3_-_34069637 | 0.41 |
ENSDART00000151588
|
ighv9-1
|
immunoglobulin heavy variable 9-1 |
| chr24_+_38522254 | 0.41 |
ENSDART00000156189
|
si:ch1073-66l23.1
|
si:ch1073-66l23.1 |
| chr6_-_24143923 | 0.41 |
ENSDART00000157948
|
si:ch73-389b16.1
|
si:ch73-389b16.1 |
| chr7_+_15736230 | 0.40 |
ENSDART00000109942
|
mctp2b
|
multiple C2 domains, transmembrane 2b |
| chr1_+_59073436 | 0.40 |
ENSDART00000161642
|
MFAP4 (1 of many)
|
si:zfos-2330d3.3 |
| chr13_-_33134611 | 0.40 |
ENSDART00000026280
|
plek2
|
pleckstrin 2 |
| chr8_+_6576940 | 0.40 |
ENSDART00000138135
|
vsig8b
|
V-set and immunoglobulin domain containing 8b |
| chr4_-_13518381 | 0.39 |
ENSDART00000067153
|
ifng1-1
|
interferon, gamma 1-1 |
| chr5_-_29531948 | 0.39 |
ENSDART00000098360
|
arrdc1a
|
arrestin domain containing 1a |
| chr7_+_33172066 | 0.39 |
ENSDART00000174013
|
si:ch211-194p6.12
|
si:ch211-194p6.12 |
| chr11_+_13629528 | 0.39 |
ENSDART00000186509
|
si:ch211-1a19.3
|
si:ch211-1a19.3 |
| chr24_+_30215475 | 0.38 |
ENSDART00000164717
|
si:ch73-358j7.2
|
si:ch73-358j7.2 |
| chr3_-_48259289 | 0.38 |
ENSDART00000160717
|
znf750
|
zinc finger protein 750 |
| chr21_+_11778823 | 0.38 |
ENSDART00000031786
|
glrx
|
glutaredoxin (thioltransferase) |
| chr17_+_21452258 | 0.38 |
ENSDART00000157098
|
pla2g4f.1
|
phospholipase A2, group IVF, tandem duplicate 1 |
| chr7_-_8417315 | 0.37 |
ENSDART00000173046
|
jac1
|
jacalin 1 |
| chr11_+_13630107 | 0.37 |
ENSDART00000172220
|
si:ch211-1a19.3
|
si:ch211-1a19.3 |
| chr3_-_34100700 | 0.37 |
ENSDART00000151628
|
ighv6-1
|
immunoglobulin heavy variable 6-1 |
| chr19_-_7420867 | 0.37 |
ENSDART00000081741
|
rab25a
|
RAB25, member RAS oncogene family a |
| chr14_-_1538600 | 0.37 |
ENSDART00000180925
|
CABZ01109480.1
|
|
| chr20_-_26532167 | 0.37 |
ENSDART00000061914
|
mthfd1l
|
methylenetetrahydrofolate dehydrogenase (NADP+ dependent) 1 like |
| chr16_-_51288178 | 0.36 |
ENSDART00000079864
|
zgc:173729
|
zgc:173729 |
| chr8_+_44358443 | 0.36 |
ENSDART00000189130
ENSDART00000189212 |
CU914776.2
|
|
| chr13_+_18533005 | 0.36 |
ENSDART00000136024
|
ftr14l
|
finTRIM family, member 14-like |
| chr14_-_34605607 | 0.36 |
ENSDART00000191608
|
afap1l1a
|
actin filament associated protein 1-like 1a |
| chr1_+_57787642 | 0.36 |
ENSDART00000127091
|
si:dkey-1c7.2
|
si:dkey-1c7.2 |
| chr2_-_41861040 | 0.35 |
ENSDART00000045763
|
keap1a
|
kelch-like ECH-associated protein 1a |
| chr2_+_31671545 | 0.35 |
ENSDART00000145446
|
ackr4a
|
atypical chemokine receptor 4a |
| chr4_+_77933084 | 0.35 |
ENSDART00000148728
|
pacsin2
|
protein kinase C and casein kinase substrate in neurons 2 |
| chr25_-_22191733 | 0.35 |
ENSDART00000067478
|
pkp3a
|
plakophilin 3a |
| chr22_-_17489040 | 0.35 |
ENSDART00000141286
|
si:ch211-197g15.6
|
si:ch211-197g15.6 |
| chr10_+_28428222 | 0.34 |
ENSDART00000135003
|
si:ch211-222e20.4
|
si:ch211-222e20.4 |
| chr4_+_16715267 | 0.34 |
ENSDART00000143849
|
pkp2
|
plakophilin 2 |
| chr13_-_51846224 | 0.34 |
ENSDART00000184663
|
LT631684.2
|
|
| chr22_+_19218733 | 0.33 |
ENSDART00000183212
ENSDART00000133595 |
si:dkey-21e2.7
|
si:dkey-21e2.7 |
| chr20_-_26531850 | 0.33 |
ENSDART00000183317
ENSDART00000131994 |
mthfd1l
|
methylenetetrahydrofolate dehydrogenase (NADP+ dependent) 1 like |
| chr8_+_28695914 | 0.33 |
ENSDART00000033386
|
ocstamp
|
osteoclast stimulatory transmembrane protein |
| chr1_-_52498146 | 0.33 |
ENSDART00000122217
|
acy3.2
|
aspartoacylase (aminocyclase) 3, tandem duplicate 2 |
| chr22_-_10110959 | 0.33 |
ENSDART00000031005
ENSDART00000147580 |
gls2b
|
glutaminase 2b (liver, mitochondrial) |
| chr3_+_32118670 | 0.33 |
ENSDART00000055287
ENSDART00000111688 |
zgc:109934
|
zgc:109934 |
| chr23_+_19213472 | 0.33 |
ENSDART00000185985
|
apobec2b
|
apolipoprotein B mRNA editing enzyme, catalytic polypeptide-like 2b |
| chr16_-_21785261 | 0.32 |
ENSDART00000078858
|
si:ch73-86n18.1
|
si:ch73-86n18.1 |
| chr2_-_3437862 | 0.32 |
ENSDART00000053012
|
cyp8b1
|
cytochrome P450, family 8, subfamily B, polypeptide 1 |
| chr11_-_24681292 | 0.32 |
ENSDART00000089601
|
olfml3b
|
olfactomedin-like 3b |
| chr6_-_442163 | 0.32 |
ENSDART00000163564
ENSDART00000189134 ENSDART00000169789 |
grap2b
|
GRB2-related adaptor protein 2b |
| chr15_+_1137337 | 0.31 |
ENSDART00000191508
|
p2ry13
|
purinergic receptor P2Y13 |
| chr21_-_21020708 | 0.30 |
ENSDART00000064032
|
eif4ebp1
|
eukaryotic translation initiation factor 4E binding protein 1 |
| chr4_+_77948970 | 0.30 |
ENSDART00000149636
|
pacsin2
|
protein kinase C and casein kinase substrate in neurons 2 |
| chr15_-_4568154 | 0.30 |
ENSDART00000155254
|
rffl
|
ring finger and FYVE-like domain containing E3 ubiquitin protein ligase |
| chr3_+_1179601 | 0.30 |
ENSDART00000173378
|
triobpb
|
TRIO and F-actin binding protein b |
| chr25_+_23336310 | 0.30 |
ENSDART00000156457
|
ptprjb.2
|
protein tyrosine phosphatase, receptor type, Jb, tandem duplicate 2 |
| chr22_-_26595027 | 0.29 |
ENSDART00000184162
|
CABZ01072309.1
|
|
| chr22_+_19247255 | 0.29 |
ENSDART00000144053
|
si:dkey-21e2.10
|
si:dkey-21e2.10 |
| chr19_+_15440841 | 0.29 |
ENSDART00000182329
|
lin28a
|
lin-28 homolog A (C. elegans) |
| chr25_+_13205878 | 0.29 |
ENSDART00000162319
ENSDART00000162283 |
si:ch211-147m6.2
|
si:ch211-147m6.2 |
| chr22_+_16497670 | 0.29 |
ENSDART00000014330
|
ier5
|
immediate early response 5 |
| chr19_+_2670130 | 0.29 |
ENSDART00000152152
|
si:ch73-24k9.2
|
si:ch73-24k9.2 |
| chr9_+_11293830 | 0.29 |
ENSDART00000144440
|
wnt6b
|
wingless-type MMTV integration site family, member 6b |
| chr19_+_348729 | 0.28 |
ENSDART00000114284
|
mcl1a
|
MCL1, BCL2 family apoptosis regulator a |
| chr23_+_16807785 | 0.28 |
ENSDART00000146992
|
zgc:114081
|
zgc:114081 |
| chr4_-_77216726 | 0.28 |
ENSDART00000099943
|
psmb10
|
proteasome subunit beta 10 |
| chr24_+_264839 | 0.27 |
ENSDART00000066872
|
ccr12b.2
|
chemokine (C-C motif) receptor 12b, tandem duplicate 2 |
| chr1_-_52497834 | 0.27 |
ENSDART00000136469
ENSDART00000004233 |
acy3.2
|
aspartoacylase (aminocyclase) 3, tandem duplicate 2 |
| chr20_+_7084154 | 0.27 |
ENSDART00000136448
|
ftr85
|
finTRIM family, member 85 |
| chr7_-_8490886 | 0.27 |
ENSDART00000159012
|
jac6
|
jacalin 6 |
| chr22_-_5006801 | 0.27 |
ENSDART00000106166
|
rx1
|
retinal homeobox gene 1 |
| chr22_+_19266995 | 0.27 |
ENSDART00000133995
ENSDART00000144963 |
si:dkey-21e2.12
|
si:dkey-21e2.12 |
| chr22_+_8979955 | 0.27 |
ENSDART00000144005
|
si:ch211-213a13.1
|
si:ch211-213a13.1 |
| chr12_-_22355430 | 0.27 |
ENSDART00000153296
ENSDART00000056919 ENSDART00000159036 |
nsfb
|
N-ethylmaleimide-sensitive factor b |
| chr23_-_30045661 | 0.27 |
ENSDART00000122239
ENSDART00000103480 |
ccdc187
|
coiled-coil domain containing 187 |
| chr5_-_1047504 | 0.27 |
ENSDART00000159346
|
mbd2
|
methyl-CpG binding domain protein 2 |
| chr7_-_8438657 | 0.26 |
ENSDART00000173054
|
si:dkeyp-32g11.8
|
si:dkeyp-32g11.8 |
| chr24_-_23998897 | 0.26 |
ENSDART00000130053
|
zmp:0000000991
|
zmp:0000000991 |
| chr2_-_985417 | 0.26 |
ENSDART00000140540
|
si:ch211-241e1.3
|
si:ch211-241e1.3 |
| chr2_-_49997055 | 0.26 |
ENSDART00000140294
|
si:ch211-106n13.3
|
si:ch211-106n13.3 |
| chr7_+_14005111 | 0.26 |
ENSDART00000187365
|
furina
|
furin (paired basic amino acid cleaving enzyme) a |
| chr8_+_8671229 | 0.26 |
ENSDART00000131963
|
usp11
|
ubiquitin specific peptidase 11 |
| chr8_-_18010735 | 0.26 |
ENSDART00000125014
|
acot11b
|
acyl-CoA thioesterase 11b |
| chr7_+_17443567 | 0.26 |
ENSDART00000060383
|
nitr2b
|
novel immune-type receptor 2b |
| chr18_+_30421528 | 0.26 |
ENSDART00000140908
|
gse1
|
Gse1 coiled-coil protein |
| chr25_-_29072162 | 0.26 |
ENSDART00000169269
|
arid3b
|
AT rich interactive domain 3B (BRIGHT-like) |
| chr24_+_260902 | 0.26 |
ENSDART00000151992
|
ccr12b.1
|
chemokine (C-C motif) receptor 12b, tandem duplicate 1 |
| chr6_+_56147812 | 0.25 |
ENSDART00000150219
|
tfap2c
|
transcription factor AP-2 gamma (activating enhancer binding protein 2 gamma) |
| chr6_+_612594 | 0.25 |
ENSDART00000150903
|
kynu
|
kynureninase |
| chr4_-_53047883 | 0.25 |
ENSDART00000157496
|
si:dkey-201g16.5
|
si:dkey-201g16.5 |
| chr14_-_34605804 | 0.25 |
ENSDART00000144547
|
afap1l1a
|
actin filament associated protein 1-like 1a |
| chr4_-_76488581 | 0.25 |
ENSDART00000174291
|
ftr51
|
finTRIM family, member 51 |
| chr11_-_27821 | 0.25 |
ENSDART00000158769
ENSDART00000172970 ENSDART00000173118 ENSDART00000168674 ENSDART00000163545 ENSDART00000173411 ENSDART00000172132 |
sp1
|
sp1 transcription factor |
| chr2_+_17524278 | 0.25 |
ENSDART00000165633
|
pimr196
|
Pim proto-oncogene, serine/threonine kinase, related 196 |
| chr3_+_16976095 | 0.25 |
ENSDART00000112450
|
cavin1a
|
caveolae associated protein 1a |
| chr6_+_39108858 | 0.25 |
ENSDART00000154232
ENSDART00000192092 |
prss60.3
|
protease, serine, 60.3 |
| chr11_-_23501467 | 0.25 |
ENSDART00000169066
|
plekha6
|
pleckstrin homology domain containing, family A member 6 |
| chr1_-_40914752 | 0.24 |
ENSDART00000113087
|
hmx1
|
H6 family homeobox 1 |
| chr9_-_9992697 | 0.24 |
ENSDART00000123415
|
ugt1ab
|
UDP glucuronosyltransferase 1 family a, b |
| chr14_+_21699414 | 0.24 |
ENSDART00000169942
|
stx3a
|
syntaxin 3A |
| chr7_-_8504355 | 0.24 |
ENSDART00000173067
|
loc564660
|
hypothetical protein LOC564660 |
| chr23_+_1730663 | 0.24 |
ENSDART00000149545
|
tgm1
|
transglutaminase 1, K polypeptide |
| chr23_+_25856541 | 0.24 |
ENSDART00000145426
ENSDART00000028236 |
hnf4a
|
hepatocyte nuclear factor 4, alpha |
| chr3_-_8765165 | 0.24 |
ENSDART00000191131
|
CABZ01058333.1
|
|
| chr2_+_30379650 | 0.24 |
ENSDART00000129542
|
crispld1b
|
cysteine-rich secretory protein LCCL domain containing 1b |
| chr19_+_37118547 | 0.24 |
ENSDART00000103163
|
cx30.9
|
connexin 30.9 |
| chr3_-_32859335 | 0.24 |
ENSDART00000158916
|
si:dkey-16l2.20
|
si:dkey-16l2.20 |
| chr23_+_5524247 | 0.24 |
ENSDART00000189679
ENSDART00000083622 |
tead3a
|
TEA domain family member 3 a |
| chr14_+_29941445 | 0.23 |
ENSDART00000181761
|
fam149a
|
family with sequence similarity 149 member A |
| chr4_-_11132617 | 0.23 |
ENSDART00000150250
|
si:dkey-21h14.9
|
si:dkey-21h14.9 |
| chr4_-_41582875 | 0.23 |
ENSDART00000182308
ENSDART00000172173 ENSDART00000163135 |
si:dkeyp-44d3.5
|
si:dkeyp-44d3.5 |
| chr3_-_584950 | 0.23 |
ENSDART00000164752
|
dicp1.1
|
diverse immunoglobulin domain-containing protein 1.1 |
| chr3_-_34082283 | 0.23 |
ENSDART00000151264
|
ighv4-3
|
immunoglobulin heavy variable 4-3 |
| chr8_-_20291922 | 0.23 |
ENSDART00000148304
|
myo1f
|
myosin IF |
| chr11_+_37638873 | 0.23 |
ENSDART00000186384
ENSDART00000184291 ENSDART00000131782 ENSDART00000140502 |
sh2d5
|
SH2 domain containing 5 |
| chr3_+_29476085 | 0.23 |
ENSDART00000184495
ENSDART00000181058 |
fam83fa
|
family with sequence similarity 83, member Fa |
| chr1_+_55452892 | 0.22 |
ENSDART00000122508
|
CR788255.1
|
|
| chr25_+_3549401 | 0.22 |
ENSDART00000166312
|
ccdc77
|
coiled-coil domain containing 77 |
| chr21_+_45316330 | 0.22 |
ENSDART00000056474
ENSDART00000149314 ENSDART00000149272 ENSDART00000149156 ENSDART00000099497 |
tcf7
|
transcription factor 7 |
| chr9_+_20483846 | 0.22 |
ENSDART00000192067
ENSDART00000145111 |
parp4
|
poly (ADP-ribose) polymerase family, member 4 |
| chr3_-_43954343 | 0.22 |
ENSDART00000157580
|
ubfd1
|
ubiquitin family domain containing 1 |
| chr24_-_38121920 | 0.22 |
ENSDART00000137056
|
si:ch211-234p6.9
|
si:ch211-234p6.9 |
| chr2_+_29995590 | 0.22 |
ENSDART00000151906
|
rbm33b
|
RNA binding motif protein 33b |
| chr23_-_4933508 | 0.22 |
ENSDART00000137578
ENSDART00000141196 |
ngfa
|
nerve growth factor a (beta polypeptide) |
| chr1_+_41588170 | 0.22 |
ENSDART00000139175
|
si:dkey-56e3.2
|
si:dkey-56e3.2 |
| chr4_+_54618332 | 0.22 |
ENSDART00000171824
|
si:ch211-227e10.2
|
si:ch211-227e10.2 |
| chr20_-_25626198 | 0.21 |
ENSDART00000126716
|
paics
|
phosphoribosylaminoimidazole carboxylase, phosphoribosylaminoimidazole succinocarboxamide synthetase |
| chr17_-_28707898 | 0.21 |
ENSDART00000135752
ENSDART00000061853 |
ap4s1
|
adaptor-related protein complex 4, sigma 1 subunit |
| chr8_-_38105053 | 0.21 |
ENSDART00000131546
|
adgra2
|
adhesion G protein-coupled receptor A2 |
| chr9_-_21838045 | 0.21 |
ENSDART00000147471
|
acod1
|
aconitate decarboxylase 1 |
| chr11_+_43401592 | 0.21 |
ENSDART00000112468
|
vipb
|
vasoactive intestinal peptide b |
| chr16_+_41517188 | 0.21 |
ENSDART00000049976
|
si:dkey-11p23.7
|
si:dkey-11p23.7 |
| chr10_+_4987766 | 0.21 |
ENSDART00000121959
|
si:ch73-234b20.5
|
si:ch73-234b20.5 |
| chr6_+_29791164 | 0.21 |
ENSDART00000017424
|
ptmaa
|
prothymosin, alpha a |
| chr4_+_25181572 | 0.21 |
ENSDART00000078529
ENSDART00000136643 |
kin
|
Kin17 DNA and RNA binding protein |
| chr17_-_26868169 | 0.21 |
ENSDART00000157204
|
si:dkey-221l4.10
|
si:dkey-221l4.10 |
| chr25_+_36292465 | 0.21 |
ENSDART00000152649
|
bmb
|
brambleberry |
| chr17_-_15600455 | 0.20 |
ENSDART00000110272
ENSDART00000156911 |
si:ch211-266g18.9
|
si:ch211-266g18.9 |
| chr5_+_15819651 | 0.20 |
ENSDART00000081230
ENSDART00000186969 ENSDART00000134206 |
hspb8
|
heat shock protein b8 |
| chr18_+_40993196 | 0.20 |
ENSDART00000115111
|
si:dkey-283j8.1
|
si:dkey-283j8.1 |
| chr22_-_22337382 | 0.20 |
ENSDART00000144684
|
si:ch211-129c21.1
|
si:ch211-129c21.1 |
| chr22_+_9862466 | 0.20 |
ENSDART00000146864
|
si:dkey-253d23.3
|
si:dkey-253d23.3 |
| chr11_+_10975012 | 0.20 |
ENSDART00000192872
|
itgb6
|
integrin, beta 6 |
| chr5_+_1933131 | 0.20 |
ENSDART00000061693
|
si:ch73-55i23.1
|
si:ch73-55i23.1 |
| chr25_+_29474583 | 0.20 |
ENSDART00000191189
|
il17rel
|
interleukin 17 receptor E-like |
| chr23_-_1017605 | 0.20 |
ENSDART00000138290
|
cdh26.1
|
cadherin 26, tandem duplicate 1 |
| chr2_+_38731696 | 0.20 |
ENSDART00000181733
|
FQ377660.1
|
|
| chr2_+_13710439 | 0.19 |
ENSDART00000155712
|
ebna1bp2
|
EBNA1 binding protein 2 |
| chr14_+_33882973 | 0.19 |
ENSDART00000019396
|
clic2
|
chloride intracellular channel 2 |
| chr24_-_6078222 | 0.19 |
ENSDART00000146830
|
apbb1ip
|
amyloid beta (A4) precursor protein-binding, family B, member 1 interacting protein |
| chr3_-_34816893 | 0.19 |
ENSDART00000084448
ENSDART00000154696 |
psmd11a
|
proteasome 26S subunit, non-ATPase 11a |
| chr25_+_8955530 | 0.19 |
ENSDART00000156444
|
si:ch211-256a21.4
|
si:ch211-256a21.4 |
| chr16_-_28878080 | 0.19 |
ENSDART00000149501
|
si:dkey-239n17.4
|
si:dkey-239n17.4 |
| chr11_+_24251141 | 0.19 |
ENSDART00000182684
|
pnp4a
|
purine nucleoside phosphorylase 4a |
| chr11_-_12198765 | 0.19 |
ENSDART00000104203
ENSDART00000128364 ENSDART00000166887 ENSDART00000041533 |
krt95
|
kertain 95 |
| chr23_-_33738945 | 0.19 |
ENSDART00000136386
|
si:ch211-210c8.7
|
si:ch211-210c8.7 |
| chr14_-_33613794 | 0.19 |
ENSDART00000010022
|
xpnpep2
|
X-prolyl aminopeptidase (aminopeptidase P) 2, membrane-bound |
| chr23_-_1017428 | 0.18 |
ENSDART00000110588
ENSDART00000183158 |
cdh26.1
|
cadherin 26, tandem duplicate 1 |
| chr19_-_27550768 | 0.18 |
ENSDART00000142313
|
si:dkeyp-46h3.8
|
si:dkeyp-46h3.8 |
| chr19_-_20403318 | 0.18 |
ENSDART00000136826
|
dazl
|
deleted in azoospermia-like |
| chr4_-_19742300 | 0.18 |
ENSDART00000066964
ENSDART00000100952 |
hgfa
|
hepatocyte growth factor a |
| chr7_-_5125799 | 0.17 |
ENSDART00000173390
|
ltb4r2a
|
leukotriene B4 receptor 2a |
| chr10_+_9092815 | 0.17 |
ENSDART00000064966
|
snx18b
|
sorting nexin 18b |
| chr7_-_25697285 | 0.17 |
ENSDART00000082620
|
dysf
|
dysferlin, limb girdle muscular dystrophy 2B (autosomal recessive) |
| chr7_-_8602864 | 0.17 |
ENSDART00000173291
|
jac2
|
jacalin 2 |
| chr3_-_21094437 | 0.17 |
ENSDART00000153739
ENSDART00000109790 |
nlk1
|
nemo-like kinase, type 1 |
| chr3_-_38918697 | 0.17 |
ENSDART00000145630
|
si:dkey-106c17.2
|
si:dkey-106c17.2 |
| chr5_-_9760173 | 0.17 |
ENSDART00000172523
|
ddr2l
|
discoidin domain receptor family, member 2, like |
Network of associatons between targets according to the STRING database.
First level regulatory network of fosb+fosl2+si:ch211-153j24.3
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.9 | GO:0071962 | mitotic sister chromatid cohesion, centromeric(GO:0071962) |
| 0.2 | 1.0 | GO:0035988 | chondrocyte proliferation(GO:0035988) |
| 0.2 | 0.5 | GO:0010525 | regulation of transposition, RNA-mediated(GO:0010525) negative regulation of transposition, RNA-mediated(GO:0010526) transposition, RNA-mediated(GO:0032197) positive regulation of neuron apoptotic process(GO:0043525) positive regulation of neuron death(GO:1901216) |
| 0.1 | 0.4 | GO:0002676 | granuloma formation(GO:0002432) chronic inflammatory response(GO:0002544) regulation of granuloma formation(GO:0002631) regulation of chronic inflammatory response(GO:0002676) |
| 0.1 | 1.0 | GO:1902975 | cell cycle DNA replication initiation(GO:1902292) nuclear cell cycle DNA replication initiation(GO:1902315) mitotic DNA replication initiation(GO:1902975) |
| 0.1 | 0.3 | GO:0043388 | cytidine to uridine editing(GO:0016554) positive regulation of DNA binding(GO:0043388) |
| 0.1 | 0.5 | GO:1901842 | regulation of high voltage-gated calcium channel activity(GO:1901841) negative regulation of high voltage-gated calcium channel activity(GO:1901842) |
| 0.1 | 0.5 | GO:0045723 | neutrophil apoptotic process(GO:0001781) regulation of T-helper 1 type immune response(GO:0002825) positive regulation of T-helper 1 type immune response(GO:0002827) negative regulation of type 2 immune response(GO:0002829) inflammatory cell apoptotic process(GO:0006925) regulation of prostaglandin biosynthetic process(GO:0031392) positive regulation of prostaglandin biosynthetic process(GO:0031394) regulation of phospholipase A2 activity(GO:0032429) interleukin-2 production(GO:0032623) regulation of interleukin-2 production(GO:0032663) positive regulation of interleukin-2 production(GO:0032743) myeloid cell apoptotic process(GO:0033028) regulation of neutrophil apoptotic process(GO:0033029) positive regulation of neutrophil apoptotic process(GO:0033031) regulation of myeloid cell apoptotic process(GO:0033032) positive regulation of myeloid cell apoptotic process(GO:0033034) T-helper 1 type immune response(GO:0042088) positive regulation of CD4-positive, alpha-beta T cell differentiation(GO:0043372) T-helper 1 cell differentiation(GO:0045063) positive regulation of T-helper cell differentiation(GO:0045624) regulation of T-helper 1 cell differentiation(GO:0045625) positive regulation of T-helper 1 cell differentiation(GO:0045627) regulation of T-helper 2 cell differentiation(GO:0045628) negative regulation of T-helper 2 cell differentiation(GO:0045629) positive regulation of fatty acid biosynthetic process(GO:0045723) positive regulation of alpha-beta T cell differentiation(GO:0046638) neutrophil clearance(GO:0097350) negative regulation of phospholipase A2 activity(GO:1900138) positive regulation of CD4-positive, alpha-beta T cell activation(GO:2000516) regulation of unsaturated fatty acid biosynthetic process(GO:2001279) positive regulation of unsaturated fatty acid biosynthetic process(GO:2001280) |
| 0.1 | 0.3 | GO:0097053 | L-kynurenine metabolic process(GO:0097052) L-kynurenine catabolic process(GO:0097053) |
| 0.1 | 0.2 | GO:0022009 | central nervous system vasculogenesis(GO:0022009) |
| 0.1 | 0.3 | GO:0048211 | Golgi vesicle docking(GO:0048211) |
| 0.1 | 0.7 | GO:0001765 | membrane raft assembly(GO:0001765) plasma membrane raft assembly(GO:0044854) plasma membrane raft organization(GO:0044857) caveola assembly(GO:0070836) |
| 0.1 | 0.7 | GO:0031294 | lymphocyte costimulation(GO:0031294) T cell costimulation(GO:0031295) |
| 0.1 | 0.2 | GO:0002568 | somatic diversification of T cell receptor genes(GO:0002568) somatic recombination of T cell receptor gene segments(GO:0002681) T cell receptor V(D)J recombination(GO:0033153) |
| 0.1 | 0.5 | GO:0006450 | regulation of translational fidelity(GO:0006450) |
| 0.1 | 0.3 | GO:2001204 | regulation of osteoclast development(GO:2001204) |
| 0.1 | 0.2 | GO:0043084 | copulation(GO:0007620) regulation of epinephrine secretion(GO:0014060) positive regulation of epinephrine secretion(GO:0032812) positive regulation of catecholamine secretion(GO:0033605) penile erection(GO:0043084) epinephrine transport(GO:0048241) epinephrine secretion(GO:0048242) regulation of penile erection(GO:0060405) positive regulation of penile erection(GO:0060406) prolactin secretion(GO:0070459) |
| 0.0 | 0.2 | GO:0098967 | exocytic insertion of neurotransmitter receptor to plasma membrane(GO:0098881) exocytic insertion of neurotransmitter receptor to postsynaptic membrane(GO:0098967) |
| 0.0 | 0.3 | GO:0045110 | intermediate filament bundle assembly(GO:0045110) |
| 0.0 | 0.6 | GO:0051121 | lipoxygenase pathway(GO:0019372) linoleic acid metabolic process(GO:0043651) hepoxilin metabolic process(GO:0051121) hepoxilin biosynthetic process(GO:0051122) |
| 0.0 | 0.7 | GO:0035999 | tetrahydrofolate interconversion(GO:0035999) |
| 0.0 | 0.5 | GO:0071379 | cellular response to prostaglandin stimulus(GO:0071379) |
| 0.0 | 0.2 | GO:0001778 | plasma membrane repair(GO:0001778) monocyte activation(GO:0042117) |
| 0.0 | 0.3 | GO:2001270 | regulation of execution phase of apoptosis(GO:1900117) negative regulation of execution phase of apoptosis(GO:1900118) regulation of cysteine-type endopeptidase activity involved in execution phase of apoptosis(GO:2001270) negative regulation of cysteine-type endopeptidase activity involved in execution phase of apoptosis(GO:2001271) |
| 0.0 | 0.3 | GO:0006537 | glutamate biosynthetic process(GO:0006537) glutamine catabolic process(GO:0006543) |
| 0.0 | 0.2 | GO:0007344 | karyogamy(GO:0000741) pronuclear fusion(GO:0007344) |
| 0.0 | 0.2 | GO:0010693 | regulation of alkaline phosphatase activity(GO:0010692) negative regulation of alkaline phosphatase activity(GO:0010693) |
| 0.0 | 0.2 | GO:0036344 | platelet formation(GO:0030220) platelet morphogenesis(GO:0036344) |
| 0.0 | 0.3 | GO:1900026 | positive regulation of substrate adhesion-dependent cell spreading(GO:1900026) |
| 0.0 | 0.3 | GO:0043154 | negative regulation of cysteine-type endopeptidase activity involved in apoptotic process(GO:0043154) |
| 0.0 | 0.2 | GO:0070254 | mucus secretion(GO:0070254) |
| 0.0 | 0.2 | GO:0070365 | hepatocyte differentiation(GO:0070365) |
| 0.0 | 0.1 | GO:0097101 | blood vessel endothelial cell fate specification(GO:0097101) |
| 0.0 | 0.2 | GO:0006361 | transcription initiation from RNA polymerase I promoter(GO:0006361) |
| 0.0 | 0.3 | GO:0045947 | negative regulation of translational initiation(GO:0045947) |
| 0.0 | 0.2 | GO:1903426 | regulation of reactive oxygen species biosynthetic process(GO:1903426) |
| 0.0 | 0.3 | GO:0000002 | mitochondrial genome maintenance(GO:0000002) |
| 0.0 | 0.2 | GO:0060149 | negative regulation of posttranscriptional gene silencing(GO:0060149) negative regulation of gene silencing by miRNA(GO:0060965) negative regulation of gene silencing by RNA(GO:0060967) |
| 0.0 | 0.1 | GO:0097095 | frontonasal suture morphogenesis(GO:0097095) |
| 0.0 | 0.3 | GO:0070498 | interleukin-1-mediated signaling pathway(GO:0070498) |
| 0.0 | 0.4 | GO:0045740 | positive regulation of DNA replication(GO:0045740) |
| 0.0 | 0.1 | GO:0019418 | sulfide oxidation(GO:0019418) sulfide oxidation, using sulfide:quinone oxidoreductase(GO:0070221) |
| 0.0 | 0.5 | GO:0071594 | T cell differentiation in thymus(GO:0033077) thymocyte aggregation(GO:0071594) |
| 0.0 | 0.2 | GO:0050957 | equilibrioception(GO:0050957) |
| 0.0 | 0.1 | GO:0009298 | GDP-mannose biosynthetic process(GO:0009298) |
| 0.0 | 0.6 | GO:0046475 | glycerophospholipid catabolic process(GO:0046475) |
| 0.0 | 0.2 | GO:0006189 | 'de novo' IMP biosynthetic process(GO:0006189) |
| 0.0 | 1.0 | GO:0006910 | phagocytosis, recognition(GO:0006910) |
| 0.0 | 0.8 | GO:0061318 | renal filtration cell differentiation(GO:0061318) glomerular epithelium development(GO:0072010) glomerular visceral epithelial cell differentiation(GO:0072112) glomerular epithelial cell differentiation(GO:0072311) |
| 0.0 | 0.1 | GO:0098773 | trabecula formation(GO:0060343) heart trabecula formation(GO:0060347) skin epidermis development(GO:0098773) |
| 0.0 | 0.1 | GO:0003242 | growth involved in heart morphogenesis(GO:0003241) cardiac chamber ballooning(GO:0003242) cardiac muscle tissue growth involved in heart morphogenesis(GO:0003245) |
| 0.0 | 0.1 | GO:1990511 | piRNA biosynthetic process(GO:1990511) |
| 0.0 | 0.4 | GO:0002430 | complement receptor mediated signaling pathway(GO:0002430) |
| 0.0 | 0.3 | GO:0014034 | neural crest cell fate commitment(GO:0014034) neural crest cell fate specification(GO:0014036) |
| 0.0 | 0.3 | GO:0046549 | retinal cone cell development(GO:0046549) |
| 0.0 | 0.2 | GO:0034058 | endosomal vesicle fusion(GO:0034058) |
| 0.0 | 0.3 | GO:0060971 | embryonic heart tube left/right pattern formation(GO:0060971) |
| 0.0 | 0.2 | GO:0048672 | positive regulation of collateral sprouting(GO:0048672) |
| 0.0 | 0.2 | GO:0021684 | cerebellar granular layer development(GO:0021681) cerebellar granular layer morphogenesis(GO:0021683) cerebellar granular layer formation(GO:0021684) cerebellar granule cell differentiation(GO:0021707) |
| 0.0 | 0.5 | GO:0055003 | cardiac myofibril assembly(GO:0055003) |
| 0.0 | 0.2 | GO:0034312 | diol biosynthetic process(GO:0034312) sphingosine biosynthetic process(GO:0046512) |
| 0.0 | 0.1 | GO:0001774 | microglial cell activation(GO:0001774) |
| 0.0 | 0.2 | GO:0031641 | regulation of myelination(GO:0031641) |
| 0.0 | 0.3 | GO:0006516 | glycoprotein catabolic process(GO:0006516) |
| 0.0 | 0.1 | GO:0030579 | ubiquitin-dependent SMAD protein catabolic process(GO:0030579) |
| 0.0 | 0.2 | GO:0071350 | interleukin-15-mediated signaling pathway(GO:0035723) response to interleukin-15(GO:0070672) cellular response to interleukin-15(GO:0071350) |
| 0.0 | 0.3 | GO:0060219 | camera-type eye photoreceptor cell differentiation(GO:0060219) |
| 0.0 | 0.2 | GO:0043248 | proteasome assembly(GO:0043248) |
| 0.0 | 0.1 | GO:0009954 | proximal/distal pattern formation(GO:0009954) |
| 0.0 | 0.1 | GO:1990592 | protein polyufmylation(GO:1990564) protein K69-linked ufmylation(GO:1990592) |
| 0.0 | 0.1 | GO:0035372 | protein localization to microtubule(GO:0035372) protein localization to microtubule plus-end(GO:1904825) |
| 0.0 | 0.1 | GO:0030325 | adrenal gland development(GO:0030325) |
| 0.0 | 1.0 | GO:0019722 | calcium-mediated signaling(GO:0019722) |
| 0.0 | 0.3 | GO:0043407 | negative regulation of MAP kinase activity(GO:0043407) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 1.0 | GO:0000811 | GINS complex(GO:0000811) |
| 0.1 | 0.3 | GO:0030289 | protein phosphatase 4 complex(GO:0030289) |
| 0.1 | 1.7 | GO:0030057 | desmosome(GO:0030057) |
| 0.1 | 0.2 | GO:0043218 | compact myelin(GO:0043218) Schmidt-Lanterman incisure(GO:0043220) |
| 0.1 | 0.2 | GO:0034685 | integrin alphav-beta6 complex(GO:0034685) |
| 0.1 | 0.2 | GO:1990909 | Wnt signalosome(GO:1990909) |
| 0.1 | 0.4 | GO:0016234 | inclusion body(GO:0016234) |
| 0.0 | 0.8 | GO:0005869 | dynactin complex(GO:0005869) |
| 0.0 | 0.6 | GO:0002102 | podosome(GO:0002102) |
| 0.0 | 0.5 | GO:0016328 | lateral plasma membrane(GO:0016328) |
| 0.0 | 0.3 | GO:0014704 | intercalated disc(GO:0014704) |
| 0.0 | 0.1 | GO:0035327 | transcriptionally active chromatin(GO:0035327) |
| 0.0 | 0.3 | GO:0008541 | proteasome regulatory particle, lid subcomplex(GO:0008541) |
| 0.0 | 1.0 | GO:0042571 | immunoglobulin complex, circulating(GO:0042571) |
| 0.0 | 0.9 | GO:0005901 | caveola(GO:0005901) plasma membrane raft(GO:0044853) |
| 0.0 | 0.3 | GO:0090545 | NuRD complex(GO:0016581) CHD-type complex(GO:0090545) |
| 0.0 | 0.1 | GO:0016589 | NURF complex(GO:0016589) |
| 0.0 | 0.1 | GO:0032300 | mismatch repair complex(GO:0032300) |
| 0.0 | 0.2 | GO:0030315 | T-tubule(GO:0030315) |
| 0.0 | 0.1 | GO:0030915 | Smc5-Smc6 complex(GO:0030915) |
| 0.0 | 0.2 | GO:1990907 | beta-catenin-TCF complex(GO:1990907) |
| 0.0 | 0.3 | GO:0005839 | proteasome core complex(GO:0005839) |
| 0.0 | 0.1 | GO:0097651 | phosphatidylinositol 3-kinase complex, class IB(GO:0005944) phosphatidylinositol 3-kinase complex, class I(GO:0097651) |
| 0.0 | 0.1 | GO:0000177 | nuclear exosome (RNase complex)(GO:0000176) cytoplasmic exosome (RNase complex)(GO:0000177) |
| 0.0 | 0.2 | GO:0048787 | presynaptic active zone membrane(GO:0048787) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.3 | GO:0008397 | sterol 12-alpha-hydroxylase activity(GO:0008397) |
| 0.1 | 0.4 | GO:0005153 | interleukin-8 receptor binding(GO:0005153) |
| 0.1 | 0.7 | GO:0004329 | formate-tetrahydrofolate ligase activity(GO:0004329) |
| 0.1 | 0.5 | GO:0070330 | aromatase activity(GO:0070330) |
| 0.1 | 0.5 | GO:0019834 | phospholipase A2 inhibitor activity(GO:0019834) |
| 0.1 | 0.6 | GO:0004051 | arachidonate 5-lipoxygenase activity(GO:0004051) |
| 0.1 | 0.4 | GO:1990756 | protein binding, bridging involved in substrate recognition for ubiquitination(GO:1990756) |
| 0.1 | 0.3 | GO:0043139 | 5'-3' DNA helicase activity(GO:0043139) |
| 0.1 | 0.2 | GO:0042134 | rRNA primary transcript binding(GO:0042134) |
| 0.1 | 0.3 | GO:0019215 | intermediate filament binding(GO:0019215) |
| 0.1 | 0.6 | GO:0004046 | aminoacylase activity(GO:0004046) |
| 0.1 | 0.3 | GO:0004126 | cytidine deaminase activity(GO:0004126) |
| 0.1 | 0.2 | GO:0004639 | phosphoribosylaminoimidazolesuccinocarboxamide synthase activity(GO:0004639) |
| 0.1 | 0.6 | GO:0047498 | calcium-dependent phospholipase A2 activity(GO:0047498) |
| 0.1 | 0.3 | GO:0016823 | hydrolase activity, acting on acid carbon-carbon bonds(GO:0016822) hydrolase activity, acting on acid carbon-carbon bonds, in ketonic substances(GO:0016823) |
| 0.0 | 0.3 | GO:0008190 | eukaryotic initiation factor 4E binding(GO:0008190) |
| 0.0 | 0.1 | GO:0004466 | long-chain-acyl-CoA dehydrogenase activity(GO:0004466) |
| 0.0 | 0.2 | GO:0001596 | angiotensin type I receptor activity(GO:0001596) |
| 0.0 | 0.9 | GO:1990841 | promoter-specific chromatin binding(GO:1990841) |
| 0.0 | 0.1 | GO:0004475 | mannose-1-phosphate guanylyltransferase activity(GO:0004475) mannose-phosphate guanylyltransferase activity(GO:0008905) |
| 0.0 | 0.3 | GO:0036042 | long-chain fatty acyl-CoA binding(GO:0036042) |
| 0.0 | 0.3 | GO:0004359 | glutaminase activity(GO:0004359) |
| 0.0 | 0.1 | GO:0033149 | FFAT motif binding(GO:0033149) |
| 0.0 | 0.1 | GO:0070224 | sulfide:quinone oxidoreductase activity(GO:0070224) |
| 0.0 | 0.8 | GO:0048306 | calcium-dependent protein binding(GO:0048306) |
| 0.0 | 0.2 | GO:0017040 | ceramidase activity(GO:0017040) |
| 0.0 | 0.1 | GO:0042288 | MHC class I protein binding(GO:0042288) |
| 0.0 | 0.2 | GO:0004974 | leukotriene receptor activity(GO:0004974) |
| 0.0 | 0.4 | GO:0001608 | G-protein coupled nucleotide receptor activity(GO:0001608) G-protein coupled purinergic nucleotide receptor activity(GO:0045028) |
| 0.0 | 0.2 | GO:0004459 | L-lactate dehydrogenase activity(GO:0004459) |
| 0.0 | 1.0 | GO:0034987 | immunoglobulin receptor binding(GO:0034987) |
| 0.0 | 0.9 | GO:0019957 | C-C chemokine binding(GO:0019957) |
| 0.0 | 0.2 | GO:0004731 | purine-nucleoside phosphorylase activity(GO:0004731) |
| 0.0 | 0.2 | GO:0008494 | translation activator activity(GO:0008494) |
| 0.0 | 0.2 | GO:0038062 | protein tyrosine kinase collagen receptor activity(GO:0038062) collagen receptor activity(GO:0038064) |
| 0.0 | 0.3 | GO:0008327 | methyl-CpG binding(GO:0008327) |
| 0.0 | 0.4 | GO:0004875 | complement receptor activity(GO:0004875) |
| 0.0 | 0.1 | GO:0004366 | glycerol-3-phosphate O-acyltransferase activity(GO:0004366) |
| 0.0 | 0.2 | GO:0005163 | nerve growth factor receptor binding(GO:0005163) neurotrophin receptor binding(GO:0005165) |
| 0.0 | 0.4 | GO:0070492 | oligosaccharide binding(GO:0070492) |
| 0.0 | 0.5 | GO:0005246 | calcium channel regulator activity(GO:0005246) |
| 0.0 | 1.6 | GO:0045296 | cadherin binding(GO:0045296) |
| 0.0 | 0.2 | GO:0030368 | interleukin-17 receptor activity(GO:0030368) |
| 0.0 | 0.3 | GO:0004298 | threonine-type endopeptidase activity(GO:0004298) threonine-type peptidase activity(GO:0070003) |
| 0.0 | 0.2 | GO:0019911 | structural constituent of myelin sheath(GO:0019911) |
| 0.0 | 0.1 | GO:0016316 | phosphatidylinositol-3,4-bisphosphate 4-phosphatase activity(GO:0016316) |
| 0.0 | 0.4 | GO:0015035 | protein disulfide oxidoreductase activity(GO:0015035) |
| 0.0 | 0.5 | GO:0000049 | tRNA binding(GO:0000049) |
| 0.0 | 0.1 | GO:0051185 | coenzyme transporter activity(GO:0051185) |
| 0.0 | 0.2 | GO:0042289 | MHC class II protein binding(GO:0042289) |
| 0.0 | 0.1 | GO:0034450 | ubiquitin-ubiquitin ligase activity(GO:0034450) |
| 0.0 | 0.6 | GO:0017124 | SH3 domain binding(GO:0017124) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.5 | PID HIF1A PATHWAY | Hypoxic and oxygen homeostasis regulation of HIF-1-alpha |
| 0.0 | 0.6 | PID NFAT TFPATHWAY | Calcineurin-regulated NFAT-dependent transcription in lymphocytes |
| 0.0 | 0.2 | PID THROMBIN PAR4 PATHWAY | PAR4-mediated thrombin signaling events |
| 0.0 | 0.2 | PID INTEGRIN5 PATHWAY | Beta5 beta6 beta7 and beta8 integrin cell surface interactions |
| 0.0 | 0.6 | PID HNF3B PATHWAY | FOXA2 and FOXA3 transcription factor networks |
| 0.0 | 0.3 | PID TOLL ENDOGENOUS PATHWAY | Endogenous TLR signaling |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.5 | REACTOME ACTIVATION OF BH3 ONLY PROTEINS | Genes involved in Activation of BH3-only proteins |
| 0.0 | 0.3 | REACTOME SYNTHESIS OF BILE ACIDS AND BILE SALTS VIA 7ALPHA HYDROXYCHOLESTEROL | Genes involved in Synthesis of bile acids and bile salts via 7alpha-hydroxycholesterol |
| 0.0 | 0.4 | REACTOME P2Y RECEPTORS | Genes involved in P2Y receptors |
| 0.0 | 0.3 | REACTOME MTORC1 MEDIATED SIGNALLING | Genes involved in mTORC1-mediated signalling |
| 0.0 | 0.3 | REACTOME NFKB IS ACTIVATED AND SIGNALS SURVIVAL | Genes involved in NF-kB is activated and signals survival |
| 0.0 | 0.3 | REACTOME RNA POL I PROMOTER OPENING | Genes involved in RNA Polymerase I Promoter Opening |
| 0.0 | 0.3 | REACTOME TRYPTOPHAN CATABOLISM | Genes involved in Tryptophan catabolism |
| 0.0 | 0.4 | REACTOME SYNTHESIS AND INTERCONVERSION OF NUCLEOTIDE DI AND TRIPHOSPHATES | Genes involved in Synthesis and interconversion of nucleotide di- and triphosphates |
| 0.0 | 0.2 | REACTOME PURINE RIBONUCLEOSIDE MONOPHOSPHATE BIOSYNTHESIS | Genes involved in Purine ribonucleoside monophosphate biosynthesis |
| 0.0 | 0.2 | REACTOME REGULATION OF GENE EXPRESSION IN BETA CELLS | Genes involved in Regulation of gene expression in beta cells |
| 0.0 | 0.2 | REACTOME P130CAS LINKAGE TO MAPK SIGNALING FOR INTEGRINS | Genes involved in p130Cas linkage to MAPK signaling for integrins |
| 0.0 | 0.1 | REACTOME MRNA DECAY BY 3 TO 5 EXORIBONUCLEASE | Genes involved in mRNA Decay by 3' to 5' Exoribonuclease |