Project
PRJNA207719: Tissue specific transcriptome profiling
Navigation
Downloads
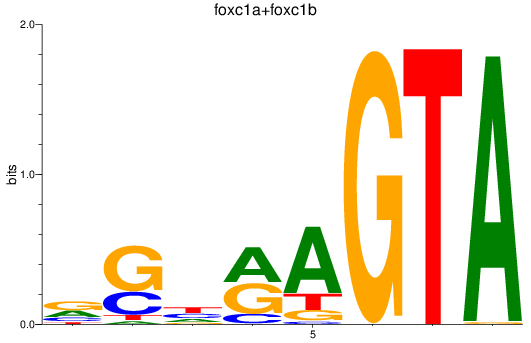
Results for foxc1a+foxc1b
Z-value: 1.25
Transcription factors associated with foxc1a+foxc1b
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
foxc1b
|
ENSDARG00000055398 | forkhead box C1b |
|
foxc1a
|
ENSDARG00000091481 | forkhead box C1a |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| foxc1a | dr11_v1_chr2_-_689047_689047 | -0.22 | 7.3e-01 | Click! |
| foxc1b | dr11_v1_chr20_+_26702377_26702377 | -0.06 | 9.2e-01 | Click! |
Activity profile of foxc1a+foxc1b motif
Sorted Z-values of foxc1a+foxc1b motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr11_+_11200550 | 1.20 |
ENSDART00000181339
ENSDART00000187116 |
myom2a
|
myomesin 2a |
| chr1_-_45177373 | 1.12 |
ENSDART00000143142
ENSDART00000034549 |
zgc:111983
|
zgc:111983 |
| chr22_+_20720808 | 0.97 |
ENSDART00000171321
|
si:dkey-211f22.5
|
si:dkey-211f22.5 |
| chr1_+_7540978 | 0.85 |
ENSDART00000147770
|
mylz3
|
myosin, light polypeptide 3, skeletal muscle |
| chr12_-_26064105 | 0.82 |
ENSDART00000168825
|
ldb3b
|
LIM domain binding 3b |
| chr12_-_26064480 | 0.80 |
ENSDART00000158215
ENSDART00000171206 ENSDART00000171212 ENSDART00000182956 ENSDART00000186779 |
ldb3b
|
LIM domain binding 3b |
| chr18_-_16792561 | 0.79 |
ENSDART00000145546
|
ampd3b
|
adenosine monophosphate deaminase 3b |
| chr20_-_27311675 | 0.75 |
ENSDART00000026088
ENSDART00000148361 |
asb2a.1
|
ankyrin repeat and SOCS box containing 2a, tandem duplicate 1 |
| chr24_-_40726073 | 0.70 |
ENSDART00000168100
|
smyhc2
|
slow myosin heavy chain 2 |
| chr12_+_20641471 | 0.67 |
ENSDART00000133654
|
calcoco2
|
calcium binding and coiled-coil domain 2 |
| chr16_-_12723324 | 0.62 |
ENSDART00000131915
|
sbk3
|
SH3 domain binding kinase family, member 3 |
| chr19_-_10730488 | 0.61 |
ENSDART00000126033
|
slc9a3.1
|
solute carrier family 9, subfamily A (NHE3, cation proton antiporter 3), member 3, tandem duplicate 1 |
| chr21_-_22928214 | 0.60 |
ENSDART00000182760
|
dub
|
duboraya |
| chr1_-_14332283 | 0.60 |
ENSDART00000090025
|
wfs1a
|
Wolfram syndrome 1a (wolframin) |
| chr10_+_44956660 | 0.57 |
ENSDART00000169225
ENSDART00000189298 |
il1b
|
interleukin 1, beta |
| chr11_-_29946927 | 0.56 |
ENSDART00000165182
|
zgc:113276
|
zgc:113276 |
| chr16_-_13992646 | 0.56 |
ENSDART00000139623
|
si:dkey-85k15.6
|
si:dkey-85k15.6 |
| chr21_+_6556635 | 0.54 |
ENSDART00000139598
|
col5a1
|
procollagen, type V, alpha 1 |
| chr6_+_40629066 | 0.50 |
ENSDART00000103757
|
slc6a11a
|
solute carrier family 6 (neurotransmitter transporter), member 11a |
| chr6_-_10828880 | 0.49 |
ENSDART00000131458
ENSDART00000020261 |
chrna1
|
cholinergic receptor, nicotinic, alpha 1 (muscle) |
| chr16_-_24815091 | 0.48 |
ENSDART00000154269
ENSDART00000131025 |
kcnn4
|
potassium intermediate/small conductance calcium-activated channel, subfamily N, member 4 |
| chr16_+_7662609 | 0.45 |
ENSDART00000184895
ENSDART00000149404 ENSDART00000081418 ENSDART00000081422 |
bves
|
blood vessel epicardial substance |
| chr18_+_16986903 | 0.43 |
ENSDART00000142088
|
si:ch211-218c6.8
|
si:ch211-218c6.8 |
| chr9_-_21918963 | 0.43 |
ENSDART00000090782
|
lmo7a
|
LIM domain 7a |
| chr23_-_16737161 | 0.41 |
ENSDART00000132573
|
si:ch211-224l10.4
|
si:ch211-224l10.4 |
| chr17_+_45404758 | 0.41 |
ENSDART00000147557
|
tagapa
|
T cell activation RhoGTPase activating protein a |
| chr7_-_38790341 | 0.41 |
ENSDART00000159884
|
si:dkey-23n7.10
|
si:dkey-23n7.10 |
| chr13_+_28417297 | 0.41 |
ENSDART00000043658
|
slc2a15a
|
solute carrier family 2 (facilitated glucose transporter), member 15a |
| chr1_-_45146834 | 0.41 |
ENSDART00000144997
|
si:ch211-239f4.6
|
si:ch211-239f4.6 |
| chr11_+_18183220 | 0.41 |
ENSDART00000113468
|
LO018315.10
|
|
| chr25_+_20216159 | 0.38 |
ENSDART00000048642
|
tnnt2d
|
troponin T2d, cardiac |
| chr6_-_51771634 | 0.38 |
ENSDART00000073847
|
blcap
|
bladder cancer associated protein |
| chr10_+_15107886 | 0.37 |
ENSDART00000188047
ENSDART00000164095 |
scpp8
|
secretory calcium-binding phosphoprotein 8 |
| chr22_-_22147375 | 0.36 |
ENSDART00000149304
|
cdc34a
|
cell division cycle 34 homolog (S. cerevisiae) a |
| chr3_-_18030938 | 0.36 |
ENSDART00000013540
|
si:ch73-141c7.1
|
si:ch73-141c7.1 |
| chr25_+_26798673 | 0.35 |
ENSDART00000157235
|
ca12
|
carbonic anhydrase XII |
| chr1_+_18550864 | 0.34 |
ENSDART00000142515
|
si:dkey-192k22.2
|
si:dkey-192k22.2 |
| chr23_+_2185397 | 0.34 |
ENSDART00000109373
|
c1qtnf7
|
C1q and TNF related 7 |
| chr7_-_18691637 | 0.34 |
ENSDART00000183715
|
si:ch211-119e14.9
|
si:ch211-119e14.9 |
| chr7_-_16598212 | 0.34 |
ENSDART00000128488
|
e2f8
|
E2F transcription factor 8 |
| chr4_-_13502549 | 0.34 |
ENSDART00000140366
|
si:ch211-266a5.12
|
si:ch211-266a5.12 |
| chr2_-_22152797 | 0.34 |
ENSDART00000145188
|
cyp7a1
|
cytochrome P450, family 7, subfamily A, polypeptide 1 |
| chr24_-_25428176 | 0.33 |
ENSDART00000090010
|
phex
|
phosphate regulating endopeptidase homolog, X-linked |
| chr16_-_42004544 | 0.33 |
ENSDART00000034544
|
caspa
|
caspase a |
| chr1_-_23274393 | 0.33 |
ENSDART00000147800
ENSDART00000130277 ENSDART00000054340 ENSDART00000054338 |
rpl9
|
ribosomal protein L9 |
| chr22_-_23000815 | 0.33 |
ENSDART00000137111
|
ptprc
|
protein tyrosine phosphatase, receptor type, C |
| chr5_+_39087364 | 0.32 |
ENSDART00000004286
|
anxa3a
|
annexin A3a |
| chr16_-_13818061 | 0.32 |
ENSDART00000132982
ENSDART00000144856 |
leng9
|
leukocyte receptor cluster (LRC) member 9 |
| chr14_+_30285613 | 0.32 |
ENSDART00000173090
|
mtus1a
|
microtubule associated tumor suppressor 1a |
| chr3_+_29641181 | 0.32 |
ENSDART00000151517
|
eif3d
|
eukaryotic translation initiation factor 3, subunit D |
| chr20_-_35578435 | 0.30 |
ENSDART00000142444
|
adgrf6
|
adhesion G protein-coupled receptor F6 |
| chr3_+_59411956 | 0.30 |
ENSDART00000166982
|
sec14l1
|
SEC14-like lipid binding 1 |
| chr16_-_5721386 | 0.29 |
ENSDART00000136655
|
ndufa3
|
NADH dehydrogenase (ubiquinone) 1 alpha subcomplex, 3 |
| chr5_-_4219741 | 0.29 |
ENSDART00000100093
|
si:ch211-283g2.2
|
si:ch211-283g2.2 |
| chr7_+_19482877 | 0.29 |
ENSDART00000077868
|
si:ch211-212k18.7
|
si:ch211-212k18.7 |
| chr9_+_15893093 | 0.28 |
ENSDART00000099483
ENSDART00000134657 |
si:dkey-14o1.20
|
si:dkey-14o1.20 |
| chr3_-_43770876 | 0.28 |
ENSDART00000160162
|
zgc:92162
|
zgc:92162 |
| chr22_+_34616151 | 0.28 |
ENSDART00000155399
ENSDART00000104705 |
si:ch1073-214b20.2
|
si:ch1073-214b20.2 |
| chr23_-_36316352 | 0.28 |
ENSDART00000014840
|
nfe2
|
nuclear factor, erythroid 2 |
| chr9_-_24244383 | 0.27 |
ENSDART00000182407
|
cavin2a
|
caveolae associated protein 2a |
| chr15_+_15456910 | 0.27 |
ENSDART00000155708
|
zgc:174895
|
zgc:174895 |
| chr3_+_12784460 | 0.27 |
ENSDART00000168382
|
cyp2k8
|
cytochrome P450, family 2, subfamily K, polypeptide 8 |
| chr6_-_8480815 | 0.27 |
ENSDART00000162300
|
rasal3
|
RAS protein activator like 3 |
| chr4_-_11132617 | 0.27 |
ENSDART00000150250
|
si:dkey-21h14.9
|
si:dkey-21h14.9 |
| chr14_-_2602445 | 0.25 |
ENSDART00000166910
|
etf1a
|
eukaryotic translation termination factor 1a |
| chr23_-_42752387 | 0.25 |
ENSDART00000149781
|
si:ch73-217n20.1
|
si:ch73-217n20.1 |
| chr1_+_45056371 | 0.25 |
ENSDART00000073689
ENSDART00000167309 |
btr01
|
bloodthirsty-related gene family, member 1 |
| chr20_+_38276690 | 0.25 |
ENSDART00000061437
|
ccl38.6
|
chemokine (C-C motif) ligand 38, duplicate 6 |
| chr5_-_65000312 | 0.23 |
ENSDART00000192893
|
ANXA1 (1 of many)
|
zgc:110283 |
| chr10_+_22034477 | 0.23 |
ENSDART00000133304
ENSDART00000134189 ENSDART00000021240 ENSDART00000100526 |
npm1a
|
nucleophosmin 1a |
| chr20_+_38458084 | 0.23 |
ENSDART00000020153
ENSDART00000135912 |
coq8a
|
coenzyme Q8A |
| chr15_+_37973197 | 0.23 |
ENSDART00000156661
|
si:dkey-238d18.9
|
si:dkey-238d18.9 |
| chr19_+_7930004 | 0.23 |
ENSDART00000160410
|
si:dkey-266f7.4
|
si:dkey-266f7.4 |
| chr17_-_53439866 | 0.23 |
ENSDART00000154826
|
mycbp
|
c-myc binding protein |
| chr20_+_10723292 | 0.23 |
ENSDART00000152805
|
si:ch211-182e10.4
|
si:ch211-182e10.4 |
| chr19_+_7929704 | 0.23 |
ENSDART00000147015
|
si:dkey-266f7.4
|
si:dkey-266f7.4 |
| chr8_-_20243389 | 0.22 |
ENSDART00000184904
|
acer1
|
alkaline ceramidase 1 |
| chr2_-_2642476 | 0.22 |
ENSDART00000124032
|
serbp1b
|
SERPINE1 mRNA binding protein 1b |
| chr19_+_4051695 | 0.22 |
ENSDART00000166368
|
btr24
|
bloodthirsty-related gene family, member 24 |
| chr22_-_10486477 | 0.22 |
ENSDART00000184366
|
aspn
|
asporin (LRR class 1) |
| chr2_-_27473645 | 0.22 |
ENSDART00000186152
|
CU469530.2
|
|
| chr2_+_49644803 | 0.21 |
ENSDART00000160342
|
si:ch211-209f23.7
|
si:ch211-209f23.7 |
| chr9_+_24192370 | 0.21 |
ENSDART00000003482
|
stk17b
|
serine/threonine kinase 17b (apoptosis-inducing) |
| chr4_-_4387012 | 0.21 |
ENSDART00000191836
|
CU468826.3
|
Danio rerio U2 small nuclear ribonucleoprotein auxiliary factor 35 kDa subunit-related protein 1-like (LOC100331497), mRNA. |
| chr7_+_19483277 | 0.21 |
ENSDART00000173750
|
si:ch211-212k18.7
|
si:ch211-212k18.7 |
| chr1_-_56717813 | 0.21 |
ENSDART00000159196
|
FO681314.1
|
|
| chr22_+_6198742 | 0.20 |
ENSDART00000132889
|
si:dkey-19a16.11
|
si:dkey-19a16.11 |
| chr15_-_1590858 | 0.20 |
ENSDART00000081875
|
nnr
|
nanor |
| chr13_-_301309 | 0.20 |
ENSDART00000131747
|
chs1
|
chitin synthase 1 |
| chr12_-_22524388 | 0.20 |
ENSDART00000020942
|
shbg
|
sex hormone-binding globulin |
| chr21_-_17290941 | 0.20 |
ENSDART00000147993
|
gfi1b
|
growth factor independent 1B transcription repressor |
| chr6_-_39275793 | 0.20 |
ENSDART00000180477
ENSDART00000148531 |
arhgef25b
|
Rho guanine nucleotide exchange factor (GEF) 25b |
| chr22_-_10487490 | 0.20 |
ENSDART00000064798
|
aspn
|
asporin (LRR class 1) |
| chr8_-_36370552 | 0.20 |
ENSDART00000097932
ENSDART00000148323 |
si:busm1-104n07.3
|
si:busm1-104n07.3 |
| chr23_+_24926407 | 0.20 |
ENSDART00000137486
|
klhl21
|
kelch-like family member 21 |
| chr6_+_33931740 | 0.19 |
ENSDART00000130492
ENSDART00000151213 |
orc1
|
origin recognition complex, subunit 1 |
| chr23_+_16910071 | 0.19 |
ENSDART00000104793
|
si:dkey-147f3.8
|
si:dkey-147f3.8 |
| chr9_+_13985567 | 0.19 |
ENSDART00000102296
|
cd28
|
CD28 molecule |
| chr8_+_19514294 | 0.18 |
ENSDART00000170622
|
si:ch73-281k2.5
|
si:ch73-281k2.5 |
| chr18_+_50669456 | 0.18 |
ENSDART00000163005
|
si:dkey-151j17.4
|
si:dkey-151j17.4 |
| chr14_-_29858883 | 0.17 |
ENSDART00000141034
|
sorbs2b
|
sorbin and SH3 domain containing 2b |
| chr7_-_24838367 | 0.17 |
ENSDART00000139455
ENSDART00000012483 ENSDART00000131530 |
fam113
|
family with sequence similarity 113 |
| chr13_+_21600946 | 0.17 |
ENSDART00000144045
|
sh2d4ba
|
SH2 domain containing 4Ba |
| chr17_-_33415740 | 0.17 |
ENSDART00000135218
|
ccdc28a
|
coiled-coil domain containing 28A |
| chr25_+_31958911 | 0.17 |
ENSDART00000191394
ENSDART00000090727 ENSDART00000185893 |
duox
|
dual oxidase |
| chr15_+_47903864 | 0.16 |
ENSDART00000063835
|
otx5
|
orthodenticle homolog 5 |
| chr9_-_23994225 | 0.16 |
ENSDART00000140346
|
col6a3
|
collagen, type VI, alpha 3 |
| chr19_-_33370271 | 0.16 |
ENSDART00000132628
|
nkd3l
|
naked cuticle homolog 3, like |
| chr15_-_38009344 | 0.16 |
ENSDART00000157094
|
si:dkey-238d18.6
|
si:dkey-238d18.6 |
| chr21_-_39931285 | 0.16 |
ENSDART00000180010
ENSDART00000024407 |
tmigd1
|
transmembrane and immunoglobulin domain containing 1 |
| chr14_+_15484544 | 0.16 |
ENSDART00000188649
|
CR382326.1
|
|
| chr24_+_9372292 | 0.16 |
ENSDART00000082422
ENSDART00000191127 ENSDART00000180510 |
si:ch211-285f17.1
|
si:ch211-285f17.1 |
| chr5_-_57624425 | 0.16 |
ENSDART00000167892
|
zgc:193711
|
zgc:193711 |
| chr17_+_21477892 | 0.16 |
ENSDART00000155309
|
pla2g4f.2
|
phospholipase A2, group IVF, tandem duplicate 2 |
| chr5_+_37890521 | 0.16 |
ENSDART00000140207
|
tmprss4b
|
transmembrane protease, serine 4b |
| chr14_-_46310204 | 0.16 |
ENSDART00000188713
ENSDART00000184930 |
crybb1l1
|
crystallin, beta B1, like 1 |
| chr9_+_12890161 | 0.16 |
ENSDART00000146477
|
si:ch211-167j6.4
|
si:ch211-167j6.4 |
| chr25_+_19238175 | 0.16 |
ENSDART00000110730
ENSDART00000193619 ENSDART00000154420 |
ppip5k1b
|
diphosphoinositol pentakisphosphate kinase 1b |
| chr9_+_18576047 | 0.16 |
ENSDART00000145174
|
lacc1
|
laccase (multicopper oxidoreductase) domain containing 1 |
| chr7_-_31321027 | 0.15 |
ENSDART00000186878
|
CR356242.1
|
|
| chr15_-_20468302 | 0.15 |
ENSDART00000018514
|
dlc
|
deltaC |
| chr20_+_36812368 | 0.15 |
ENSDART00000062931
|
abracl
|
ABRA C-terminal like |
| chr9_+_51180508 | 0.15 |
ENSDART00000138990
|
ifih1
|
interferon induced with helicase C domain 1 |
| chr17_-_17764801 | 0.15 |
ENSDART00000155261
|
slirp
|
SRA stem-loop interacting RNA binding protein |
| chr3_-_34586403 | 0.15 |
ENSDART00000151515
|
sept9a
|
septin 9a |
| chr2_-_43635777 | 0.15 |
ENSDART00000148633
|
itgb1b.1
|
integrin, beta 1b.1 |
| chr12_-_9438227 | 0.15 |
ENSDART00000003932
|
erbb2
|
erb-b2 receptor tyrosine kinase 2 |
| chr4_-_17353100 | 0.15 |
ENSDART00000134467
ENSDART00000189019 |
parpbp
|
PARP1 binding protein |
| chr22_+_26793389 | 0.15 |
ENSDART00000165381
|
pimr69
|
Pim proto-oncogene, serine/threonine kinase, related 69 |
| chr7_+_49695904 | 0.14 |
ENSDART00000183550
ENSDART00000126991 |
ascl1b
|
achaete-scute family bHLH transcription factor 1b |
| chr1_-_23293261 | 0.14 |
ENSDART00000122648
|
ugdh
|
UDP-glucose 6-dehydrogenase |
| chr9_+_17309195 | 0.14 |
ENSDART00000048548
|
scel
|
sciellin |
| chr8_-_13419049 | 0.14 |
ENSDART00000133656
|
pimr101
|
Pim proto-oncogene, serine/threonine kinase, related 101 |
| chr22_-_20289948 | 0.14 |
ENSDART00000132951
|
si:dkey-110c1.10
|
si:dkey-110c1.10 |
| chr4_+_12358822 | 0.14 |
ENSDART00000172557
ENSDART00000150627 |
pimr168
|
Pim proto-oncogene, serine/threonine kinase, related 168 |
| chr8_-_13471916 | 0.13 |
ENSDART00000146558
|
pimr105
|
Pim proto-oncogene, serine/threonine kinase, related 105 |
| chr7_-_17591007 | 0.13 |
ENSDART00000171023
|
CU672228.1
|
|
| chr13_+_21601149 | 0.13 |
ENSDART00000179369
|
sh2d4ba
|
SH2 domain containing 4Ba |
| chr22_-_8860869 | 0.13 |
ENSDART00000166188
|
CABZ01046427.2
|
|
| chr2_-_51500957 | 0.13 |
ENSDART00000172481
|
pigrl3.5
|
polymeric immunoglobulin receptor-like 3.5 |
| chr15_-_37921998 | 0.13 |
ENSDART00000193597
ENSDART00000181443 ENSDART00000168790 |
si:dkey-238d18.5
|
si:dkey-238d18.5 |
| chr6_+_18441331 | 0.13 |
ENSDART00000171005
|
si:dkey-31g6.4
|
si:dkey-31g6.4 |
| chr18_+_15706160 | 0.13 |
ENSDART00000131524
|
si:ch211-264e16.1
|
si:ch211-264e16.1 |
| chr8_-_18200003 | 0.12 |
ENSDART00000080014
|
rps8b
|
ribosomal protein S8b |
| chr1_-_2061419 | 0.12 |
ENSDART00000058878
|
oxgr1a.3
|
oxoglutarate (alpha-ketoglutarate) receptor 1a, tandem duplicate 3 |
| chr1_+_40158146 | 0.12 |
ENSDART00000145694
|
si:ch211-113e8.9
|
si:ch211-113e8.9 |
| chr25_-_35101673 | 0.12 |
ENSDART00000140864
|
zgc:162611
|
zgc:162611 |
| chr11_+_16138065 | 0.12 |
ENSDART00000188616
|
pimr204
|
Pim proto-oncogene, serine/threonine kinase, related 204 |
| chr4_+_12333168 | 0.12 |
ENSDART00000172455
ENSDART00000146166 |
pimr214
pimr171
|
Pim proto-oncogene, serine/threonine kinase, related 214 Pim proto-oncogene, serine/threonine kinase, related 171 |
| chr12_+_29236274 | 0.12 |
ENSDART00000006505
|
mxtx2
|
mix-type homeobox gene 2 |
| chr15_-_3976035 | 0.12 |
ENSDART00000168061
|
si:ch73-309g22.1
|
si:ch73-309g22.1 |
| chr24_-_36270855 | 0.12 |
ENSDART00000154858
|
si:ch211-40k21.5
|
si:ch211-40k21.5 |
| chr7_-_57509447 | 0.12 |
ENSDART00000051973
ENSDART00000147036 |
sirt3
|
sirtuin 3 |
| chr20_+_51479263 | 0.12 |
ENSDART00000148798
|
tlr5a
|
toll-like receptor 5a |
| chr2_-_37537887 | 0.12 |
ENSDART00000143496
ENSDART00000025841 |
arhgef18a
|
rho/rac guanine nucleotide exchange factor (GEF) 18a |
| chr8_-_46525092 | 0.12 |
ENSDART00000030482
|
sult1st2
|
sulfotransferase family 1, cytosolic sulfotransferase 2 |
| chr9_-_7089303 | 0.11 |
ENSDART00000146609
|
coa5
|
cytochrome C oxidase assembly factor 5 |
| chr4_+_12342173 | 0.11 |
ENSDART00000161518
|
pimr214
|
Pim proto-oncogene, serine/threonine kinase, related 214 |
| chr14_-_38865800 | 0.11 |
ENSDART00000173047
|
gsr
|
glutathione reductase |
| chr11_+_42765963 | 0.11 |
ENSDART00000156080
ENSDART00000179888 |
tdrd3
|
tudor domain containing 3 |
| chr5_-_23705828 | 0.11 |
ENSDART00000189419
|
gbgt1l1
|
globoside alpha-1,3-N-acetylgalactosaminyltransferase 1, like 1 |
| chr7_-_16214889 | 0.11 |
ENSDART00000076318
ENSDART00000173946 |
btr06
|
bloodthirsty-related gene family, member 6 |
| chr24_-_12958668 | 0.11 |
ENSDART00000178982
|
FITM1 (1 of many)
|
Danio rerio fat storage inducing transmembrane protein 1 (LOC792443), mRNA. |
| chr23_-_44819100 | 0.11 |
ENSDART00000076373
|
st8sia7.1
|
ST8 alpha-N-acetyl-neuraminide alpha-2,8-sialyltransferase 7.1 |
| chr15_-_37921702 | 0.10 |
ENSDART00000153491
|
si:dkey-238d18.5
|
si:dkey-238d18.5 |
| chr24_-_36271352 | 0.10 |
ENSDART00000153682
ENSDART00000155892 |
si:ch211-40k21.5
|
si:ch211-40k21.5 |
| chr12_-_22355430 | 0.10 |
ENSDART00000153296
ENSDART00000056919 ENSDART00000159036 |
nsfb
|
N-ethylmaleimide-sensitive factor b |
| chr1_-_25679339 | 0.10 |
ENSDART00000161703
ENSDART00000054230 |
fgg
|
fibrinogen gamma chain |
| chr17_+_12265095 | 0.10 |
ENSDART00000153812
|
pimr168
|
Pim proto-oncogene, serine/threonine kinase, related 168 |
| chr25_-_31953516 | 0.10 |
ENSDART00000110180
|
duox2
|
dual oxidase 2 |
| chr12_+_695619 | 0.10 |
ENSDART00000161691
|
abcc3
|
ATP-binding cassette, sub-family C (CFTR/MRP), member 3 |
| chr23_-_22113455 | 0.10 |
ENSDART00000142474
|
phc2a
|
polyhomeotic homolog 2a (Drosophila) |
| chr3_-_34052882 | 0.10 |
ENSDART00000151463
|
ighv11-1
|
immunoglobulin heavy variable 11-1 |
| chr6_-_34008827 | 0.10 |
ENSDART00000191183
ENSDART00000003701 ENSDART00000192502 |
tie1
|
tyrosine kinase with immunoglobulin-like and EGF-like domains 1 |
| chr16_-_31974482 | 0.09 |
ENSDART00000189073
|
styk1
|
serine/threonine/tyrosine kinase 1 |
| chr10_+_8554929 | 0.09 |
ENSDART00000190849
|
tbc1d10ab
|
TBC1 domain family, member 10Ab |
| chr18_+_808911 | 0.09 |
ENSDART00000172518
|
cox5ab
|
cytochrome c oxidase subunit Vab |
| chr4_-_67980261 | 0.09 |
ENSDART00000182305
|
si:ch211-223k15.1
|
si:ch211-223k15.1 |
| chr15_-_36347858 | 0.09 |
ENSDART00000155274
ENSDART00000157936 |
si:dkey-23k10.2
|
si:dkey-23k10.2 |
| chr16_+_33938227 | 0.09 |
ENSDART00000166254
|
gpn2
|
GPN-loop GTPase 2 |
| chr11_+_17984354 | 0.09 |
ENSDART00000179986
|
rpusd4
|
RNA pseudouridylate synthase domain containing 4 |
| chr12_-_20665164 | 0.09 |
ENSDART00000105352
|
gip
|
gastric inhibitory polypeptide |
| chr7_+_53254234 | 0.09 |
ENSDART00000169830
|
trip4
|
thyroid hormone receptor interactor 4 |
| chr18_+_25752592 | 0.09 |
ENSDART00000111767
|
si:ch211-39k3.2
|
si:ch211-39k3.2 |
| chr7_-_32981559 | 0.09 |
ENSDART00000175614
|
pkp3b
|
plakophilin 3b |
| chr13_+_36680564 | 0.09 |
ENSDART00000136030
ENSDART00000006923 |
atp5s
|
ATP synthase, H+ transporting, mitochondrial F0 complex subunit s |
| chr21_+_18274825 | 0.08 |
ENSDART00000144322
ENSDART00000147768 |
wdr5
|
WD repeat domain 5 |
| chr20_-_46817223 | 0.08 |
ENSDART00000100336
|
esrrgb
|
estrogen-related receptor gamma b |
| chr16_-_4203022 | 0.08 |
ENSDART00000018686
|
rrp15
|
ribosomal RNA processing 15 homolog |
| chr13_-_36680531 | 0.08 |
ENSDART00000085298
|
l2hgdh
|
L-2-hydroxyglutarate dehydrogenase |
| chr7_+_9981757 | 0.07 |
ENSDART00000113429
ENSDART00000173233 |
adamts17
|
ADAM metallopeptidase with thrombospondin type 1 motif, 17 |
| chr10_+_38512270 | 0.07 |
ENSDART00000109752
|
serpinh1a
|
serpin peptidase inhibitor, clade H (heat shock protein 47), member 1a |
| chr2_-_24554416 | 0.07 |
ENSDART00000052061
|
cnn2
|
calponin 2 |
| chr2_-_11101329 | 0.07 |
ENSDART00000164416
|
lrrc53
|
leucine rich repeat containing 53 |
| chr6_-_40448416 | 0.07 |
ENSDART00000187019
ENSDART00000179469 |
tatdn2
|
TatD DNase domain containing 2 |
| chr3_+_25825043 | 0.07 |
ENSDART00000153749
|
pik3r6b
|
phosphoinositide-3-kinase, regulatory subunit 6b |
| chr21_-_30111134 | 0.07 |
ENSDART00000014223
|
slc23a1
|
solute carrier family 23 (ascorbic acid transporter), member 1 |
| chr1_-_35113974 | 0.07 |
ENSDART00000192811
ENSDART00000167461 |
BX897691.1
|
|
| chr13_+_8696825 | 0.07 |
ENSDART00000109059
|
ttc7a
|
tetratricopeptide repeat domain 7A |
| chr10_+_28160265 | 0.07 |
ENSDART00000022484
|
rnft1
|
ring finger protein, transmembrane 1 |
Network of associatons between targets according to the STRING database.
First level regulatory network of foxc1a+foxc1b
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.6 | GO:0061341 | non-canonical Wnt signaling pathway involved in heart development(GO:0061341) |
| 0.1 | 0.6 | GO:2000660 | regulation of interleukin-1-mediated signaling pathway(GO:2000659) negative regulation of interleukin-1-mediated signaling pathway(GO:2000660) |
| 0.1 | 0.3 | GO:0002191 | cap-dependent translational initiation(GO:0002191) |
| 0.1 | 0.7 | GO:0031444 | slow-twitch skeletal muscle fiber contraction(GO:0031444) |
| 0.1 | 0.4 | GO:0050891 | multicellular organismal water homeostasis(GO:0050891) |
| 0.1 | 0.3 | GO:0033301 | cell cycle comprising mitosis without cytokinesis(GO:0033301) |
| 0.1 | 0.5 | GO:0086009 | membrane repolarization(GO:0086009) |
| 0.1 | 0.8 | GO:0032264 | IMP salvage(GO:0032264) |
| 0.1 | 0.3 | GO:0008206 | bile acid biosynthetic process(GO:0006699) bile acid metabolic process(GO:0008206) |
| 0.1 | 0.6 | GO:0097369 | sodium ion import(GO:0097369) sodium ion import across plasma membrane(GO:0098719) sodium ion import into cell(GO:1990118) |
| 0.1 | 0.3 | GO:0002184 | cytoplasmic translational termination(GO:0002184) |
| 0.0 | 0.2 | GO:0006590 | thyroid hormone generation(GO:0006590) |
| 0.0 | 0.2 | GO:1901073 | chitin biosynthetic process(GO:0006031) glucosamine-containing compound biosynthetic process(GO:1901073) |
| 0.0 | 0.2 | GO:0034146 | toll-like receptor 5 signaling pathway(GO:0034146) |
| 0.0 | 0.1 | GO:0048939 | anterior lateral line nerve glial cell differentiation(GO:0048913) myelination of anterior lateral line nerve axons(GO:0048914) anterior lateral line nerve glial cell development(GO:0048939) anterior lateral line nerve glial cell morphogenesis involved in differentiation(GO:0048940) |
| 0.0 | 0.1 | GO:0048211 | Golgi vesicle docking(GO:0048211) |
| 0.0 | 0.2 | GO:1902369 | negative regulation of RNA catabolic process(GO:1902369) |
| 0.0 | 0.4 | GO:0045880 | positive regulation of smoothened signaling pathway(GO:0045880) |
| 0.0 | 0.2 | GO:0000056 | ribosomal small subunit export from nucleus(GO:0000056) |
| 0.0 | 0.2 | GO:0060842 | arterial endothelial cell differentiation(GO:0060842) |
| 0.0 | 0.2 | GO:0000455 | enzyme-directed rRNA pseudouridine synthesis(GO:0000455) |
| 0.0 | 0.2 | GO:0032728 | interferon-beta production(GO:0032608) regulation of interferon-beta production(GO:0032648) positive regulation of interferon-beta production(GO:0032728) |
| 0.0 | 0.1 | GO:2000042 | negative regulation of double-strand break repair via homologous recombination(GO:2000042) |
| 0.0 | 0.2 | GO:0033314 | mitotic DNA replication checkpoint(GO:0033314) |
| 0.0 | 0.3 | GO:0045646 | regulation of erythrocyte differentiation(GO:0045646) |
| 0.0 | 0.1 | GO:0015882 | L-ascorbic acid transport(GO:0015882) |
| 0.0 | 0.1 | GO:0042304 | regulation of fatty acid biosynthetic process(GO:0042304) |
| 0.0 | 0.1 | GO:1904292 | regulation of ERAD pathway(GO:1904292) |
| 0.0 | 0.2 | GO:0046512 | diol biosynthetic process(GO:0034312) sphingosine biosynthetic process(GO:0046512) |
| 0.0 | 0.3 | GO:1903963 | icosanoid secretion(GO:0032309) arachidonic acid secretion(GO:0050482) arachidonate transport(GO:1903963) |
| 0.0 | 0.4 | GO:0006743 | ubiquinone metabolic process(GO:0006743) ubiquinone biosynthetic process(GO:0006744) quinone biosynthetic process(GO:1901663) |
| 0.0 | 0.6 | GO:0030199 | collagen fibril organization(GO:0030199) |
| 0.0 | 0.1 | GO:0008210 | estrogen metabolic process(GO:0008210) |
| 0.0 | 0.2 | GO:0034629 | cellular protein complex localization(GO:0034629) |
| 0.0 | 0.1 | GO:0090136 | epithelial cell-cell adhesion(GO:0090136) |
| 0.0 | 0.6 | GO:0030968 | endoplasmic reticulum unfolded protein response(GO:0030968) |
| 0.0 | 0.1 | GO:0032475 | otolith formation(GO:0032475) |
| 0.0 | 0.1 | GO:0032776 | DNA methylation on cytosine(GO:0032776) |
| 0.0 | 0.6 | GO:0000187 | activation of MAPK activity(GO:0000187) |
| 0.0 | 0.2 | GO:0006020 | inositol metabolic process(GO:0006020) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.5 | GO:0005588 | collagen type V trimer(GO:0005588) |
| 0.1 | 0.3 | GO:0061702 | inflammasome complex(GO:0061702) |
| 0.1 | 0.6 | GO:0031526 | brush border membrane(GO:0031526) |
| 0.1 | 1.2 | GO:0031430 | M band(GO:0031430) |
| 0.1 | 0.3 | GO:0018444 | translation release factor complex(GO:0018444) |
| 0.0 | 0.3 | GO:0030428 | cell septum(GO:0030428) |
| 0.0 | 1.6 | GO:0031941 | filamentous actin(GO:0031941) |
| 0.0 | 0.1 | GO:0005577 | fibrinogen complex(GO:0005577) |
| 0.0 | 0.4 | GO:0016328 | lateral plasma membrane(GO:0016328) |
| 0.0 | 0.2 | GO:0043020 | NADPH oxidase complex(GO:0043020) |
| 0.0 | 0.2 | GO:0005664 | origin recognition complex(GO:0000808) nuclear origin of replication recognition complex(GO:0005664) |
| 0.0 | 0.3 | GO:0016282 | eukaryotic 43S preinitiation complex(GO:0016282) |
| 0.0 | 0.1 | GO:0035145 | exon-exon junction complex(GO:0035145) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.3 | GO:0098808 | mRNA cap binding(GO:0098808) |
| 0.1 | 0.3 | GO:0003999 | adenine phosphoribosyltransferase activity(GO:0003999) |
| 0.1 | 0.3 | GO:1990825 | sequence-specific mRNA binding(GO:1990825) |
| 0.1 | 1.6 | GO:0051371 | muscle alpha-actinin binding(GO:0051371) alpha-actinin binding(GO:0051393) |
| 0.1 | 0.8 | GO:0047623 | AMP deaminase activity(GO:0003876) adenosine-phosphate deaminase activity(GO:0047623) |
| 0.1 | 0.6 | GO:0048019 | receptor antagonist activity(GO:0048019) |
| 0.1 | 0.6 | GO:0015386 | monovalent cation:proton antiporter activity(GO:0005451) sodium:proton antiporter activity(GO:0015385) potassium:proton antiporter activity(GO:0015386) |
| 0.1 | 0.5 | GO:0016286 | small conductance calcium-activated potassium channel activity(GO:0016286) |
| 0.0 | 0.3 | GO:0001217 | bacterial-type RNA polymerase transcription factor activity, sequence-specific DNA binding(GO:0001130) bacterial-type RNA polymerase transcriptional repressor activity, sequence-specific DNA binding(GO:0001217) |
| 0.0 | 0.3 | GO:0004100 | chitin synthase activity(GO:0004100) |
| 0.0 | 0.4 | GO:0047498 | calcium-dependent phospholipase A2 activity(GO:0047498) |
| 0.0 | 0.4 | GO:0048039 | ubiquinone binding(GO:0048039) |
| 0.0 | 0.2 | GO:0033857 | inositol heptakisphosphate kinase activity(GO:0000829) diphosphoinositol-pentakisphosphate kinase activity(GO:0033857) |
| 0.0 | 0.5 | GO:0005332 | gamma-aminobutyric acid:sodium symporter activity(GO:0005332) |
| 0.0 | 0.2 | GO:0017040 | ceramidase activity(GO:0017040) |
| 0.0 | 0.1 | GO:0003979 | UDP-glucose 6-dehydrogenase activity(GO:0003979) |
| 0.0 | 0.4 | GO:0030172 | troponin C binding(GO:0030172) troponin I binding(GO:0031013) |
| 0.0 | 0.1 | GO:0098639 | C-X3-C chemokine binding(GO:0019960) protein binding involved in cell-matrix adhesion(GO:0098634) collagen binding involved in cell-matrix adhesion(GO:0098639) |
| 0.0 | 0.6 | GO:0004708 | MAP kinase kinase activity(GO:0004708) |
| 0.0 | 0.5 | GO:0033691 | sialic acid binding(GO:0033691) |
| 0.0 | 0.4 | GO:0030552 | cAMP binding(GO:0030552) |
| 0.0 | 0.3 | GO:0055102 | phospholipase inhibitor activity(GO:0004859) lipase inhibitor activity(GO:0055102) |
| 0.0 | 0.1 | GO:0004362 | glutathione-disulfide reductase activity(GO:0004362) |
| 0.0 | 0.3 | GO:0097199 | cysteine-type endopeptidase activity involved in apoptotic signaling pathway(GO:0097199) |
| 0.0 | 0.5 | GO:0022848 | acetylcholine-gated cation channel activity(GO:0022848) |
| 0.0 | 0.2 | GO:0016175 | superoxide-generating NADPH oxidase activity(GO:0016175) |
| 0.0 | 0.1 | GO:0031769 | glucagon receptor binding(GO:0031769) |
| 0.0 | 0.0 | GO:0030251 | cyclase inhibitor activity(GO:0010852) guanylate cyclase inhibitor activity(GO:0030251) |
| 0.0 | 0.3 | GO:0005080 | protein kinase C binding(GO:0005080) |
| 0.0 | 0.5 | GO:0005518 | collagen binding(GO:0005518) |
| 0.0 | 0.3 | GO:0005112 | Notch binding(GO:0005112) |
| 0.0 | 0.1 | GO:0003828 | alpha-N-acetylneuraminate alpha-2,8-sialyltransferase activity(GO:0003828) |
| 0.0 | 0.1 | GO:0004062 | aryl sulfotransferase activity(GO:0004062) |
| 0.0 | 0.3 | GO:0004089 | carbonate dehydratase activity(GO:0004089) |
| 0.0 | 0.2 | GO:0070403 | NAD+ binding(GO:0070403) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.6 | SA MMP CYTOKINE CONNECTION | Cytokines can induce activation of matrix metalloproteinases, which degrade extracellular matrix. |
| 0.0 | 0.7 | NABA COLLAGENS | Genes encoding collagen proteins |
| 0.0 | 0.1 | PID ERBB NETWORK PATHWAY | ErbB receptor signaling network |
| 0.0 | 0.2 | PID AR NONGENOMIC PATHWAY | Nongenotropic Androgen signaling |
| 0.0 | 0.4 | NABA PROTEOGLYCANS | Genes encoding proteoglycans |
| 0.0 | 0.2 | PID IL12 STAT4 PATHWAY | IL12 signaling mediated by STAT4 |
| 0.0 | 0.6 | PID ERBB2 ERBB3 PATHWAY | ErbB2/ErbB3 signaling events |
| 0.0 | 0.3 | SIG BCR SIGNALING PATHWAY | Members of the BCR signaling pathway |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.5 | REACTOME PRESYNAPTIC NICOTINIC ACETYLCHOLINE RECEPTORS | Genes involved in Presynaptic nicotinic acetylcholine receptors |
| 0.0 | 0.3 | REACTOME SYNTHESIS OF BILE ACIDS AND BILE SALTS VIA 7ALPHA HYDROXYCHOLESTEROL | Genes involved in Synthesis of bile acids and bile salts via 7alpha-hydroxycholesterol |
| 0.0 | 0.3 | REACTOME PHOSPHORYLATION OF CD3 AND TCR ZETA CHAINS | Genes involved in Phosphorylation of CD3 and TCR zeta chains |
| 0.0 | 0.2 | REACTOME CD28 DEPENDENT VAV1 PATHWAY | Genes involved in CD28 dependent Vav1 pathway |
| 0.0 | 0.3 | REACTOME REVERSIBLE HYDRATION OF CARBON DIOXIDE | Genes involved in Reversible Hydration of Carbon Dioxide |
| 0.0 | 0.1 | REACTOME RECYCLING OF BILE ACIDS AND SALTS | Genes involved in Recycling of bile acids and salts |
| 0.0 | 0.2 | REACTOME NFKB ACTIVATION THROUGH FADD RIP1 PATHWAY MEDIATED BY CASPASE 8 AND10 | Genes involved in NF-kB activation through FADD/RIP-1 pathway mediated by caspase-8 and -10 |
| 0.0 | 0.1 | REACTOME DOWNREGULATION OF ERBB2 ERBB3 SIGNALING | Genes involved in Downregulation of ERBB2:ERBB3 signaling |
| 0.0 | 0.7 | REACTOME COLLAGEN FORMATION | Genes involved in Collagen formation |
| 0.0 | 0.6 | REACTOME IL1 SIGNALING | Genes involved in Interleukin-1 signaling |
| 0.0 | 0.2 | REACTOME ASSOCIATION OF LICENSING FACTORS WITH THE PRE REPLICATIVE COMPLEX | Genes involved in Association of licensing factors with the pre-replicative complex |