Project
PRJNA207719: Tissue specific transcriptome profiling
Navigation
Downloads
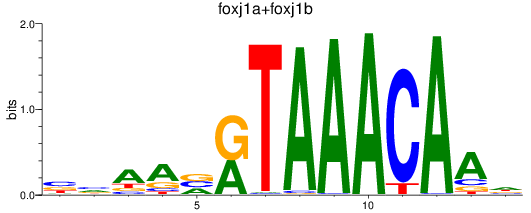
Results for foxj1a+foxj1b
Z-value: 1.22
Transcription factors associated with foxj1a+foxj1b
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
foxj1b
|
ENSDARG00000088290 | forkhead box J1b |
|
foxj1a
|
ENSDARG00000101919 | forkhead box J1a |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| foxj1b | dr11_v1_chr12_+_20149305_20149311 | -0.92 | 2.6e-02 | Click! |
| foxj1a | dr11_v1_chr3_+_60721342_60721342 | -0.90 | 3.9e-02 | Click! |
Activity profile of foxj1a+foxj1b motif
Sorted Z-values of foxj1a+foxj1b motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr4_+_7391400 | 1.60 |
ENSDART00000169111
ENSDART00000186395 |
tnni4a
|
troponin I4a |
| chr4_+_7391110 | 1.48 |
ENSDART00000160708
ENSDART00000187823 |
tnni4a
|
troponin I4a |
| chr1_+_53945934 | 1.39 |
ENSDART00000052838
|
acta1a
|
actin, alpha 1a, skeletal muscle |
| chr23_-_5683147 | 0.94 |
ENSDART00000102766
ENSDART00000067351 |
tnnt2a
|
troponin T type 2a (cardiac) |
| chr21_-_22951604 | 0.89 |
ENSDART00000083449
ENSDART00000180129 |
dub
|
duboraya |
| chr21_+_10577527 | 0.84 |
ENSDART00000165070
ENSDART00000127963 |
ccbe1
|
collagen and calcium binding EGF domains 1 |
| chr25_-_13188678 | 0.72 |
ENSDART00000125754
|
si:ch211-147m6.1
|
si:ch211-147m6.1 |
| chr22_+_19553390 | 0.68 |
ENSDART00000061739
|
hsd11b1la
|
hydroxysteroid (11-beta) dehydrogenase 1-like a |
| chr12_+_17100021 | 0.63 |
ENSDART00000177923
|
acta2
|
actin, alpha 2, smooth muscle, aorta |
| chr18_+_35842933 | 0.58 |
ENSDART00000151587
ENSDART00000131121 |
ppp1r13l
|
protein phosphatase 1, regulatory subunit 13 like |
| chr16_-_26731928 | 0.55 |
ENSDART00000135860
|
rnf41l
|
ring finger protein 41, like |
| chr10_+_22775253 | 0.52 |
ENSDART00000190141
|
tmem88a
|
transmembrane protein 88 a |
| chr25_-_27843066 | 0.50 |
ENSDART00000179684
ENSDART00000186000 ENSDART00000190065 |
asb15a
|
ankyrin repeat and SOCS box containing 15a |
| chr17_+_15297398 | 0.50 |
ENSDART00000156574
|
si:ch211-270g19.5
|
si:ch211-270g19.5 |
| chr24_-_37640705 | 0.49 |
ENSDART00000066583
|
zgc:112496
|
zgc:112496 |
| chr21_-_20840714 | 0.49 |
ENSDART00000144861
ENSDART00000139430 |
c6
|
complement component 6 |
| chr21_-_5881344 | 0.49 |
ENSDART00000009241
|
rpl35
|
ribosomal protein L35 |
| chr16_+_26732086 | 0.48 |
ENSDART00000138496
|
rad54b
|
RAD54 homolog B (S. cerevisiae) |
| chr3_+_32671146 | 0.48 |
ENSDART00000039466
|
rnf25
|
ring finger protein 25 |
| chr8_-_30791266 | 0.48 |
ENSDART00000062220
|
gstt1a
|
glutathione S-transferase theta 1a |
| chr15_+_6661343 | 0.47 |
ENSDART00000160136
|
nop53
|
NOP53 ribosome biogenesis factor |
| chr8_-_30791089 | 0.47 |
ENSDART00000147332
|
gstt1a
|
glutathione S-transferase theta 1a |
| chr4_-_6373735 | 0.47 |
ENSDART00000140100
|
MDFIC
|
si:ch73-156e19.1 |
| chr2_+_55365727 | 0.46 |
ENSDART00000162943
|
FP245456.1
|
|
| chr14_+_15155684 | 0.45 |
ENSDART00000167966
|
zgc:158852
|
zgc:158852 |
| chr17_-_20167206 | 0.45 |
ENSDART00000104874
ENSDART00000191995 |
p4ha1b
|
prolyl 4-hydroxylase, alpha polypeptide I b |
| chr15_-_30816370 | 0.45 |
ENSDART00000142982
ENSDART00000050649 ENSDART00000136901 ENSDART00000100194 |
msi2b
|
musashi RNA-binding protein 2b |
| chr21_-_3796461 | 0.45 |
ENSDART00000021976
|
nsa2
|
NSA2 ribosome biogenesis homolog (S. cerevisiae) |
| chr4_-_16353733 | 0.44 |
ENSDART00000186785
|
lum
|
lumican |
| chr19_+_30990815 | 0.43 |
ENSDART00000134645
|
sync
|
syncoilin, intermediate filament protein |
| chr4_-_12795436 | 0.43 |
ENSDART00000131026
ENSDART00000075127 |
b2m
|
beta-2-microglobulin |
| chr2_-_38035235 | 0.42 |
ENSDART00000075904
|
cbln5
|
cerebellin 5 |
| chr15_+_34069746 | 0.42 |
ENSDART00000163513
|
arl4aa
|
ADP-ribosylation factor-like 4aa |
| chr7_-_28658143 | 0.41 |
ENSDART00000173556
|
adgrg1
|
adhesion G protein-coupled receptor G1 |
| chr7_-_30127082 | 0.41 |
ENSDART00000173749
|
alpk3b
|
alpha-kinase 3b |
| chr3_-_13546610 | 0.40 |
ENSDART00000159647
|
amdhd2
|
amidohydrolase domain containing 2 |
| chr11_+_7264457 | 0.40 |
ENSDART00000154182
|
reep6
|
receptor accessory protein 6 |
| chr14_+_26439227 | 0.39 |
ENSDART00000054183
|
gpr137
|
G protein-coupled receptor 137 |
| chr16_-_24195252 | 0.39 |
ENSDART00000136205
ENSDART00000048599 ENSDART00000161547 |
rps19
|
ribosomal protein S19 |
| chr6_-_55864687 | 0.39 |
ENSDART00000160991
|
cyp24a1
|
cytochrome P450, family 24, subfamily A, polypeptide 1 |
| chr5_-_34993242 | 0.39 |
ENSDART00000134516
ENSDART00000051295 |
btf3
|
basic transcription factor 3 |
| chr10_-_17501528 | 0.38 |
ENSDART00000144847
|
slc2a11l
|
solute carrier family 2 (facilitated glucose transporter), member 11-like |
| chr4_+_5842433 | 0.38 |
ENSDART00000124085
ENSDART00000179848 |
usp18
|
ubiquitin specific peptidase 18 |
| chr15_-_28805493 | 0.38 |
ENSDART00000179617
|
cd3eap
|
CD3e molecule, epsilon associated protein |
| chr25_-_27842654 | 0.38 |
ENSDART00000154852
ENSDART00000156906 |
asb15a
|
ankyrin repeat and SOCS box containing 15a |
| chr14_-_29799993 | 0.37 |
ENSDART00000133775
ENSDART00000005568 |
pdlim3b
|
PDZ and LIM domain 3b |
| chr2_+_21000334 | 0.37 |
ENSDART00000062563
ENSDART00000147809 |
rreb1b
|
ras responsive element binding protein 1b |
| chr7_-_60831082 | 0.37 |
ENSDART00000073654
ENSDART00000136999 |
pcxb
|
pyruvate carboxylase b |
| chr18_-_17399291 | 0.37 |
ENSDART00000192075
ENSDART00000060949 ENSDART00000188506 |
zfpm1
|
zinc finger protein, FOG family member 1 |
| chr19_+_18739085 | 0.37 |
ENSDART00000188868
|
spaca4l
|
sperm acrosome associated 4 like |
| chr23_+_7692042 | 0.36 |
ENSDART00000018512
|
pofut1
|
protein O-fucosyltransferase 1 |
| chr25_+_28555584 | 0.36 |
ENSDART00000157046
|
SLC15A5
|
si:ch211-190o6.3 |
| chr4_-_17725008 | 0.36 |
ENSDART00000016658
|
chpt1
|
choline phosphotransferase 1 |
| chr16_+_33143503 | 0.35 |
ENSDART00000058471
ENSDART00000179385 |
rhbdl2
|
rhomboid, veinlet-like 2 (Drosophila) |
| chr10_-_3416258 | 0.35 |
ENSDART00000005168
|
ddx55
|
DEAD (Asp-Glu-Ala-Asp) box polypeptide 55 |
| chr11_-_6048490 | 0.35 |
ENSDART00000066164
|
plvapb
|
plasmalemma vesicle associated protein b |
| chr19_+_33476557 | 0.34 |
ENSDART00000181800
|
triqk
|
triple QxxK/R motif containing |
| chr2_+_38271392 | 0.34 |
ENSDART00000042100
|
homeza
|
homeobox and leucine zipper encoding a |
| chr15_-_4011013 | 0.33 |
ENSDART00000158029
|
CR626884.3
|
|
| chr14_-_35462148 | 0.33 |
ENSDART00000045809
|
srpx2
|
sushi-repeat containing protein, X-linked 2 |
| chr10_+_28428222 | 0.33 |
ENSDART00000135003
|
si:ch211-222e20.4
|
si:ch211-222e20.4 |
| chr13_+_42602406 | 0.33 |
ENSDART00000133388
ENSDART00000147996 |
mlh1
|
mutL homolog 1, colon cancer, nonpolyposis type 2 (E. coli) |
| chr17_+_24111392 | 0.32 |
ENSDART00000180123
ENSDART00000182787 ENSDART00000189752 ENSDART00000184940 ENSDART00000185363 ENSDART00000064067 |
ehbp1
|
EH domain binding protein 1 |
| chr16_+_22587661 | 0.32 |
ENSDART00000129612
ENSDART00000142241 |
she
|
Src homology 2 domain containing E |
| chr13_-_31647323 | 0.32 |
ENSDART00000135381
|
six4a
|
SIX homeobox 4a |
| chr25_-_13188214 | 0.31 |
ENSDART00000187298
|
si:ch211-147m6.1
|
si:ch211-147m6.1 |
| chr23_-_31266586 | 0.31 |
ENSDART00000139746
|
si:dkey-261l7.2
|
si:dkey-261l7.2 |
| chr21_-_11632403 | 0.31 |
ENSDART00000171708
ENSDART00000138619 ENSDART00000136308 ENSDART00000144770 |
cast
|
calpastatin |
| chr22_-_28653074 | 0.31 |
ENSDART00000154717
|
col8a1b
|
collagen, type VIII, alpha 1b |
| chr5_-_19014589 | 0.31 |
ENSDART00000002624
|
ranbp1
|
RAN binding protein 1 |
| chr4_-_12795030 | 0.30 |
ENSDART00000150427
|
b2m
|
beta-2-microglobulin |
| chr14_-_41478265 | 0.30 |
ENSDART00000149886
ENSDART00000016002 |
tspan7
|
tetraspanin 7 |
| chr22_-_2959005 | 0.29 |
ENSDART00000040701
|
ing5a
|
inhibitor of growth family, member 5a |
| chr17_-_15747296 | 0.29 |
ENSDART00000153970
|
cx52.7
|
connexin 52.7 |
| chr14_+_30910114 | 0.29 |
ENSDART00000187166
ENSDART00000078187 |
foxo4
|
forkhead box O4 |
| chr9_+_11293830 | 0.29 |
ENSDART00000144440
|
wnt6b
|
wingless-type MMTV integration site family, member 6b |
| chr17_+_43843536 | 0.29 |
ENSDART00000164097
|
FO704777.1
|
|
| chr2_+_19633493 | 0.29 |
ENSDART00000147989
|
pimr54
|
Pim proto-oncogene, serine/threonine kinase, related 54 |
| chr7_+_9904627 | 0.28 |
ENSDART00000172824
|
cers3a
|
ceramide synthase 3a |
| chr20_-_54014539 | 0.28 |
ENSDART00000060466
|
si:dkey-241l7.6
|
si:dkey-241l7.6 |
| chr5_+_29726428 | 0.28 |
ENSDART00000143183
|
ddx31
|
DEAD (Asp-Glu-Ala-Asp) box polypeptide 31 |
| chr25_-_26893006 | 0.27 |
ENSDART00000006709
|
dldh
|
dihydrolipoamide dehydrogenase |
| chr3_+_25216790 | 0.27 |
ENSDART00000156544
|
c1qtnf6a
|
C1q and TNF related 6a |
| chr5_-_66160415 | 0.27 |
ENSDART00000073895
|
mboat4
|
membrane bound O-acyltransferase domain containing 4 |
| chr25_-_35360096 | 0.27 |
ENSDART00000154053
ENSDART00000171917 |
si:ch73-147o17.1
|
si:ch73-147o17.1 |
| chr15_-_44465679 | 0.27 |
ENSDART00000054605
|
aasdhppt
|
aminoadipate-semialdehyde dehydrogenase-phosphopantetheinyl transferase |
| chr13_+_35635672 | 0.27 |
ENSDART00000148481
|
thbs2a
|
thrombospondin 2a |
| chr8_+_10305400 | 0.26 |
ENSDART00000172400
|
pim1
|
Pim-1 proto-oncogene, serine/threonine kinase |
| chr10_+_11260170 | 0.25 |
ENSDART00000155742
|
hsdl2
|
hydroxysteroid dehydrogenase like 2 |
| chr14_-_36799280 | 0.25 |
ENSDART00000168615
|
rnf130
|
ring finger protein 130 |
| chr13_-_43149063 | 0.25 |
ENSDART00000099601
|
vsir
|
V-set immunoregulatory receptor |
| chr12_+_28117365 | 0.25 |
ENSDART00000066290
|
UTS2R
|
urotensin 2 receptor |
| chr20_+_10723292 | 0.25 |
ENSDART00000152805
|
si:ch211-182e10.4
|
si:ch211-182e10.4 |
| chr17_+_25519089 | 0.24 |
ENSDART00000041721
|
aim1a
|
crystallin beta-gamma domain containing 1a |
| chr8_+_49570884 | 0.24 |
ENSDART00000182117
ENSDART00000108613 |
rasef
|
RAS and EF-hand domain containing |
| chr17_-_2690083 | 0.24 |
ENSDART00000135374
|
ptpn21
|
protein tyrosine phosphatase, non-receptor type 21 |
| chr17_-_23709347 | 0.24 |
ENSDART00000124661
|
papss2a
|
3'-phosphoadenosine 5'-phosphosulfate synthase 2a |
| chr13_+_18533005 | 0.24 |
ENSDART00000136024
|
ftr14l
|
finTRIM family, member 14-like |
| chr21_+_3796620 | 0.24 |
ENSDART00000099535
ENSDART00000144515 |
spout1
|
SPOUT domain containing methyltransferase 1 |
| chr16_-_24605969 | 0.24 |
ENSDART00000163305
ENSDART00000167121 |
fxyd6l
|
FXYD domain containing ion transport regulator 6 like |
| chr5_+_44944778 | 0.24 |
ENSDART00000130428
ENSDART00000044361 ENSDART00000128825 ENSDART00000124637 ENSDART00000126066 ENSDART00000177635 |
dmrt1
|
doublesex and mab-3 related transcription factor 1 |
| chr20_-_54014373 | 0.24 |
ENSDART00000152934
|
si:dkey-241l7.6
|
si:dkey-241l7.6 |
| chr7_+_4694924 | 0.23 |
ENSDART00000144873
|
si:ch211-225k7.3
|
si:ch211-225k7.3 |
| chr23_+_38159715 | 0.23 |
ENSDART00000137969
|
zgc:112994
|
zgc:112994 |
| chr20_-_1314355 | 0.23 |
ENSDART00000152436
|
pcmt
|
protein-L-isoaspartate (D-aspartate) O-methyltransferase |
| chr23_+_33296588 | 0.23 |
ENSDART00000030368
|
si:ch211-226m16.3
|
si:ch211-226m16.3 |
| chr17_-_26867725 | 0.23 |
ENSDART00000153590
|
si:dkey-221l4.10
|
si:dkey-221l4.10 |
| chr10_+_40836378 | 0.23 |
ENSDART00000085792
|
trim69
|
tripartite motif containing 69 |
| chr3_-_18410968 | 0.23 |
ENSDART00000041842
|
ndufb10
|
NADH dehydrogenase (ubiquinone) 1 beta subcomplex, 10 |
| chr7_-_28549361 | 0.22 |
ENSDART00000173918
ENSDART00000054368 ENSDART00000113313 |
st5
|
suppression of tumorigenicity 5 |
| chr12_-_11457625 | 0.22 |
ENSDART00000012318
|
htra1b
|
HtrA serine peptidase 1b |
| chr7_+_24165371 | 0.22 |
ENSDART00000173472
|
si:ch211-216p19.5
|
si:ch211-216p19.5 |
| chr15_-_44052927 | 0.22 |
ENSDART00000166209
|
wu:fb44b02
|
wu:fb44b02 |
| chr10_-_23099809 | 0.22 |
ENSDART00000148333
ENSDART00000079703 ENSDART00000162444 |
nle1
|
notchless homolog 1 (Drosophila) |
| chr25_-_13202208 | 0.22 |
ENSDART00000168155
|
si:ch211-194m7.4
|
si:ch211-194m7.4 |
| chr22_+_3184500 | 0.22 |
ENSDART00000176409
ENSDART00000160604 |
ftsj3
|
FtsJ RNA methyltransferase homolog 3 |
| chr10_-_42237304 | 0.22 |
ENSDART00000140341
|
tcf7l1a
|
transcription factor 7 like 1a |
| chr4_-_1908179 | 0.21 |
ENSDART00000139586
|
ano6
|
anoctamin 6 |
| chr15_+_14856307 | 0.21 |
ENSDART00000167213
|
diabloa
|
diablo, IAP-binding mitochondrial protein a |
| chr22_+_11520249 | 0.21 |
ENSDART00000063147
|
sgsh
|
N-sulfoglucosamine sulfohydrolase (sulfamidase) |
| chr16_-_7228276 | 0.21 |
ENSDART00000149030
|
nt5c3a
|
5'-nucleotidase, cytosolic IIIA |
| chr20_+_26881600 | 0.21 |
ENSDART00000174799
|
serpinb1
|
serpin peptidase inhibitor, clade B (ovalbumin), member 1 |
| chr7_-_36096582 | 0.21 |
ENSDART00000188507
|
CR848715.1
|
|
| chr5_+_12528693 | 0.21 |
ENSDART00000051670
|
rfc5
|
replication factor C (activator 1) 5 |
| chr15_-_7337148 | 0.21 |
ENSDART00000182568
|
SLC7A1 (1 of many)
|
high affinity cationic amino acid transporter 1 |
| chr8_+_49065348 | 0.21 |
ENSDART00000032277
|
ddx51
|
DEAD (Asp-Glu-Ala-Asp) box polypeptide 51 |
| chr19_+_4051535 | 0.21 |
ENSDART00000167084
|
btr24
|
bloodthirsty-related gene family, member 24 |
| chr1_-_30689004 | 0.21 |
ENSDART00000018827
|
dachc
|
dachshund c |
| chr23_-_27123171 | 0.21 |
ENSDART00000023218
ENSDART00000146467 |
stat6
|
signal transducer and activator of transcription 6, interleukin-4 induced |
| chr25_-_31423493 | 0.21 |
ENSDART00000027661
|
myod1
|
myogenic differentiation 1 |
| chr24_-_31306724 | 0.20 |
ENSDART00000165399
|
acp5b
|
acid phosphatase 5b, tartrate resistant |
| chr11_-_183328 | 0.20 |
ENSDART00000168562
|
avil
|
advillin |
| chr1_+_9153141 | 0.20 |
ENSDART00000081343
|
plk1
|
polo-like kinase 1 (Drosophila) |
| chr17_+_36627099 | 0.20 |
ENSDART00000154104
|
impg1b
|
interphotoreceptor matrix proteoglycan 1b |
| chr7_+_4732597 | 0.20 |
ENSDART00000134882
|
si:dkey-28d5.1
|
si:dkey-28d5.1 |
| chr25_-_37435060 | 0.20 |
ENSDART00000102855
ENSDART00000148566 |
pdhx
|
pyruvate dehydrogenase complex, component X |
| chr3_+_46762703 | 0.20 |
ENSDART00000133283
|
prkcsh
|
protein kinase C substrate 80K-H |
| chr24_+_37640626 | 0.20 |
ENSDART00000008047
|
wdr24
|
WD repeat domain 24 |
| chr20_+_6142433 | 0.20 |
ENSDART00000054084
ENSDART00000136986 |
ttr
|
transthyretin (prealbumin, amyloidosis type I) |
| chr22_-_9714593 | 0.20 |
ENSDART00000188282
|
si:dkey-286j17.4
|
si:dkey-286j17.4 |
| chr1_+_51537250 | 0.20 |
ENSDART00000152789
|
etaa1
|
ETAA1, ATR kinase activator |
| chr1_-_26702930 | 0.19 |
ENSDART00000109297
ENSDART00000152389 |
foxe1
|
forkhead box E1 |
| chr7_-_50367326 | 0.19 |
ENSDART00000141926
|
prc1b
|
protein regulator of cytokinesis 1b |
| chr3_-_40664868 | 0.19 |
ENSDART00000138783
ENSDART00000178567 |
rnf216
|
ring finger protein 216 |
| chr13_+_30951155 | 0.19 |
ENSDART00000057469
ENSDART00000162254 |
vstm4a
|
V-set and transmembrane domain containing 4a |
| chr10_-_35257458 | 0.19 |
ENSDART00000143890
ENSDART00000139107 ENSDART00000082445 |
prr11
|
proline rich 11 |
| chr23_-_27701361 | 0.19 |
ENSDART00000186688
ENSDART00000183985 |
dnajc22
|
DnaJ (Hsp40) homolog, subfamily C, member 22 |
| chr1_+_36651059 | 0.19 |
ENSDART00000187475
|
ednraa
|
endothelin receptor type Aa |
| chr20_+_37383034 | 0.19 |
ENSDART00000134997
ENSDART00000145927 |
adgrg6
|
adhesion G protein-coupled receptor G6 |
| chr2_+_19522082 | 0.19 |
ENSDART00000146098
|
pimr49
|
Pim proto-oncogene, serine/threonine kinase, related 49 |
| chr8_+_23034718 | 0.19 |
ENSDART00000184512
|
ythdf1
|
YTH N(6)-methyladenosine RNA binding protein 1 |
| chr8_-_42624454 | 0.18 |
ENSDART00000075550
|
kazald3
|
Kazal-type serine peptidase inhibitor domain 3 |
| chr19_-_24958393 | 0.18 |
ENSDART00000098592
|
si:ch211-195b13.6
|
si:ch211-195b13.6 |
| chr17_+_53284040 | 0.18 |
ENSDART00000165368
|
atxn3
|
ataxin 3 |
| chr20_+_25552057 | 0.18 |
ENSDART00000102913
|
cyp2v1
|
cytochrome P450, family 2, subfamily V, polypeptide 1 |
| chr5_+_39563301 | 0.18 |
ENSDART00000131245
ENSDART00000097872 |
si:ch211-57h10.1
|
si:ch211-57h10.1 |
| chr18_-_48550426 | 0.18 |
ENSDART00000145189
|
kcnj1a.1
|
potassium inwardly-rectifying channel, subfamily J, member 1a, tandem duplicate 1 |
| chr6_-_18228358 | 0.18 |
ENSDART00000167937
|
p4hb
|
prolyl 4-hydroxylase, beta polypeptide |
| chr10_-_34089779 | 0.18 |
ENSDART00000140070
|
pimr144
|
Pim proto-oncogene, serine/threonine kinase, related 144 |
| chr6_+_32834760 | 0.18 |
ENSDART00000121562
|
cyldl
|
cylindromatosis (turban tumor syndrome), like |
| chr11_+_25430851 | 0.18 |
ENSDART00000164999
ENSDART00000126403 |
si:dkey-13a21.4
|
si:dkey-13a21.4 |
| chr14_-_28566238 | 0.18 |
ENSDART00000172547
|
insb
|
preproinsulin b |
| chr11_-_16394971 | 0.18 |
ENSDART00000180981
ENSDART00000179925 |
lrig1
|
leucine-rich repeats and immunoglobulin-like domains 1 |
| chr16_+_11699760 | 0.18 |
ENSDART00000156132
|
si:dkey-250k15.7
|
si:dkey-250k15.7 |
| chr7_+_4694762 | 0.18 |
ENSDART00000132862
|
si:ch211-225k7.3
|
si:ch211-225k7.3 |
| chr15_-_21849300 | 0.17 |
ENSDART00000183603
|
laynb
|
layilin b |
| chr13_-_21650404 | 0.17 |
ENSDART00000078352
|
tspan14
|
tetraspanin 14 |
| chr20_+_53368611 | 0.17 |
ENSDART00000060432
|
cdc40
|
cell division cycle 40 homolog (S. cerevisiae) |
| chr9_+_31222026 | 0.17 |
ENSDART00000145573
|
clybl
|
citrate lyase beta like |
| chr9_+_2041535 | 0.17 |
ENSDART00000093187
|
lnpa
|
limb and neural patterns a |
| chr16_-_13595027 | 0.17 |
ENSDART00000060004
|
ntd5
|
ntl-dependent gene 5 |
| chr8_-_50133064 | 0.17 |
ENSDART00000030064
|
antxr1a
|
anthrax toxin receptor 1a |
| chr1_+_55239160 | 0.17 |
ENSDART00000152318
|
si:ch211-286b5.2
|
si:ch211-286b5.2 |
| chr3_+_29510818 | 0.17 |
ENSDART00000055407
ENSDART00000193743 ENSDART00000123619 |
rac2
|
Rac family small GTPase 2 |
| chr18_-_46280578 | 0.17 |
ENSDART00000131724
|
pld3
|
phospholipase D family, member 3 |
| chr23_+_13533885 | 0.17 |
ENSDART00000144386
|
uckl1b
|
uridine-cytidine kinase 1-like 1b |
| chr23_+_24085531 | 0.17 |
ENSDART00000139710
|
ttll10
|
tubulin tyrosine ligase-like family, member 10 |
| chr4_-_49312747 | 0.17 |
ENSDART00000181057
|
BX942819.2
|
|
| chr1_-_54191626 | 0.16 |
ENSDART00000062941
|
nthl1
|
nth-like DNA glycosylase 1 |
| chr4_+_72578548 | 0.16 |
ENSDART00000174035
|
si:cabz01054394.5
|
si:cabz01054394.5 |
| chr22_+_7462997 | 0.16 |
ENSDART00000106082
|
zgc:112368
|
zgc:112368 |
| chr22_-_37686966 | 0.16 |
ENSDART00000192217
|
htr2b
|
5-hydroxytryptamine (serotonin) receptor 2B, G protein-coupled |
| chr9_-_19161982 | 0.16 |
ENSDART00000081878
|
pou1f1
|
POU class 1 homeobox 1 |
| chr7_-_26270014 | 0.16 |
ENSDART00000079347
|
serpine1
|
serpin peptidase inhibitor, clade E (nexin, plasminogen activator inhibitor type 1), member 1 |
| chr23_+_24598910 | 0.16 |
ENSDART00000126510
ENSDART00000078796 |
kansl2
|
KAT8 regulatory NSL complex subunit 2 |
| chr21_+_15704556 | 0.16 |
ENSDART00000024858
ENSDART00000146909 |
chchd10
|
coiled-coil-helix-coiled-coil-helix domain containing 10 |
| chr7_-_24112484 | 0.16 |
ENSDART00000111923
|
ajuba
|
ajuba LIM protein |
| chr16_+_10413551 | 0.16 |
ENSDART00000032877
ENSDART00000172827 |
ino80e
|
INO80 complex subunit E |
| chr17_+_14965570 | 0.16 |
ENSDART00000066604
|
gpr137c
|
G protein-coupled receptor 137c |
| chr11_+_18612421 | 0.16 |
ENSDART00000110621
|
ncoa3
|
nuclear receptor coactivator 3 |
| chr7_-_51300277 | 0.16 |
ENSDART00000174174
|
gc2
|
guanylyl cyclase 2 |
| chr2_+_14992879 | 0.16 |
ENSDART00000137546
|
pimr55
|
Pim proto-oncogene, serine/threonine kinase, related 55 |
| chr6_+_29791164 | 0.16 |
ENSDART00000017424
|
ptmaa
|
prothymosin, alpha a |
| chr24_+_9298198 | 0.15 |
ENSDART00000165780
|
otud1
|
OTU deubiquitinase 1 |
| chr8_-_10048059 | 0.15 |
ENSDART00000138411
|
si:dkey-8e10.3
|
si:dkey-8e10.3 |
| chr7_-_3912905 | 0.15 |
ENSDART00000143864
|
si:dkey-88n24.10
|
si:dkey-88n24.10 |
| chr25_+_19734038 | 0.15 |
ENSDART00000067354
|
zgc:101783
|
zgc:101783 |
| chr15_+_30158652 | 0.15 |
ENSDART00000190682
|
nlk2
|
nemo-like kinase, type 2 |
| chr23_-_43424510 | 0.15 |
ENSDART00000055564
|
rprd1b
|
regulation of nuclear pre-mRNA domain containing 1B |
| chr7_+_46003449 | 0.15 |
ENSDART00000159700
ENSDART00000173625 |
si:ch211-260e23.9
|
si:ch211-260e23.9 |
Network of associatons between targets according to the STRING database.
First level regulatory network of foxj1a+foxj1b
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.7 | 2.0 | GO:0090131 | mesenchyme migration(GO:0090131) |
| 0.3 | 0.9 | GO:0061341 | non-canonical Wnt signaling pathway involved in heart development(GO:0061341) |
| 0.2 | 0.9 | GO:0003228 | atrial cardiac muscle tissue development(GO:0003228) |
| 0.2 | 0.5 | GO:2000726 | negative regulation of striated muscle cell differentiation(GO:0051154) negative regulation of cardiac muscle tissue development(GO:0055026) cardiac muscle cell fate commitment(GO:0060923) canonical Wnt signaling pathway involved in cardiac muscle cell fate commitment(GO:0061317) regulation of canonical Wnt signaling pathway involved in cardiac muscle cell fate commitment(GO:1901295) negative regulation of canonical Wnt signaling pathway involved in cardiac muscle cell fate commitment(GO:1901296) negative regulation of cardiocyte differentiation(GO:1905208) negative regulation of cardiac muscle cell differentiation(GO:2000726) |
| 0.1 | 0.4 | GO:0010430 | fatty acid omega-oxidation(GO:0010430) |
| 0.1 | 0.8 | GO:0060855 | venous endothelial cell migration involved in lymph vessel development(GO:0060855) |
| 0.1 | 0.3 | GO:0051026 | chiasma assembly(GO:0051026) |
| 0.1 | 0.4 | GO:0006046 | N-acetylglucosamine catabolic process(GO:0006046) |
| 0.1 | 0.4 | GO:0051661 | maintenance of centrosome location(GO:0051661) |
| 0.1 | 0.3 | GO:1903069 | regulation of ER-associated ubiquitin-dependent protein catabolic process(GO:1903069) |
| 0.1 | 0.3 | GO:0009085 | lysine biosynthetic process(GO:0009085) lysine biosynthetic process via aminoadipic acid(GO:0019878) |
| 0.1 | 0.5 | GO:0006004 | fucose metabolic process(GO:0006004) |
| 0.1 | 0.2 | GO:0034035 | sulfate assimilation(GO:0000103) purine ribonucleoside bisphosphate metabolic process(GO:0034035) purine ribonucleoside bisphosphate biosynthetic process(GO:0034036) 3'-phosphoadenosine 5'-phosphosulfate metabolic process(GO:0050427) 3'-phosphoadenosine 5'-phosphosulfate biosynthetic process(GO:0050428) |
| 0.1 | 0.2 | GO:0030238 | male sex determination(GO:0030238) |
| 0.1 | 0.4 | GO:0030219 | megakaryocyte differentiation(GO:0030219) |
| 0.1 | 0.2 | GO:0002407 | dendritic cell chemotaxis(GO:0002407) dendritic cell migration(GO:0036336) |
| 0.1 | 0.2 | GO:0002074 | extraocular skeletal muscle development(GO:0002074) |
| 0.1 | 0.4 | GO:0070445 | regulation of oligodendrocyte progenitor proliferation(GO:0070445) |
| 0.1 | 0.2 | GO:0032290 | peripheral nervous system myelin formation(GO:0032290) semicircular canal fusion(GO:0060879) |
| 0.1 | 0.3 | GO:0046607 | positive regulation of centrosome cycle(GO:0046607) |
| 0.1 | 3.1 | GO:0003009 | skeletal muscle contraction(GO:0003009) |
| 0.1 | 0.2 | GO:1903371 | regulation of endoplasmic reticulum tubular network organization(GO:1903371) positive regulation of endoplasmic reticulum tubular network organization(GO:1903373) |
| 0.1 | 0.2 | GO:0060829 | negative regulation of canonical Wnt signaling pathway involved in neural plate anterior/posterior pattern formation(GO:0060829) |
| 0.1 | 0.2 | GO:0060148 | negative regulation of hippo signaling(GO:0035331) positive regulation of posttranscriptional gene silencing(GO:0060148) positive regulation of gene silencing by miRNA(GO:2000637) |
| 0.1 | 0.7 | GO:0045824 | negative regulation of toll-like receptor signaling pathway(GO:0034122) negative regulation of innate immune response(GO:0045824) |
| 0.0 | 0.5 | GO:0010955 | negative regulation of protein processing(GO:0010955) negative regulation of protein maturation(GO:1903318) |
| 0.0 | 0.6 | GO:1904263 | positive regulation of TORC1 signaling(GO:1904263) |
| 0.0 | 0.2 | GO:0008635 | activation of cysteine-type endopeptidase activity involved in apoptotic process by cytochrome c(GO:0008635) |
| 0.0 | 0.2 | GO:0006296 | nucleotide-excision repair, DNA incision, 5'-to lesion(GO:0006296) |
| 0.0 | 0.7 | GO:0000463 | maturation of LSU-rRNA from tricistronic rRNA transcript (SSU-rRNA, 5.8S rRNA, LSU-rRNA)(GO:0000463) |
| 0.0 | 0.2 | GO:2000001 | regulation of DNA damage checkpoint(GO:2000001) |
| 0.0 | 0.1 | GO:0050881 | multicellular organismal movement(GO:0050879) musculoskeletal movement(GO:0050881) |
| 0.0 | 0.4 | GO:0035284 | central nervous system segmentation(GO:0035283) brain segmentation(GO:0035284) |
| 0.0 | 0.2 | GO:0060397 | JAK-STAT cascade involved in growth hormone signaling pathway(GO:0060397) |
| 0.0 | 0.2 | GO:2000389 | neutrophil extravasation(GO:0072672) regulation of neutrophil extravasation(GO:2000389) positive regulation of neutrophil extravasation(GO:2000391) |
| 0.0 | 0.5 | GO:0043462 | regulation of ATPase activity(GO:0043462) |
| 0.0 | 0.1 | GO:0043504 | mitochondrial DNA repair(GO:0043504) |
| 0.0 | 0.2 | GO:0051703 | social behavior(GO:0035176) intraspecies interaction between organisms(GO:0051703) |
| 0.0 | 0.7 | GO:0000027 | ribosomal large subunit assembly(GO:0000027) |
| 0.0 | 0.3 | GO:0051256 | mitotic spindle elongation(GO:0000022) spindle elongation(GO:0051231) mitotic spindle midzone assembly(GO:0051256) |
| 0.0 | 0.3 | GO:1901998 | toxin transport(GO:1901998) |
| 0.0 | 0.4 | GO:0021772 | olfactory bulb development(GO:0021772) olfactory lobe development(GO:0021988) |
| 0.0 | 0.4 | GO:0000470 | maturation of LSU-rRNA(GO:0000470) |
| 0.0 | 0.1 | GO:1904184 | regulation of pyruvate dehydrogenase activity(GO:1904182) positive regulation of pyruvate dehydrogenase activity(GO:1904184) |
| 0.0 | 0.1 | GO:0032637 | interleukin-8 production(GO:0032637) regulation of interleukin-8 production(GO:0032677) positive regulation of interleukin-8 production(GO:0032757) |
| 0.0 | 0.2 | GO:0045721 | negative regulation of gluconeogenesis(GO:0045721) |
| 0.0 | 0.1 | GO:0002025 | vasodilation by norepinephrine-epinephrine involved in regulation of systemic arterial blood pressure(GO:0002025) |
| 0.0 | 0.4 | GO:0018401 | peptidyl-proline hydroxylation to 4-hydroxy-L-proline(GO:0018401) 4-hydroxyproline metabolic process(GO:0019471) |
| 0.0 | 0.9 | GO:0006749 | glutathione metabolic process(GO:0006749) |
| 0.0 | 0.2 | GO:0050779 | RNA destabilization(GO:0050779) mRNA destabilization(GO:0061157) |
| 0.0 | 0.3 | GO:0043114 | regulation of vascular permeability(GO:0043114) |
| 0.0 | 0.2 | GO:0045453 | bone resorption(GO:0045453) |
| 0.0 | 0.2 | GO:0090385 | phagosome-lysosome fusion(GO:0090385) |
| 0.0 | 0.2 | GO:0072337 | modified amino acid transport(GO:0072337) |
| 0.0 | 0.2 | GO:0044273 | sulfur compound catabolic process(GO:0044273) |
| 0.0 | 0.4 | GO:0046686 | response to cadmium ion(GO:0046686) |
| 0.0 | 0.2 | GO:0044211 | CTP salvage(GO:0044211) |
| 0.0 | 0.1 | GO:0051988 | regulation of attachment of spindle microtubules to kinetochore(GO:0051988) |
| 0.0 | 0.3 | GO:0032094 | response to food(GO:0032094) |
| 0.0 | 0.2 | GO:0070782 | phosphatidylserine exposure on apoptotic cell surface(GO:0070782) |
| 0.0 | 0.5 | GO:0019835 | cytolysis(GO:0019835) |
| 0.0 | 0.5 | GO:0051092 | positive regulation of NF-kappaB transcription factor activity(GO:0051092) |
| 0.0 | 0.1 | GO:0021910 | smoothened signaling pathway involved in ventral spinal cord patterning(GO:0021910) regulation of myotome development(GO:2000290) |
| 0.0 | 0.2 | GO:0007076 | mitotic chromosome condensation(GO:0007076) |
| 0.0 | 0.2 | GO:0031641 | regulation of myelination(GO:0031641) |
| 0.0 | 0.3 | GO:2001235 | positive regulation of apoptotic signaling pathway(GO:2001235) |
| 0.0 | 0.1 | GO:0034454 | microtubule anchoring at centrosome(GO:0034454) |
| 0.0 | 0.3 | GO:0045454 | cell redox homeostasis(GO:0045454) |
| 0.0 | 0.0 | GO:0051574 | positive regulation of histone H3-K9 methylation(GO:0051574) |
| 0.0 | 0.1 | GO:0006488 | dolichol-linked oligosaccharide biosynthetic process(GO:0006488) |
| 0.0 | 0.3 | GO:0070536 | protein K63-linked deubiquitination(GO:0070536) |
| 0.0 | 0.2 | GO:0032008 | positive regulation of TOR signaling(GO:0032008) |
| 0.0 | 0.1 | GO:0043330 | response to exogenous dsRNA(GO:0043330) |
| 0.0 | 0.2 | GO:0098868 | endochondral bone growth(GO:0003416) growth plate cartilage development(GO:0003417) bone growth(GO:0098868) |
| 0.0 | 0.1 | GO:0051754 | meiotic sister chromatid cohesion, centromeric(GO:0051754) |
| 0.0 | 0.4 | GO:0030199 | collagen fibril organization(GO:0030199) |
| 0.0 | 0.3 | GO:0007286 | spermatid development(GO:0007286) |
| 0.0 | 0.3 | GO:0007634 | optokinetic behavior(GO:0007634) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 2.0 | GO:0005869 | dynactin complex(GO:0005869) |
| 0.1 | 0.7 | GO:0042612 | MHC class I protein complex(GO:0042612) |
| 0.1 | 4.0 | GO:0005861 | troponin complex(GO:0005861) |
| 0.1 | 0.2 | GO:0043218 | compact myelin(GO:0043218) Schmidt-Lanterman incisure(GO:0043220) |
| 0.1 | 0.4 | GO:0005854 | nascent polypeptide-associated complex(GO:0005854) |
| 0.1 | 0.2 | GO:0098826 | endoplasmic reticulum tubular network membrane(GO:0098826) |
| 0.0 | 0.2 | GO:0017177 | glucosidase II complex(GO:0017177) |
| 0.0 | 0.4 | GO:0031616 | spindle pole centrosome(GO:0031616) |
| 0.0 | 0.2 | GO:0033165 | interphotoreceptor matrix(GO:0033165) |
| 0.0 | 0.3 | GO:0032300 | mismatch repair complex(GO:0032300) |
| 0.0 | 0.5 | GO:0005579 | membrane attack complex(GO:0005579) |
| 0.0 | 0.2 | GO:0005663 | DNA replication factor C complex(GO:0005663) |
| 0.0 | 0.2 | GO:0045254 | pyruvate dehydrogenase complex(GO:0045254) |
| 0.0 | 0.2 | GO:0001669 | acrosomal vesicle(GO:0001669) |
| 0.0 | 0.7 | GO:0030687 | preribosome, large subunit precursor(GO:0030687) |
| 0.0 | 0.3 | GO:1990023 | mitotic spindle midzone(GO:1990023) |
| 0.0 | 0.2 | GO:0005614 | interstitial matrix(GO:0005614) |
| 0.0 | 0.4 | GO:0033017 | sarcoplasmic reticulum membrane(GO:0033017) |
| 0.0 | 0.1 | GO:0070209 | ASTRA complex(GO:0070209) |
| 0.0 | 0.1 | GO:0031428 | box C/D snoRNP complex(GO:0031428) |
| 0.0 | 0.3 | GO:0070776 | H3 histone acetyltransferase complex(GO:0070775) MOZ/MORF histone acetyltransferase complex(GO:0070776) |
| 0.0 | 0.2 | GO:0000815 | ESCRT III complex(GO:0000815) |
| 0.0 | 0.4 | GO:0005844 | polysome(GO:0005844) |
| 0.0 | 0.7 | GO:0031941 | filamentous actin(GO:0031941) |
| 0.0 | 0.8 | GO:0005788 | endoplasmic reticulum lumen(GO:0005788) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.4 | GO:0004736 | pyruvate carboxylase activity(GO:0004736) |
| 0.1 | 0.5 | GO:0042030 | ATPase inhibitor activity(GO:0042030) |
| 0.1 | 0.4 | GO:0070643 | vitamin D 25-hydroxylase activity(GO:0070643) |
| 0.1 | 0.5 | GO:0008097 | 5S rRNA binding(GO:0008097) |
| 0.1 | 0.9 | GO:0030172 | troponin C binding(GO:0030172) troponin I binding(GO:0031013) |
| 0.1 | 0.2 | GO:0004779 | adenylylsulfate kinase activity(GO:0004020) sulfate adenylyltransferase activity(GO:0004779) sulfate adenylyltransferase (ATP) activity(GO:0004781) |
| 0.1 | 0.5 | GO:0015616 | DNA translocase activity(GO:0015616) |
| 0.1 | 0.2 | GO:0016436 | rRNA (uridine) methyltransferase activity(GO:0016436) |
| 0.1 | 0.4 | GO:0004142 | diacylglycerol cholinephosphotransferase activity(GO:0004142) |
| 0.1 | 0.2 | GO:0047777 | (3S)-citramalyl-CoA lyase activity(GO:0047777) |
| 0.1 | 0.3 | GO:0004719 | protein-L-isoaspartate (D-aspartate) O-methyltransferase activity(GO:0004719) |
| 0.1 | 0.2 | GO:0000703 | oxidized pyrimidine nucleobase lesion DNA N-glycosylase activity(GO:0000703) |
| 0.1 | 0.3 | GO:0001784 | phosphotyrosine binding(GO:0001784) |
| 0.0 | 0.2 | GO:0070324 | thyroid hormone binding(GO:0070324) |
| 0.0 | 0.1 | GO:0004560 | alpha-L-fucosidase activity(GO:0004560) fucosidase activity(GO:0015928) |
| 0.0 | 0.3 | GO:0010859 | calcium-dependent cysteine-type endopeptidase inhibitor activity(GO:0010859) |
| 0.0 | 0.4 | GO:0035198 | miRNA binding(GO:0035198) |
| 0.0 | 0.4 | GO:0004656 | procollagen-proline 4-dioxygenase activity(GO:0004656) |
| 0.0 | 0.1 | GO:0051380 | norepinephrine binding(GO:0051380) |
| 0.0 | 0.1 | GO:0003909 | DNA ligase activity(GO:0003909) DNA ligase (ATP) activity(GO:0003910) |
| 0.0 | 0.2 | GO:1990247 | N6-methyladenosine-containing RNA binding(GO:1990247) |
| 0.0 | 0.2 | GO:0070888 | E-box binding(GO:0070888) |
| 0.0 | 0.9 | GO:0004364 | glutathione transferase activity(GO:0004364) |
| 0.0 | 0.2 | GO:0004738 | pyruvate dehydrogenase activity(GO:0004738) |
| 0.0 | 0.1 | GO:0010852 | cyclase inhibitor activity(GO:0010852) guanylate cyclase inhibitor activity(GO:0030251) |
| 0.0 | 0.2 | GO:0004962 | endothelin receptor activity(GO:0004962) |
| 0.0 | 0.1 | GO:0004741 | [pyruvate dehydrogenase (lipoamide)] phosphatase activity(GO:0004741) |
| 0.0 | 0.2 | GO:0003993 | acid phosphatase activity(GO:0003993) |
| 0.0 | 0.2 | GO:0016922 | ligand-dependent nuclear receptor binding(GO:0016922) |
| 0.0 | 0.7 | GO:0016229 | steroid dehydrogenase activity(GO:0016229) |
| 0.0 | 0.1 | GO:0017064 | fatty acid amide hydrolase activity(GO:0017064) |
| 0.0 | 0.2 | GO:0016316 | phosphatidylinositol-3,4-bisphosphate 4-phosphatase activity(GO:0016316) |
| 0.0 | 0.4 | GO:0008417 | fucosyltransferase activity(GO:0008417) |
| 0.0 | 0.2 | GO:0004849 | uridine kinase activity(GO:0004849) |
| 0.0 | 0.3 | GO:0008449 | N-acetylglucosamine-6-sulfatase activity(GO:0008449) |
| 0.0 | 0.3 | GO:0030983 | mismatched DNA binding(GO:0030983) |
| 0.0 | 0.2 | GO:0033170 | DNA clamp loader activity(GO:0003689) protein-DNA loading ATPase activity(GO:0033170) |
| 0.0 | 0.3 | GO:0050291 | sphingosine N-acyltransferase activity(GO:0050291) |
| 0.0 | 0.4 | GO:0051393 | muscle alpha-actinin binding(GO:0051371) alpha-actinin binding(GO:0051393) |
| 0.0 | 0.3 | GO:0016668 | oxidoreductase activity, acting on a sulfur group of donors, NAD(P) as acceptor(GO:0016668) |
| 0.0 | 0.5 | GO:0030165 | PDZ domain binding(GO:0030165) |
| 0.0 | 0.1 | GO:0004045 | aminoacyl-tRNA hydrolase activity(GO:0004045) |
| 0.0 | 0.2 | GO:0019911 | structural constituent of myelin sheath(GO:0019911) |
| 0.0 | 0.1 | GO:1903924 | estradiol binding(GO:1903924) |
| 0.0 | 0.2 | GO:0003756 | protein disulfide isomerase activity(GO:0003756) intramolecular oxidoreductase activity, transposing S-S bonds(GO:0016864) |
| 0.0 | 0.2 | GO:0015272 | ATP-activated inward rectifier potassium channel activity(GO:0015272) |
| 0.0 | 0.4 | GO:0005520 | insulin-like growth factor binding(GO:0005520) |
| 0.0 | 0.1 | GO:0042169 | SH2 domain binding(GO:0042169) |
| 0.0 | 0.3 | GO:0016780 | phosphotransferase activity, for other substituted phosphate groups(GO:0016780) |
| 0.0 | 0.2 | GO:0061578 | Lys63-specific deubiquitinase activity(GO:0061578) |
| 0.0 | 0.8 | GO:0003724 | RNA helicase activity(GO:0003724) |
| 0.0 | 0.2 | GO:0008253 | 5'-nucleotidase activity(GO:0008253) |
| 0.0 | 0.4 | GO:0019213 | deacetylase activity(GO:0019213) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.8 | PID CD8 TCR DOWNSTREAM PATHWAY | Downstream signaling in naïve CD8+ T cells |
| 0.0 | 0.3 | PID IL5 PATHWAY | IL5-mediated signaling events |
| 0.0 | 0.4 | PID HDAC CLASSIII PATHWAY | Signaling events mediated by HDAC Class III |
| 0.0 | 0.4 | NABA PROTEOGLYCANS | Genes encoding proteoglycans |
| 0.0 | 0.2 | PID ERB GENOMIC PATHWAY | Validated nuclear estrogen receptor beta network |
| 0.0 | 0.2 | SIG IL4RECEPTOR IN B LYPHOCYTES | Genes related to IL4 rceptor signaling in B lymphocytes |
| 0.0 | 0.6 | PID P53 REGULATION PATHWAY | p53 pathway |
| 0.0 | 0.4 | PID ATR PATHWAY | ATR signaling pathway |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.7 | REACTOME ENDOSOMAL VACUOLAR PATHWAY | Genes involved in Endosomal/Vacuolar pathway |
| 0.0 | 0.4 | REACTOME KERATAN SULFATE DEGRADATION | Genes involved in Keratan sulfate degradation |
| 0.0 | 0.4 | REACTOME REGULATION OF IFNA SIGNALING | Genes involved in Regulation of IFNA signaling |
| 0.0 | 0.6 | REACTOME SMOOTH MUSCLE CONTRACTION | Genes involved in Smooth Muscle Contraction |
| 0.0 | 0.2 | REACTOME SEROTONIN RECEPTORS | Genes involved in Serotonin receptors |
| 0.0 | 0.3 | REACTOME REGULATION OF PYRUVATE DEHYDROGENASE PDH COMPLEX | Genes involved in Regulation of pyruvate dehydrogenase (PDH) complex |
| 0.0 | 0.3 | REACTOME SYNTHESIS SECRETION AND DEACYLATION OF GHRELIN | Genes involved in Synthesis, Secretion, and Deacylation of Ghrelin |
| 0.0 | 0.2 | REACTOME ADVANCED GLYCOSYLATION ENDPRODUCT RECEPTOR SIGNALING | Genes involved in Advanced glycosylation endproduct receptor signaling |
| 0.0 | 0.3 | REACTOME VITAMIN B5 PANTOTHENATE METABOLISM | Genes involved in Vitamin B5 (pantothenate) metabolism |
| 0.0 | 0.5 | REACTOME COMPLEMENT CASCADE | Genes involved in Complement cascade |
| 0.0 | 0.2 | REACTOME CHYLOMICRON MEDIATED LIPID TRANSPORT | Genes involved in Chylomicron-mediated lipid transport |
| 0.0 | 0.4 | REACTOME CYTOCHROME P450 ARRANGED BY SUBSTRATE TYPE | Genes involved in Cytochrome P450 - arranged by substrate type |
| 0.0 | 0.2 | REACTOME RESOLUTION OF AP SITES VIA THE SINGLE NUCLEOTIDE REPLACEMENT PATHWAY | Genes involved in Resolution of AP sites via the single-nucleotide replacement pathway |
| 0.0 | 0.2 | REACTOME HS GAG DEGRADATION | Genes involved in HS-GAG degradation |
| 0.0 | 0.2 | REACTOME POL SWITCHING | Genes involved in Polymerase switching |
| 0.0 | 0.2 | REACTOME SEMA4D INDUCED CELL MIGRATION AND GROWTH CONE COLLAPSE | Genes involved in Sema4D induced cell migration and growth-cone collapse |
| 0.0 | 0.2 | REACTOME CYCLIN A B1 ASSOCIATED EVENTS DURING G2 M TRANSITION | Genes involved in Cyclin A/B1 associated events during G2/M transition |
| 0.0 | 0.2 | REACTOME AMYLOIDS | Genes involved in Amyloids |