Project
PRJNA207719: Tissue specific transcriptome profiling
Navigation
Downloads
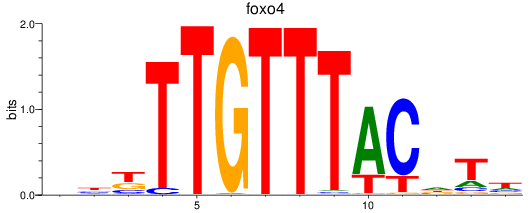
Results for foxo4
Z-value: 1.13
Transcription factors associated with foxo4
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
foxo4
|
ENSDARG00000055792 | forkhead box O4 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| foxo4 | dr11_v1_chr14_+_30910114_30910114 | 0.10 | 8.7e-01 | Click! |
Activity profile of foxo4 motif
Sorted Z-values of foxo4 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr5_-_25583125 | 0.50 |
ENSDART00000031665
ENSDART00000145353 |
anxa1a
|
annexin A1a |
| chr20_-_39273987 | 0.46 |
ENSDART00000127173
|
clu
|
clusterin |
| chr10_+_39084354 | 0.44 |
ENSDART00000158245
|
si:ch73-1a9.3
|
si:ch73-1a9.3 |
| chr22_-_29689981 | 0.42 |
ENSDART00000009223
|
pdcd4b
|
programmed cell death 4b |
| chr14_-_34044369 | 0.41 |
ENSDART00000149396
ENSDART00000123607 ENSDART00000190746 |
cyfip2
|
cytoplasmic FMR1 interacting protein 2 |
| chr7_-_24828296 | 0.40 |
ENSDART00000138193
|
otub1b
|
OTU deubiquitinase, ubiquitin aldehyde binding 1b |
| chr24_+_17007407 | 0.40 |
ENSDART00000110652
|
zfx
|
zinc finger protein, X-linked |
| chr20_-_39273505 | 0.38 |
ENSDART00000153114
|
clu
|
clusterin |
| chr16_-_38001040 | 0.38 |
ENSDART00000133861
ENSDART00000138711 ENSDART00000143846 ENSDART00000146564 |
si:ch211-198c19.3
|
si:ch211-198c19.3 |
| chr12_+_25640480 | 0.38 |
ENSDART00000105608
|
prkcea
|
protein kinase C, epsilon a |
| chr1_+_45969240 | 0.38 |
ENSDART00000042086
|
arhgef7b
|
Rho guanine nucleotide exchange factor (GEF) 7b |
| chr22_-_29689485 | 0.36 |
ENSDART00000182173
|
pdcd4b
|
programmed cell death 4b |
| chr23_-_17450746 | 0.34 |
ENSDART00000145399
ENSDART00000136457 ENSDART00000133125 ENSDART00000145719 ENSDART00000147524 ENSDART00000005366 ENSDART00000104680 |
tpd52l2b
|
tumor protein D52-like 2b |
| chr19_-_32600823 | 0.33 |
ENSDART00000134149
ENSDART00000187858 |
zgc:91944
|
zgc:91944 |
| chr11_+_25257022 | 0.33 |
ENSDART00000156052
|
tp53inp2
|
tumor protein p53 inducible nuclear protein 2 |
| chr11_-_183328 | 0.33 |
ENSDART00000168562
|
avil
|
advillin |
| chr5_-_20446082 | 0.32 |
ENSDART00000051607
|
ISCU (1 of many)
|
si:ch211-191d15.2 |
| chr5_-_46329880 | 0.32 |
ENSDART00000156577
|
si:ch211-130m23.5
|
si:ch211-130m23.5 |
| chr13_+_1100197 | 0.32 |
ENSDART00000139560
|
ppp3r1a
|
protein phosphatase 3, regulatory subunit B, alpha a |
| chr14_-_33177935 | 0.31 |
ENSDART00000180583
ENSDART00000078856 |
dlg3
|
discs, large homolog 3 (Drosophila) |
| chr18_+_17428506 | 0.31 |
ENSDART00000100223
|
zgc:91860
|
zgc:91860 |
| chr3_-_29962345 | 0.30 |
ENSDART00000136819
|
bcat2
|
branched chain amino-acid transaminase 2, mitochondrial |
| chr21_+_33459524 | 0.30 |
ENSDART00000053205
|
cd74b
|
CD74 molecule, major histocompatibility complex, class II invariant chain b |
| chr19_-_5103313 | 0.29 |
ENSDART00000037007
|
tpi1a
|
triosephosphate isomerase 1a |
| chr13_+_22675802 | 0.29 |
ENSDART00000145538
ENSDART00000143312 |
zgc:193505
|
zgc:193505 |
| chr19_+_5315987 | 0.28 |
ENSDART00000145749
|
si:dkeyp-113d7.1
|
si:dkeyp-113d7.1 |
| chr15_+_37589698 | 0.28 |
ENSDART00000076066
ENSDART00000153894 ENSDART00000156298 |
lin37
|
lin-37 DREAM MuvB core complex component |
| chr10_+_20070178 | 0.28 |
ENSDART00000027612
ENSDART00000145264 ENSDART00000172713 |
xpo7
|
exportin 7 |
| chr19_-_5103141 | 0.27 |
ENSDART00000150952
|
tpi1a
|
triosephosphate isomerase 1a |
| chr17_+_24718272 | 0.27 |
ENSDART00000007271
|
mtfr1l
|
mitochondrial fission regulator 1-like |
| chr18_+_20226843 | 0.27 |
ENSDART00000100632
|
tle3a
|
transducin-like enhancer of split 3a |
| chr6_+_51932563 | 0.26 |
ENSDART00000181778
|
angpt4
|
angiopoietin 4 |
| chr20_-_20610812 | 0.26 |
ENSDART00000181870
|
ppm1ab
|
protein phosphatase, Mg2+/Mn2+ dependent, 1Ab |
| chr15_+_19990068 | 0.26 |
ENSDART00000154033
ENSDART00000054428 |
zgc:112083
|
zgc:112083 |
| chr22_+_4707663 | 0.26 |
ENSDART00000042194
|
cers4a
|
ceramide synthase 4a |
| chr11_-_16152400 | 0.26 |
ENSDART00000123665
|
arpc4l
|
actin related protein 2/3 complex, subunit 4, like |
| chr12_-_11457625 | 0.25 |
ENSDART00000012318
|
htra1b
|
HtrA serine peptidase 1b |
| chr4_+_9612574 | 0.25 |
ENSDART00000150336
ENSDART00000041289 ENSDART00000150828 |
tmem243b
|
transmembrane protein 243, mitochondrial b |
| chr12_-_28848015 | 0.25 |
ENSDART00000153200
|
si:ch211-194k22.8
|
si:ch211-194k22.8 |
| chr11_-_12051805 | 0.24 |
ENSDART00000110117
ENSDART00000182744 |
socs7
|
suppressor of cytokine signaling 7 |
| chr23_+_3731375 | 0.24 |
ENSDART00000141782
|
smim29
|
small integral membrane protein 29 |
| chr21_-_32081552 | 0.24 |
ENSDART00000135659
|
mat2b
|
methionine adenosyltransferase II, beta |
| chr17_+_24111392 | 0.24 |
ENSDART00000180123
ENSDART00000182787 ENSDART00000189752 ENSDART00000184940 ENSDART00000185363 ENSDART00000064067 |
ehbp1
|
EH domain binding protein 1 |
| chr1_+_16127825 | 0.24 |
ENSDART00000122503
|
tusc3
|
tumor suppressor candidate 3 |
| chr9_+_32358514 | 0.24 |
ENSDART00000144608
|
plcl1
|
phospholipase C like 1 |
| chr14_-_36378494 | 0.23 |
ENSDART00000058503
|
gpm6aa
|
glycoprotein M6Aa |
| chr19_-_32600638 | 0.23 |
ENSDART00000143497
|
zgc:91944
|
zgc:91944 |
| chr13_+_22280983 | 0.23 |
ENSDART00000173258
ENSDART00000173379 |
usp54a
|
ubiquitin specific peptidase 54a |
| chr3_-_28258462 | 0.23 |
ENSDART00000191573
|
rbfox1
|
RNA binding fox-1 homolog 1 |
| chr25_+_28823952 | 0.23 |
ENSDART00000067072
|
nfybb
|
nuclear transcription factor Y, beta b |
| chr6_+_51932267 | 0.23 |
ENSDART00000156256
|
angpt4
|
angiopoietin 4 |
| chr23_-_36449111 | 0.22 |
ENSDART00000110478
|
zgc:174906
|
zgc:174906 |
| chr4_+_5156117 | 0.22 |
ENSDART00000067392
|
tigarb
|
tp53-induced glycolysis and apoptosis regulator b |
| chr17_+_38295847 | 0.22 |
ENSDART00000008532
|
mbip
|
MAP3K12 binding inhibitory protein 1 |
| chr6_+_48618512 | 0.22 |
ENSDART00000111190
|
FAM19A3
|
si:dkey-238f9.1 |
| chr18_-_25177230 | 0.21 |
ENSDART00000013363
|
slco3a1
|
solute carrier organic anion transporter family, member 3A1 |
| chr15_+_21252532 | 0.21 |
ENSDART00000162619
ENSDART00000019636 ENSDART00000144901 ENSDART00000138676 ENSDART00000133821 ENSDART00000146967 ENSDART00000143990 ENSDART00000142070 ENSDART00000132373 |
usf1
|
upstream transcription factor 1 |
| chr7_+_72279584 | 0.21 |
ENSDART00000172021
|
tollip
|
toll interacting protein |
| chr3_+_19685873 | 0.21 |
ENSDART00000006490
|
tlk2
|
tousled-like kinase 2 |
| chr21_+_19547806 | 0.21 |
ENSDART00000159707
ENSDART00000184869 ENSDART00000181321 ENSDART00000058487 ENSDART00000058485 |
rai14
|
retinoic acid induced 14 |
| chr19_+_46222428 | 0.21 |
ENSDART00000183984
|
vps28
|
vacuolar protein sorting 28 (yeast) |
| chr1_-_9940494 | 0.20 |
ENSDART00000138726
|
tmem8a
|
transmembrane protein 8A |
| chr23_-_3758637 | 0.20 |
ENSDART00000131536
ENSDART00000139408 ENSDART00000137826 |
hmga1a
|
high mobility group AT-hook 1a |
| chr12_+_25223843 | 0.20 |
ENSDART00000077180
ENSDART00000127454 ENSDART00000122665 |
mta3
|
metastasis associated 1 family, member 3 |
| chr16_+_28578352 | 0.20 |
ENSDART00000149306
|
nmt2
|
N-myristoyltransferase 2 |
| chr14_+_33427837 | 0.20 |
ENSDART00000105687
|
zbtb33
|
zinc finger and BTB domain containing 33 |
| chr1_+_25801648 | 0.20 |
ENSDART00000129471
|
gucy1b1
|
guanylate cyclase 1 soluble subunit beta 1 |
| chr7_+_60359347 | 0.19 |
ENSDART00000145201
ENSDART00000039827 |
ppp1r14bb
|
protein phosphatase 1, regulatory (inhibitor) subunit 14Bb |
| chr23_-_12158685 | 0.19 |
ENSDART00000135035
|
fam217b
|
family with sequence similarity 217, member B |
| chr15_+_30157602 | 0.19 |
ENSDART00000047248
ENSDART00000123937 |
nlk2
|
nemo-like kinase, type 2 |
| chr20_-_16156419 | 0.19 |
ENSDART00000037420
|
ralgps2
|
Ral GEF with PH domain and SH3 binding motif 2 |
| chr9_-_2572790 | 0.19 |
ENSDART00000135076
ENSDART00000016710 |
scrn3
|
secernin 3 |
| chr3_+_10152092 | 0.19 |
ENSDART00000066053
|
cbx2
|
chromobox homolog 2 (Drosophila Pc class) |
| chr11_+_16153207 | 0.19 |
ENSDART00000192356
|
tada3l
|
transcriptional adaptor 3 (NGG1 homolog, yeast)-like |
| chr13_+_30951155 | 0.19 |
ENSDART00000057469
ENSDART00000162254 |
vstm4a
|
V-set and transmembrane domain containing 4a |
| chr14_+_29769336 | 0.19 |
ENSDART00000105898
|
si:dkey-34l15.1
|
si:dkey-34l15.1 |
| chr11_-_36001495 | 0.18 |
ENSDART00000190330
|
itpr1b
|
inositol 1,4,5-trisphosphate receptor, type 1b |
| chr2_+_1988036 | 0.18 |
ENSDART00000155956
|
ssx2ipa
|
synovial sarcoma, X breakpoint 2 interacting protein a |
| chr13_-_34781984 | 0.18 |
ENSDART00000172138
|
ism1
|
isthmin 1 |
| chr10_+_26834985 | 0.18 |
ENSDART00000147518
|
fam89b
|
family with sequence similarity 89, member B |
| chr19_+_2275019 | 0.18 |
ENSDART00000136138
|
itgb8
|
integrin, beta 8 |
| chr5_+_55934129 | 0.18 |
ENSDART00000050969
|
tmem150ab
|
transmembrane protein 150Ab |
| chr4_-_4256300 | 0.18 |
ENSDART00000103319
ENSDART00000150279 |
cd9b
|
CD9 molecule b |
| chr9_-_2573121 | 0.18 |
ENSDART00000181340
|
scrn3
|
secernin 3 |
| chr21_+_45757317 | 0.18 |
ENSDART00000163152
|
h2afy
|
H2A histone family, member Y |
| chr10_-_8197049 | 0.18 |
ENSDART00000129467
|
dhx29
|
DEAH (Asp-Glu-Ala-His) box polypeptide 29 |
| chr3_+_32403758 | 0.17 |
ENSDART00000156982
|
si:ch211-195b15.8
|
si:ch211-195b15.8 |
| chr2_+_16696052 | 0.17 |
ENSDART00000022356
ENSDART00000164329 |
ppp1r7
|
protein phosphatase 1, regulatory (inhibitor) subunit 7 |
| chr3_+_31621774 | 0.17 |
ENSDART00000076636
|
fzd2
|
frizzled class receptor 2 |
| chr8_+_16990120 | 0.17 |
ENSDART00000018934
|
pde4d
|
phosphodiesterase 4D, cAMP-specific |
| chr15_+_46313082 | 0.17 |
ENSDART00000153830
|
si:ch1073-190k2.1
|
si:ch1073-190k2.1 |
| chr3_-_15496295 | 0.17 |
ENSDART00000144369
|
sgf29
|
SAGA complex associated factor 29 |
| chr14_-_7885707 | 0.17 |
ENSDART00000029981
|
ppp3cb
|
protein phosphatase 3, catalytic subunit, beta isozyme |
| chr12_+_22404108 | 0.17 |
ENSDART00000153055
|
hdlbpb
|
high density lipoprotein binding protein b |
| chr17_-_20236228 | 0.17 |
ENSDART00000136490
ENSDART00000029380 |
bnip4
|
BCL2 interacting protein 4 |
| chr7_+_24528866 | 0.17 |
ENSDART00000180552
|
si:dkeyp-75h12.2
|
si:dkeyp-75h12.2 |
| chr19_+_46222918 | 0.17 |
ENSDART00000158703
|
vps28
|
vacuolar protein sorting 28 (yeast) |
| chr25_+_17920361 | 0.17 |
ENSDART00000185644
|
borcs5
|
BLOC-1 related complex subunit 5 |
| chr5_+_12528693 | 0.16 |
ENSDART00000051670
|
rfc5
|
replication factor C (activator 1) 5 |
| chr2_-_26720854 | 0.16 |
ENSDART00000148110
|
si:dkey-181m9.8
|
si:dkey-181m9.8 |
| chr3_-_26191960 | 0.16 |
ENSDART00000113843
|
ypel3
|
yippee-like 3 |
| chr16_+_6944564 | 0.16 |
ENSDART00000104252
|
eaf1
|
ELL associated factor 1 |
| chr7_-_41013575 | 0.16 |
ENSDART00000150139
|
insig1
|
insulin induced gene 1 |
| chr5_-_40190949 | 0.16 |
ENSDART00000175588
|
wdfy3
|
WD repeat and FYVE domain containing 3 |
| chr21_-_42202792 | 0.16 |
ENSDART00000124708
|
gabra6b
|
gamma-aminobutyric acid (GABA) A receptor, alpha 6b |
| chr22_+_26798853 | 0.16 |
ENSDART00000087576
ENSDART00000179780 |
slx4
|
SLX4 structure-specific endonuclease subunit homolog (S. cerevisiae) |
| chr20_-_45062514 | 0.16 |
ENSDART00000183529
ENSDART00000182955 |
klhl29
|
kelch-like family member 29 |
| chr20_-_19365875 | 0.16 |
ENSDART00000063703
ENSDART00000187707 ENSDART00000161065 |
si:dkey-71h2.2
|
si:dkey-71h2.2 |
| chr3_-_60142530 | 0.16 |
ENSDART00000153247
|
si:ch211-120g10.1
|
si:ch211-120g10.1 |
| chr13_-_22903246 | 0.16 |
ENSDART00000089133
|
rufy2
|
RUN and FYVE domain containing 2 |
| chr25_-_25142387 | 0.16 |
ENSDART00000031814
|
tsg101a
|
tumor susceptibility 101a |
| chr18_+_6857071 | 0.16 |
ENSDART00000018735
ENSDART00000181969 |
dnaja2l
|
DnaJ (Hsp40) homolog, subfamily A, member 2, like |
| chr9_-_24209083 | 0.16 |
ENSDART00000134599
|
zgc:153521
|
zgc:153521 |
| chr5_+_28271412 | 0.16 |
ENSDART00000031727
|
vamp8
|
vesicle-associated membrane protein 8 (endobrevin) |
| chr6_-_12270226 | 0.16 |
ENSDART00000180473
|
pkp4
|
plakophilin 4 |
| chr11_+_16152316 | 0.15 |
ENSDART00000081054
|
tada3l
|
transcriptional adaptor 3 (NGG1 homolog, yeast)-like |
| chr13_-_25842074 | 0.15 |
ENSDART00000015154
|
papolg
|
poly(A) polymerase gamma |
| chr21_-_11820379 | 0.15 |
ENSDART00000126640
|
RHOBTB3
|
si:dkey-6b12.5 |
| chr14_+_41406321 | 0.15 |
ENSDART00000111480
|
bcorl1
|
BCL6 corepressor-like 1 |
| chr19_-_34063567 | 0.15 |
ENSDART00000157815
ENSDART00000183907 |
elmo1
|
engulfment and cell motility 1 (ced-12 homolog, C. elegans) |
| chr6_-_32045951 | 0.15 |
ENSDART00000016629
ENSDART00000139055 |
efcab7
|
EF-hand calcium binding domain 7 |
| chr16_+_6944311 | 0.15 |
ENSDART00000144763
|
eaf1
|
ELL associated factor 1 |
| chr23_-_5683147 | 0.15 |
ENSDART00000102766
ENSDART00000067351 |
tnnt2a
|
troponin T type 2a (cardiac) |
| chr3_+_32532645 | 0.15 |
ENSDART00000055312
ENSDART00000150981 |
nosip
|
nitric oxide synthase interacting protein |
| chr15_-_22147860 | 0.15 |
ENSDART00000149784
|
scn3b
|
sodium channel, voltage-gated, type III, beta |
| chr8_-_1267247 | 0.15 |
ENSDART00000150064
|
cdc14b
|
cell division cycle 14B |
| chr3_-_15496551 | 0.15 |
ENSDART00000124063
ENSDART00000007726 |
sgf29
|
SAGA complex associated factor 29 |
| chr5_-_51998708 | 0.15 |
ENSDART00000097194
|
serinc5
|
serine incorporator 5 |
| chr11_+_25112269 | 0.15 |
ENSDART00000147546
|
ndrg3a
|
ndrg family member 3a |
| chr16_+_10413551 | 0.15 |
ENSDART00000032877
ENSDART00000172827 |
ino80e
|
INO80 complex subunit E |
| chr10_-_9089545 | 0.15 |
ENSDART00000080781
|
arl15b
|
ADP-ribosylation factor-like 15b |
| chr16_+_5612547 | 0.15 |
ENSDART00000140226
ENSDART00000189352 |
CYTH2
|
si:dkey-283b15.4 |
| chr17_-_14966384 | 0.15 |
ENSDART00000105064
|
txndc16
|
thioredoxin domain containing 16 |
| chr20_+_34596205 | 0.15 |
ENSDART00000138338
|
si:ch211-242b18.1
|
si:ch211-242b18.1 |
| chr17_-_43399896 | 0.15 |
ENSDART00000156033
ENSDART00000156418 |
itpk1b
|
inositol-tetrakisphosphate 1-kinase b |
| chr19_+_20177887 | 0.14 |
ENSDART00000008595
|
tra2a
|
transformer 2 alpha homolog |
| chr11_+_18873619 | 0.14 |
ENSDART00000176141
|
magi1b
|
membrane associated guanylate kinase, WW and PDZ domain containing 1b |
| chr6_-_40657653 | 0.14 |
ENSDART00000154359
|
ppil1
|
peptidylprolyl isomerase (cyclophilin)-like 1 |
| chr3_+_27798094 | 0.14 |
ENSDART00000075100
ENSDART00000151437 |
carhsp1
|
calcium regulated heat stable protein 1 |
| chr3_+_52999962 | 0.14 |
ENSDART00000104683
|
pbx4
|
pre-B-cell leukemia transcription factor 4 |
| chr18_+_26899316 | 0.14 |
ENSDART00000050230
|
tspan3a
|
tetraspanin 3a |
| chr10_+_33744098 | 0.14 |
ENSDART00000147775
|
rxfp2a
|
relaxin/insulin-like family peptide receptor 2a |
| chr12_-_19346678 | 0.14 |
ENSDART00000044860
|
maff
|
v-maf avian musculoaponeurotic fibrosarcoma oncogene homolog F |
| chr19_-_22843480 | 0.14 |
ENSDART00000052503
|
nudcd1
|
NudC domain containing 1 |
| chr10_-_41156348 | 0.14 |
ENSDART00000058622
|
aak1b
|
AP2 associated kinase 1b |
| chr14_-_48939560 | 0.14 |
ENSDART00000021736
|
scocb
|
short coiled-coil protein b |
| chr19_-_10656667 | 0.14 |
ENSDART00000081379
ENSDART00000151456 ENSDART00000143271 ENSDART00000182126 |
olah
|
oleoyl-ACP hydrolase |
| chr13_+_27316934 | 0.14 |
ENSDART00000164533
|
eef1a1a
|
eukaryotic translation elongation factor 1 alpha 1a |
| chr21_-_27272657 | 0.14 |
ENSDART00000040754
ENSDART00000175009 |
mark2a
|
MAP/microtubule affinity-regulating kinase 2a |
| chr17_+_31221761 | 0.14 |
ENSDART00000155580
|
ccdc32
|
coiled-coil domain containing 32 |
| chr5_-_56924747 | 0.14 |
ENSDART00000014028
|
ppm1db
|
protein phosphatase, Mg2+/Mn2+ dependent, 1Db |
| chr3_+_15817644 | 0.14 |
ENSDART00000055787
|
natd1
|
zgc:110779 |
| chr2_-_11662851 | 0.14 |
ENSDART00000145108
|
zgc:110130
|
zgc:110130 |
| chr15_+_20543770 | 0.14 |
ENSDART00000092357
|
sgsm2
|
small G protein signaling modulator 2 |
| chr5_-_20678300 | 0.14 |
ENSDART00000088639
|
wscd2
|
WSC domain containing 2 |
| chr9_-_52386733 | 0.13 |
ENSDART00000171721
|
dap1b
|
death associated protein 1b |
| chr22_-_4769140 | 0.13 |
ENSDART00000165235
|
calr3a
|
calreticulin 3a |
| chr23_-_16682186 | 0.13 |
ENSDART00000020810
|
sdcbp2
|
syndecan binding protein (syntenin) 2 |
| chr13_-_13030851 | 0.13 |
ENSDART00000009499
|
nsd2
|
nuclear receptor binding SET domain protein 2 |
| chr14_-_36799280 | 0.13 |
ENSDART00000168615
|
rnf130
|
ring finger protein 130 |
| chr3_-_33113879 | 0.13 |
ENSDART00000044677
|
rarab
|
retinoic acid receptor, alpha b |
| chr4_-_16876281 | 0.13 |
ENSDART00000016690
ENSDART00000044005 ENSDART00000042874 ENSDART00000125762 ENSDART00000185974 |
tmpoa
|
thymopoietin a |
| chr20_-_31497300 | 0.13 |
ENSDART00000046841
|
sash1a
|
SAM and SH3 domain containing 1a |
| chr3_-_16039619 | 0.13 |
ENSDART00000143324
|
spsb3a
|
splA/ryanodine receptor domain and SOCS box containing 3a |
| chr7_+_60551133 | 0.13 |
ENSDART00000148038
|
lrfn4b
|
leucine rich repeat and fibronectin type III domain containing 4b |
| chr25_+_17920668 | 0.13 |
ENSDART00000093358
|
borcs5
|
BLOC-1 related complex subunit 5 |
| chr2_-_30200206 | 0.13 |
ENSDART00000130142
|
ube2w
|
ubiquitin-conjugating enzyme E2W (putative) |
| chr15_+_8043751 | 0.13 |
ENSDART00000193701
|
cadm2b
|
cell adhesion molecule 2b |
| chr17_+_25519089 | 0.13 |
ENSDART00000041721
|
aim1a
|
crystallin beta-gamma domain containing 1a |
| chr5_+_22579975 | 0.13 |
ENSDART00000080877
|
tnfsf10l4
|
tumor necrosis factor (ligand) superfamily, member 10 like 4 |
| chr12_+_19188542 | 0.13 |
ENSDART00000134726
ENSDART00000148011 ENSDART00000109541 |
cby1
|
chibby homolog 1 (Drosophila) |
| chr17_+_14965570 | 0.13 |
ENSDART00000066604
|
gpr137c
|
G protein-coupled receptor 137c |
| chr6_-_30839763 | 0.13 |
ENSDART00000154228
|
sgip1a
|
SH3-domain GRB2-like (endophilin) interacting protein 1a |
| chr12_-_33314894 | 0.13 |
ENSDART00000152908
|
c1qtnf1
|
C1q and TNF related 1 |
| chr18_+_16749091 | 0.13 |
ENSDART00000061265
|
rnf141
|
ring finger protein 141 |
| chr15_+_39977461 | 0.13 |
ENSDART00000063786
|
cab39
|
calcium binding protein 39 |
| chr23_+_19759128 | 0.13 |
ENSDART00000135820
|
flna
|
filamin A, alpha (actin binding protein 280) |
| chr8_+_7778770 | 0.12 |
ENSDART00000171325
|
tfe3a
|
transcription factor binding to IGHM enhancer 3a |
| chr1_-_9277986 | 0.12 |
ENSDART00000146065
ENSDART00000114876 ENSDART00000132812 |
ubn1
|
ubinuclein 1 |
| chr7_-_18547420 | 0.12 |
ENSDART00000173969
|
rgs12a
|
regulator of G protein signaling 12a |
| chr19_+_10396042 | 0.12 |
ENSDART00000028048
ENSDART00000151735 |
necap1
|
NECAP endocytosis associated 1 |
| chr16_-_15988320 | 0.12 |
ENSDART00000160883
|
CABZ01060453.1
|
|
| chr25_-_16589461 | 0.12 |
ENSDART00000064204
|
cpa4
|
carboxypeptidase A4 |
| chr11_-_34577034 | 0.12 |
ENSDART00000133302
ENSDART00000184367 |
pfkfb4a
|
6-phosphofructo-2-kinase/fructose-2,6-biphosphatase 4a |
| chr2_+_26179096 | 0.12 |
ENSDART00000024662
|
plppr3a
|
phospholipid phosphatase related 3a |
| chr18_+_17660402 | 0.12 |
ENSDART00000143475
|
cpne2
|
copine II |
| chr9_+_3388099 | 0.12 |
ENSDART00000019910
|
dlx1a
|
distal-less homeobox 1a |
| chr3_-_25369557 | 0.12 |
ENSDART00000055491
|
smurf2
|
SMAD specific E3 ubiquitin protein ligase 2 |
| chr25_-_3393705 | 0.12 |
ENSDART00000163096
|
gpr22b
|
G protein-coupled receptor 22b |
| chr4_+_14926948 | 0.12 |
ENSDART00000019647
|
psmc2
|
proteasome 26S subunit, ATPase 2 |
| chr12_+_18916285 | 0.12 |
ENSDART00000127536
|
cbx7b
|
chromobox homolog 7b |
| chr10_+_33754967 | 0.12 |
ENSDART00000153442
|
rxfp2a
|
relaxin/insulin-like family peptide receptor 2a |
| chr18_+_1703984 | 0.12 |
ENSDART00000114010
|
slitrk3a
|
SLIT and NTRK-like family, member 3a |
| chr5_+_37504309 | 0.12 |
ENSDART00000165465
|
si:ch1073-224n8.1
|
si:ch1073-224n8.1 |
| chr16_-_44900306 | 0.12 |
ENSDART00000058380
|
rbm42
|
RNA binding motif protein 42 |
| chr14_+_16937997 | 0.12 |
ENSDART00000163013
ENSDART00000167856 |
limch1b
|
LIM and calponin homology domains 1b |
| chr21_+_25071805 | 0.12 |
ENSDART00000078651
|
dixdc1b
|
DIX domain containing 1b |
| chr11_-_21586157 | 0.12 |
ENSDART00000190095
|
srgap2
|
SLIT-ROBO Rho GTPase activating protein 2 |
| chr20_-_35470891 | 0.12 |
ENSDART00000152993
ENSDART00000016090 |
pla2g7
|
phospholipase A2, group VII (platelet-activating factor acetylhydrolase, plasma) |
| chr23_+_31596441 | 0.12 |
ENSDART00000053534
|
tbpl1
|
TBP-like 1 |
Network of associatons between targets according to the STRING database.
First level regulatory network of foxo4
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.6 | GO:0019242 | methylglyoxal biosynthetic process(GO:0019242) glyceraldehyde-3-phosphate biosynthetic process(GO:0046166) |
| 0.1 | 0.4 | GO:0071896 | protein localization to adherens junction(GO:0071896) |
| 0.1 | 0.3 | GO:0010935 | macrophage cytokine production(GO:0010934) regulation of macrophage cytokine production(GO:0010935) positive regulation of macrophage cytokine production(GO:0060907) negative regulation of intrinsic apoptotic signaling pathway in response to DNA damage(GO:1902230) |
| 0.1 | 0.5 | GO:0002828 | neutrophil apoptotic process(GO:0001781) regulation of T-helper 1 type immune response(GO:0002825) positive regulation of T-helper 1 type immune response(GO:0002827) regulation of type 2 immune response(GO:0002828) negative regulation of type 2 immune response(GO:0002829) inflammatory cell apoptotic process(GO:0006925) regulation of prostaglandin biosynthetic process(GO:0031392) positive regulation of prostaglandin biosynthetic process(GO:0031394) regulation of phospholipase A2 activity(GO:0032429) interleukin-2 production(GO:0032623) regulation of interleukin-2 production(GO:0032663) positive regulation of interleukin-2 production(GO:0032743) myeloid cell apoptotic process(GO:0033028) regulation of neutrophil apoptotic process(GO:0033029) positive regulation of neutrophil apoptotic process(GO:0033031) regulation of myeloid cell apoptotic process(GO:0033032) positive regulation of myeloid cell apoptotic process(GO:0033034) T-helper 1 type immune response(GO:0042088) positive regulation of CD4-positive, alpha-beta T cell differentiation(GO:0043372) T-helper 1 cell differentiation(GO:0045063) positive regulation of T-helper cell differentiation(GO:0045624) regulation of T-helper 1 cell differentiation(GO:0045625) positive regulation of T-helper 1 cell differentiation(GO:0045627) regulation of T-helper 2 cell differentiation(GO:0045628) negative regulation of T-helper 2 cell differentiation(GO:0045629) positive regulation of fatty acid biosynthetic process(GO:0045723) positive regulation of alpha-beta T cell differentiation(GO:0046638) neutrophil clearance(GO:0097350) negative regulation of phospholipase A2 activity(GO:1900138) positive regulation of CD4-positive, alpha-beta T cell activation(GO:2000516) regulation of unsaturated fatty acid biosynthetic process(GO:2001279) positive regulation of unsaturated fatty acid biosynthetic process(GO:2001280) |
| 0.1 | 0.3 | GO:0009098 | leucine biosynthetic process(GO:0009098) |
| 0.1 | 0.2 | GO:0043455 | regulation of secondary metabolic process(GO:0043455) |
| 0.1 | 0.4 | GO:1901315 | negative regulation of histone ubiquitination(GO:0033183) regulation of protein K63-linked ubiquitination(GO:1900044) negative regulation of protein K63-linked ubiquitination(GO:1900045) regulation of histone H2A K63-linked ubiquitination(GO:1901314) negative regulation of histone H2A K63-linked ubiquitination(GO:1901315) regulation of protein polyubiquitination(GO:1902914) negative regulation of protein polyubiquitination(GO:1902915) |
| 0.1 | 0.2 | GO:0018377 | N-terminal protein myristoylation(GO:0006499) N-terminal peptidyl-glycine N-myristoylation(GO:0018008) protein myristoylation(GO:0018377) |
| 0.1 | 0.2 | GO:0036314 | response to sterol(GO:0036314) cellular response to sterol(GO:0036315) SREBP-SCAP complex retention in endoplasmic reticulum(GO:0036316) regulation of SREBP signaling pathway(GO:2000638) negative regulation of SREBP signaling pathway(GO:2000639) |
| 0.1 | 0.4 | GO:0099514 | anterograde synaptic vesicle transport(GO:0048490) synaptic vesicle cytoskeletal transport(GO:0099514) synaptic vesicle transport along microtubule(GO:0099517) |
| 0.0 | 0.2 | GO:0099548 | trans-synaptic signaling by soluble gas(GO:0099543) trans-synaptic signaling by nitric oxide(GO:0099548) |
| 0.0 | 0.1 | GO:0032957 | inositol trisphosphate metabolic process(GO:0032957) |
| 0.0 | 0.1 | GO:0097676 | histone H3-K36 dimethylation(GO:0097676) |
| 0.0 | 0.4 | GO:0043328 | protein targeting to vacuole involved in ubiquitin-dependent protein catabolic process via the multivesicular body sorting pathway(GO:0043328) |
| 0.0 | 0.2 | GO:0061010 | gall bladder development(GO:0061010) |
| 0.0 | 0.1 | GO:0061341 | non-canonical Wnt signaling pathway involved in heart development(GO:0061341) |
| 0.0 | 0.1 | GO:0055109 | invagination involved in gastrulation with mouth forming second(GO:0055109) morphogenesis of an epithelial fold(GO:0060571) |
| 0.0 | 0.1 | GO:0003228 | atrial cardiac muscle tissue development(GO:0003228) |
| 0.0 | 0.1 | GO:0086005 | ventricular cardiac muscle cell action potential(GO:0086005) |
| 0.0 | 0.1 | GO:0016116 | tetraterpenoid metabolic process(GO:0016108) tetraterpenoid biosynthetic process(GO:0016109) carotenoid metabolic process(GO:0016116) carotenoid biosynthetic process(GO:0016117) xanthophyll metabolic process(GO:0016122) xanthophyll biosynthetic process(GO:0016123) zeaxanthin metabolic process(GO:1901825) zeaxanthin biosynthetic process(GO:1901827) |
| 0.0 | 0.1 | GO:0031630 | regulation of synaptic vesicle fusion to presynaptic membrane(GO:0031630) |
| 0.0 | 0.2 | GO:0006556 | S-adenosylmethionine biosynthetic process(GO:0006556) |
| 0.0 | 0.2 | GO:1904292 | regulation of ERAD pathway(GO:1904292) |
| 0.0 | 0.4 | GO:0001964 | startle response(GO:0001964) |
| 0.0 | 0.1 | GO:0036037 | CD8-positive, alpha-beta T cell activation(GO:0036037) |
| 0.0 | 0.1 | GO:0051196 | regulation of glycolytic process(GO:0006110) regulation of nucleotide catabolic process(GO:0030811) regulation of cofactor metabolic process(GO:0051193) regulation of coenzyme metabolic process(GO:0051196) |
| 0.0 | 0.1 | GO:0061187 | regulation of chromatin silencing at rDNA(GO:0061187) negative regulation of chromatin silencing at rDNA(GO:0061188) |
| 0.0 | 0.1 | GO:0042940 | D-amino acid transport(GO:0042940) D-alanine transport(GO:0042941) D-serine transport(GO:0042942) |
| 0.0 | 0.3 | GO:0061056 | sclerotome development(GO:0061056) |
| 0.0 | 0.2 | GO:0032228 | regulation of synaptic transmission, GABAergic(GO:0032228) |
| 0.0 | 0.2 | GO:0001672 | regulation of chromatin assembly or disassembly(GO:0001672) |
| 0.0 | 0.2 | GO:0070254 | mucus secretion(GO:0070254) |
| 0.0 | 0.8 | GO:0003171 | atrioventricular valve development(GO:0003171) |
| 0.0 | 0.3 | GO:0006611 | protein export from nucleus(GO:0006611) |
| 0.0 | 0.1 | GO:1990918 | double-strand break repair involved in meiotic recombination(GO:1990918) |
| 0.0 | 0.1 | GO:0003232 | bulbus arteriosus development(GO:0003232) |
| 0.0 | 0.8 | GO:0043966 | histone H3 acetylation(GO:0043966) |
| 0.0 | 0.1 | GO:2001223 | negative regulation of neuron migration(GO:2001223) |
| 0.0 | 0.1 | GO:0045922 | negative regulation of fatty acid metabolic process(GO:0045922) |
| 0.0 | 0.1 | GO:0060623 | regulation of chromosome condensation(GO:0060623) |
| 0.0 | 0.3 | GO:0007190 | activation of adenylate cyclase activity(GO:0007190) positive regulation of cAMP metabolic process(GO:0030816) positive regulation of cAMP biosynthetic process(GO:0030819) positive regulation of adenylate cyclase activity(GO:0045762) |
| 0.0 | 0.1 | GO:0010692 | regulation of alkaline phosphatase activity(GO:0010692) negative regulation of alkaline phosphatase activity(GO:0010693) |
| 0.0 | 0.1 | GO:1903723 | negative regulation of centriole elongation(GO:1903723) |
| 0.0 | 0.2 | GO:0071715 | icosanoid transport(GO:0071715) fatty acid derivative transport(GO:1901571) |
| 0.0 | 0.1 | GO:0006296 | nucleotide-excision repair, DNA incision, 5'-to lesion(GO:0006296) |
| 0.0 | 0.1 | GO:0044342 | type B pancreatic cell proliferation(GO:0044342) |
| 0.0 | 0.1 | GO:0045604 | regulation of epidermal cell differentiation(GO:0045604) |
| 0.0 | 0.1 | GO:0061635 | regulation of protein complex stability(GO:0061635) |
| 0.0 | 0.1 | GO:0035881 | amacrine cell differentiation(GO:0035881) |
| 0.0 | 0.1 | GO:0030579 | ubiquitin-dependent SMAD protein catabolic process(GO:0030579) |
| 0.0 | 0.1 | GO:0045898 | regulation of RNA polymerase II transcriptional preinitiation complex assembly(GO:0045898) positive regulation of RNA polymerase II transcriptional preinitiation complex assembly(GO:0045899) |
| 0.0 | 0.3 | GO:0014812 | muscle cell migration(GO:0014812) |
| 0.0 | 0.2 | GO:0045948 | positive regulation of translational initiation(GO:0045948) |
| 0.0 | 0.1 | GO:1900120 | regulation of cytokine activity(GO:0060300) regulation of receptor binding(GO:1900120) regulation of chemokine activity(GO:1900136) |
| 0.0 | 0.1 | GO:0031640 | killing of cells of other organism(GO:0031640) disruption of cells of other organism(GO:0044364) disruption of cells of other organism involved in symbiotic interaction(GO:0051818) killing of cells in other organism involved in symbiotic interaction(GO:0051883) |
| 0.0 | 0.3 | GO:0032467 | positive regulation of cytokinesis(GO:0032467) |
| 0.0 | 0.1 | GO:0007084 | mitotic nuclear envelope reassembly(GO:0007084) |
| 0.0 | 0.1 | GO:0097345 | mitochondrial outer membrane permeabilization(GO:0097345) |
| 0.0 | 0.1 | GO:0010960 | magnesium ion homeostasis(GO:0010960) |
| 0.0 | 0.1 | GO:0032793 | positive regulation of CREB transcription factor activity(GO:0032793) |
| 0.0 | 0.1 | GO:0008592 | regulation of Toll signaling pathway(GO:0008592) |
| 0.0 | 0.1 | GO:0071881 | adenylate cyclase-inhibiting adrenergic receptor signaling pathway(GO:0071881) |
| 0.0 | 0.1 | GO:1901741 | positive regulation of myoblast fusion(GO:1901741) |
| 0.0 | 0.2 | GO:0015693 | magnesium ion transport(GO:0015693) |
| 0.0 | 0.0 | GO:0042416 | dopamine biosynthetic process from tyrosine(GO:0006585) dopamine biosynthetic process(GO:0042416) |
| 0.0 | 0.3 | GO:0010508 | positive regulation of autophagy(GO:0010508) |
| 0.0 | 0.1 | GO:0034122 | negative regulation of toll-like receptor signaling pathway(GO:0034122) |
| 0.0 | 0.1 | GO:0051881 | regulation of mitochondrial membrane potential(GO:0051881) |
| 0.0 | 0.8 | GO:1901214 | regulation of neuron death(GO:1901214) |
| 0.0 | 0.1 | GO:1904184 | regulation of pyruvate dehydrogenase activity(GO:1904182) positive regulation of pyruvate dehydrogenase activity(GO:1904184) |
| 0.0 | 0.1 | GO:0061031 | endodermal digestive tract morphogenesis(GO:0061031) pancreas morphogenesis(GO:0061113) |
| 0.0 | 0.2 | GO:0035970 | peptidyl-threonine dephosphorylation(GO:0035970) |
| 0.0 | 0.3 | GO:0051014 | actin filament severing(GO:0051014) |
| 0.0 | 0.5 | GO:0021854 | hypothalamus development(GO:0021854) |
| 0.0 | 0.2 | GO:0051932 | synaptic transmission, GABAergic(GO:0051932) |
| 0.0 | 0.1 | GO:0045670 | regulation of osteoclast differentiation(GO:0045670) |
| 0.0 | 0.2 | GO:0033173 | calcineurin-NFAT signaling cascade(GO:0033173) |
| 0.0 | 0.1 | GO:0003139 | secondary heart field specification(GO:0003139) |
| 0.0 | 0.3 | GO:0043124 | negative regulation of I-kappaB kinase/NF-kappaB signaling(GO:0043124) |
| 0.0 | 0.1 | GO:1902624 | positive regulation of neutrophil migration(GO:1902624) |
| 0.0 | 0.2 | GO:0071425 | hematopoietic stem cell proliferation(GO:0071425) |
| 0.0 | 0.1 | GO:0045475 | locomotor rhythm(GO:0045475) |
| 0.0 | 0.1 | GO:0006004 | fucose metabolic process(GO:0006004) |
| 0.0 | 0.1 | GO:1904263 | positive regulation of TORC1 signaling(GO:1904263) |
| 0.0 | 0.1 | GO:0099612 | protein localization to juxtaparanode region of axon(GO:0071205) protein localization to axon(GO:0099612) |
| 0.0 | 0.2 | GO:0006658 | phosphatidylserine metabolic process(GO:0006658) |
| 0.0 | 0.0 | GO:0010591 | regulation of lamellipodium assembly(GO:0010591) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.8 | GO:0042583 | chromaffin granule(GO:0042583) |
| 0.1 | 0.3 | GO:0098574 | cytoplasmic side of lysosomal membrane(GO:0098574) |
| 0.1 | 0.7 | GO:0005671 | Ada2/Gcn5/Ada3 transcription activator complex(GO:0005671) |
| 0.1 | 0.2 | GO:0048269 | methionine adenosyltransferase complex(GO:0048269) |
| 0.1 | 0.2 | GO:0032937 | SREBP-SCAP-Insig complex(GO:0032937) |
| 0.0 | 0.3 | GO:0032783 | ELL-EAF complex(GO:0032783) |
| 0.0 | 0.3 | GO:0042613 | MHC class II protein complex(GO:0042613) |
| 0.0 | 0.1 | GO:0044279 | other organism(GO:0044215) other organism cell(GO:0044216) other organism part(GO:0044217) other organism cell membrane(GO:0044218) other organism membrane(GO:0044279) |
| 0.0 | 0.5 | GO:0016328 | lateral plasma membrane(GO:0016328) |
| 0.0 | 0.1 | GO:0005828 | kinetochore microtubule(GO:0005828) |
| 0.0 | 0.1 | GO:0031213 | RSF complex(GO:0031213) |
| 0.0 | 0.1 | GO:0016460 | myosin II complex(GO:0016460) |
| 0.0 | 0.2 | GO:0005955 | calcineurin complex(GO:0005955) |
| 0.0 | 0.2 | GO:0033557 | Slx1-Slx4 complex(GO:0033557) |
| 0.0 | 0.4 | GO:0031209 | SCAR complex(GO:0031209) |
| 0.0 | 0.1 | GO:0042611 | MHC protein complex(GO:0042611) MHC class I protein complex(GO:0042612) |
| 0.0 | 0.2 | GO:0005663 | DNA replication factor C complex(GO:0005663) |
| 0.0 | 0.4 | GO:0000813 | ESCRT I complex(GO:0000813) |
| 0.0 | 0.2 | GO:0016602 | CCAAT-binding factor complex(GO:0016602) |
| 0.0 | 0.2 | GO:0044295 | axonal growth cone(GO:0044295) |
| 0.0 | 0.1 | GO:0017177 | glucosidase II complex(GO:0017177) |
| 0.0 | 0.2 | GO:0071797 | LUBAC complex(GO:0071797) |
| 0.0 | 0.1 | GO:0032983 | kainate selective glutamate receptor complex(GO:0032983) |
| 0.0 | 0.2 | GO:0008250 | oligosaccharyltransferase complex(GO:0008250) |
| 0.0 | 0.2 | GO:0032045 | guanyl-nucleotide exchange factor complex(GO:0032045) |
| 0.0 | 0.1 | GO:1990513 | CLOCK-BMAL transcription complex(GO:1990513) |
| 0.0 | 0.5 | GO:0017053 | transcriptional repressor complex(GO:0017053) |
| 0.0 | 0.3 | GO:0035102 | PRC1 complex(GO:0035102) |
| 0.0 | 0.3 | GO:0034706 | voltage-gated sodium channel complex(GO:0001518) sodium channel complex(GO:0034706) |
| 0.0 | 0.2 | GO:0030667 | secretory granule membrane(GO:0030667) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.6 | GO:0004807 | triose-phosphate isomerase activity(GO:0004807) methylglyoxal synthase activity(GO:0008929) |
| 0.1 | 0.5 | GO:0019834 | phospholipase A2 inhibitor activity(GO:0019834) |
| 0.1 | 0.4 | GO:0019784 | NEDD8-specific protease activity(GO:0019784) |
| 0.1 | 0.3 | GO:0052656 | branched-chain-amino-acid transaminase activity(GO:0004084) L-leucine transaminase activity(GO:0052654) L-valine transaminase activity(GO:0052655) L-isoleucine transaminase activity(GO:0052656) |
| 0.1 | 0.4 | GO:0070004 | cysteine-type exopeptidase activity(GO:0070004) |
| 0.1 | 0.2 | GO:0004379 | glycylpeptide N-tetradecanoyltransferase activity(GO:0004379) |
| 0.1 | 0.2 | GO:0034416 | bisphosphoglycerate phosphatase activity(GO:0034416) |
| 0.1 | 0.3 | GO:0070016 | armadillo repeat domain binding(GO:0070016) |
| 0.1 | 0.6 | GO:0003691 | double-stranded telomeric DNA binding(GO:0003691) |
| 0.0 | 0.1 | GO:0047325 | inositol tetrakisphosphate 1-kinase activity(GO:0047325) inositol-1,3,4-trisphosphate 6-kinase activity(GO:0052725) inositol-1,3,4-trisphosphate 5-kinase activity(GO:0052726) |
| 0.0 | 0.1 | GO:0045545 | syndecan binding(GO:0045545) |
| 0.0 | 0.2 | GO:0004652 | polynucleotide adenylyltransferase activity(GO:0004652) |
| 0.0 | 0.1 | GO:0052885 | retinol isomerase activity(GO:0050251) all-trans-retinyl-ester hydrolase, 11-cis retinol forming activity(GO:0052885) |
| 0.0 | 0.1 | GO:0070513 | death domain binding(GO:0070513) |
| 0.0 | 0.1 | GO:0032038 | myosin head/neck binding(GO:0032028) myosin II head/neck binding(GO:0032034) myosin II heavy chain binding(GO:0032038) |
| 0.0 | 0.1 | GO:0000703 | oxidized pyrimidine nucleobase lesion DNA N-glycosylase activity(GO:0000703) |
| 0.0 | 0.2 | GO:0004723 | calcium-dependent protein serine/threonine phosphatase activity(GO:0004723) calmodulin-dependent protein phosphatase activity(GO:0033192) |
| 0.0 | 0.3 | GO:0005049 | nuclear export signal receptor activity(GO:0005049) |
| 0.0 | 0.2 | GO:0008142 | oxysterol binding(GO:0008142) |
| 0.0 | 0.7 | GO:0051787 | misfolded protein binding(GO:0051787) |
| 0.0 | 0.4 | GO:0000340 | RNA 7-methylguanosine cap binding(GO:0000340) |
| 0.0 | 0.1 | GO:0031701 | angiotensin receptor binding(GO:0031701) |
| 0.0 | 0.1 | GO:0004466 | long-chain-acyl-CoA dehydrogenase activity(GO:0004466) |
| 0.0 | 0.4 | GO:0031492 | nucleosomal DNA binding(GO:0031492) |
| 0.0 | 0.2 | GO:0004865 | protein serine/threonine phosphatase inhibitor activity(GO:0004865) |
| 0.0 | 0.1 | GO:0035575 | histone demethylase activity (H4-K20 specific)(GO:0035575) |
| 0.0 | 0.1 | GO:0036402 | proteasome-activating ATPase activity(GO:0036402) |
| 0.0 | 0.1 | GO:0052855 | ATP-dependent NAD(P)H-hydrate dehydratase activity(GO:0047453) ADP-dependent NAD(P)H-hydrate dehydratase activity(GO:0052855) |
| 0.0 | 0.1 | GO:0004679 | AMP-activated protein kinase activity(GO:0004679) |
| 0.0 | 0.4 | GO:0051019 | mitogen-activated protein kinase binding(GO:0051019) |
| 0.0 | 0.1 | GO:0019871 | sodium channel inhibitor activity(GO:0019871) |
| 0.0 | 0.1 | GO:0070182 | DNA polymerase binding(GO:0070182) |
| 0.0 | 0.1 | GO:0015277 | kainate selective glutamate receptor activity(GO:0015277) |
| 0.0 | 0.1 | GO:0008140 | cAMP response element binding protein binding(GO:0008140) |
| 0.0 | 0.2 | GO:0005220 | inositol 1,4,5-trisphosphate-sensitive calcium-release channel activity(GO:0005220) |
| 0.0 | 0.2 | GO:0033170 | DNA clamp loader activity(GO:0003689) protein-DNA loading ATPase activity(GO:0033170) |
| 0.0 | 0.5 | GO:0035064 | methylated histone binding(GO:0035064) |
| 0.0 | 0.4 | GO:0004698 | protein kinase C activity(GO:0004697) calcium-dependent protein kinase C activity(GO:0004698) |
| 0.0 | 0.1 | GO:0031013 | troponin C binding(GO:0030172) troponin I binding(GO:0031013) |
| 0.0 | 0.0 | GO:1990226 | histone methyltransferase binding(GO:1990226) |
| 0.0 | 0.3 | GO:0042289 | MHC class II protein binding(GO:0042289) |
| 0.0 | 0.1 | GO:0004719 | protein-L-isoaspartate (D-aspartate) O-methyltransferase activity(GO:0004719) |
| 0.0 | 0.1 | GO:0004741 | [pyruvate dehydrogenase (lipoamide)] phosphatase activity(GO:0004741) |
| 0.0 | 0.4 | GO:0030971 | receptor tyrosine kinase binding(GO:0030971) protein tyrosine kinase binding(GO:1990782) |
| 0.0 | 0.1 | GO:0004938 | alpha2-adrenergic receptor activity(GO:0004938) |
| 0.0 | 0.2 | GO:0050811 | GABA receptor binding(GO:0050811) |
| 0.0 | 0.5 | GO:0070491 | repressing transcription factor binding(GO:0070491) |
| 0.0 | 0.2 | GO:0022851 | benzodiazepine receptor activity(GO:0008503) GABA-gated chloride ion channel activity(GO:0022851) |
| 0.0 | 0.6 | GO:0004402 | histone acetyltransferase activity(GO:0004402) |
| 0.0 | 0.2 | GO:0050321 | tau-protein kinase activity(GO:0050321) |
| 0.0 | 0.2 | GO:0004383 | guanylate cyclase activity(GO:0004383) |
| 0.0 | 0.2 | GO:0015347 | sodium-independent organic anion transmembrane transporter activity(GO:0015347) |
| 0.0 | 0.1 | GO:0043515 | kinetochore binding(GO:0043515) |
| 0.0 | 0.2 | GO:0050291 | sphingosine N-acyltransferase activity(GO:0050291) |
| 0.0 | 0.1 | GO:0008349 | MAP kinase kinase kinase kinase activity(GO:0008349) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.3 | PID IL12 STAT4 PATHWAY | IL12 signaling mediated by STAT4 |
| 0.0 | 0.2 | PID P38 ALPHA BETA DOWNSTREAM PATHWAY | Signaling mediated by p38-alpha and p38-beta |
| 0.0 | 0.2 | PID INTEGRIN5 PATHWAY | Beta5 beta6 beta7 and beta8 integrin cell surface interactions |
| 0.0 | 0.8 | PID MYC REPRESS PATHWAY | Validated targets of C-MYC transcriptional repression |
| 0.0 | 0.2 | PID CIRCADIAN PATHWAY | Circadian rhythm pathway |
| 0.0 | 0.2 | PID ATF2 PATHWAY | ATF-2 transcription factor network |
| 0.0 | 0.4 | PID ECADHERIN NASCENT AJ PATHWAY | E-cadherin signaling in the nascent adherens junction |
| 0.0 | 0.3 | PID WNT NONCANONICAL PATHWAY | Noncanonical Wnt signaling pathway |
| 0.0 | 0.5 | PID ANGIOPOIETIN RECEPTOR PATHWAY | Angiopoietin receptor Tie2-mediated signaling |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.6 | REACTOME MEMBRANE BINDING AND TARGETTING OF GAG PROTEINS | Genes involved in Membrane binding and targetting of GAG proteins |
| 0.0 | 0.1 | REACTOME ENDOSOMAL VACUOLAR PATHWAY | Genes involved in Endosomal/Vacuolar pathway |
| 0.0 | 0.4 | REACTOME IONOTROPIC ACTIVITY OF KAINATE RECEPTORS | Genes involved in Ionotropic activity of Kainate Receptors |
| 0.0 | 0.2 | REACTOME TRANSPORT OF ORGANIC ANIONS | Genes involved in Transport of organic anions |
| 0.0 | 0.3 | REACTOME DARPP 32 EVENTS | Genes involved in DARPP-32 events |
| 0.0 | 0.5 | REACTOME TIE2 SIGNALING | Genes involved in Tie2 Signaling |
| 0.0 | 0.2 | REACTOME REGULATION OF AMPK ACTIVITY VIA LKB1 | Genes involved in Regulation of AMPK activity via LKB1 |
| 0.0 | 0.3 | REACTOME BRANCHED CHAIN AMINO ACID CATABOLISM | Genes involved in Branched-chain amino acid catabolism |
| 0.0 | 0.2 | REACTOME BASE FREE SUGAR PHOSPHATE REMOVAL VIA THE SINGLE NUCLEOTIDE REPLACEMENT PATHWAY | Genes involved in Base-free sugar-phosphate removal via the single-nucleotide replacement pathway |
| 0.0 | 0.1 | REACTOME CD28 DEPENDENT VAV1 PATHWAY | Genes involved in CD28 dependent Vav1 pathway |
| 0.0 | 0.8 | REACTOME RESPONSE TO ELEVATED PLATELET CYTOSOLIC CA2 | Genes involved in Response to elevated platelet cytosolic Ca2+ |
| 0.0 | 0.1 | REACTOME REGULATION OF THE FANCONI ANEMIA PATHWAY | Genes involved in Regulation of the Fanconi anemia pathway |
| 0.0 | 0.1 | REACTOME ENOS ACTIVATION AND REGULATION | Genes involved in eNOS activation and regulation |
| 0.0 | 0.1 | REACTOME SYNTHESIS SECRETION AND DEACYLATION OF GHRELIN | Genes involved in Synthesis, Secretion, and Deacylation of Ghrelin |