Project
PRJNA207719: Tissue specific transcriptome profiling
Navigation
Downloads
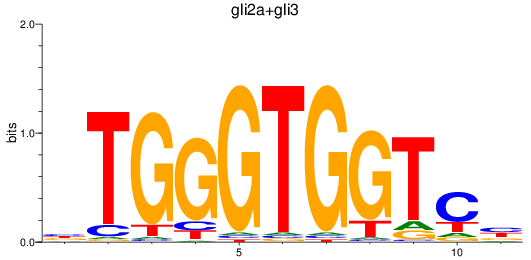
Results for gli2a+gli3
Z-value: 1.09
Transcription factors associated with gli2a+gli3
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
gli2a
|
ENSDARG00000025641 | GLI family zinc finger 2a |
|
gli3
|
ENSDARG00000052131 | GLI family zinc finger 3 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| gli2a | dr11_v1_chr9_+_37141836_37141836 | -0.93 | 2.2e-02 | Click! |
| gli3 | dr11_v1_chr24_-_11821505_11821505 | -0.68 | 2.0e-01 | Click! |
Activity profile of gli2a+gli3 motif
Sorted Z-values of gli2a+gli3 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr22_-_24858042 | 1.50 |
ENSDART00000137998
ENSDART00000078216 ENSDART00000138378 |
vtg7
|
vitellogenin 7 |
| chr1_-_19845378 | 1.26 |
ENSDART00000139314
ENSDART00000132958 ENSDART00000147502 |
grhprb
|
glyoxylate reductase/hydroxypyruvate reductase b |
| chr22_+_38173960 | 1.01 |
ENSDART00000010537
|
cp
|
ceruloplasmin |
| chr22_-_24818066 | 1.00 |
ENSDART00000143443
|
vtg6
|
vitellogenin 6 |
| chr16_+_52512025 | 0.99 |
ENSDART00000056095
|
fabp10a
|
fatty acid binding protein 10a, liver basic |
| chr10_+_26747755 | 0.98 |
ENSDART00000100329
|
f9b
|
coagulation factor IXb |
| chr25_-_17403598 | 0.88 |
ENSDART00000104216
|
cyp2x9
|
cytochrome P450, family 2, subfamily X, polypeptide 9 |
| chr22_-_24791505 | 0.86 |
ENSDART00000136837
|
vtg4
|
vitellogenin 4 |
| chr5_-_43959972 | 0.70 |
ENSDART00000180517
|
si:ch211-204c21.1
|
si:ch211-204c21.1 |
| chr4_+_72797711 | 0.67 |
ENSDART00000190934
ENSDART00000163236 |
MYRFL
|
myelin regulatory factor-like |
| chr8_-_40075983 | 0.67 |
ENSDART00000141455
|
ggt1a
|
gamma-glutamyltransferase 1a |
| chr10_+_19595009 | 0.55 |
ENSDART00000112276
|
zgc:173837
|
zgc:173837 |
| chr22_-_188102 | 0.55 |
ENSDART00000125391
ENSDART00000170463 |
CABZ01052268.1
|
|
| chr1_+_7956030 | 0.54 |
ENSDART00000159655
|
CR855320.1
|
|
| chr4_-_77114795 | 0.51 |
ENSDART00000144849
|
CU467646.2
|
|
| chr6_+_10094061 | 0.48 |
ENSDART00000150998
ENSDART00000162236 |
atp7b
|
ATPase copper transporting beta |
| chr4_-_52165969 | 0.46 |
ENSDART00000171130
|
si:dkeyp-44b5.4
|
si:dkeyp-44b5.4 |
| chr3_-_46410387 | 0.39 |
ENSDART00000156822
|
cdip1
|
cell death-inducing p53 target 1 |
| chr20_+_27194833 | 0.39 |
ENSDART00000150072
|
si:dkey-85n7.8
|
si:dkey-85n7.8 |
| chr23_+_44374041 | 0.38 |
ENSDART00000136056
|
ephb4b
|
eph receptor B4b |
| chr10_+_28428222 | 0.38 |
ENSDART00000135003
|
si:ch211-222e20.4
|
si:ch211-222e20.4 |
| chr19_-_46566430 | 0.37 |
ENSDART00000166668
|
ext1b
|
exostosin glycosyltransferase 1b |
| chr10_-_24868581 | 0.35 |
ENSDART00000193055
|
CABZ01058107.1
|
|
| chr4_-_55785014 | 0.34 |
ENSDART00000181348
|
CT583728.24
|
|
| chr4_-_55801210 | 0.34 |
ENSDART00000186943
|
CT583728.13
|
|
| chr4_-_55829342 | 0.34 |
ENSDART00000185611
|
CT583728.26
|
|
| chr18_+_5273953 | 0.33 |
ENSDART00000165073
|
slc12a1
|
solute carrier family 12 (sodium/potassium/chloride transporter), member 1 |
| chr4_+_57881965 | 0.31 |
ENSDART00000162234
|
si:dkeyp-44b5.4
|
si:dkeyp-44b5.4 |
| chr13_-_40726865 | 0.30 |
ENSDART00000099847
|
st3gal7
|
ST3 beta-galactoside alpha-2,3-sialyltransferase 7 |
| chr5_-_25621386 | 0.30 |
ENSDART00000051565
|
cyp1d1
|
cytochrome P450, family 1, subfamily D, polypeptide 1 |
| chr18_-_14836600 | 0.29 |
ENSDART00000045232
|
mtss1la
|
metastasis suppressor 1-like a |
| chr3_+_23692462 | 0.27 |
ENSDART00000145934
|
hoxb7a
|
homeobox B7a |
| chr8_-_49201258 | 0.27 |
ENSDART00000114584
ENSDART00000142172 |
zgc:171501
|
zgc:171501 |
| chr1_+_54737353 | 0.27 |
ENSDART00000130675
ENSDART00000162075 |
pi4k2a
|
phosphatidylinositol 4-kinase type 2 alpha |
| chr4_-_75175407 | 0.27 |
ENSDART00000180125
|
CABZ01043953.1
|
|
| chr6_-_10693111 | 0.26 |
ENSDART00000187206
ENSDART00000192319 |
dnah9l
|
dynein, axonemal, heavy polypeptide 9 like |
| chr13_-_38039871 | 0.25 |
ENSDART00000140645
|
CR456624.1
|
|
| chr11_+_440305 | 0.25 |
ENSDART00000082517
|
rab43
|
RAB43, member RAS oncogene family |
| chr5_-_15948833 | 0.24 |
ENSDART00000051649
ENSDART00000124467 |
xbp1
|
X-box binding protein 1 |
| chr12_+_27117609 | 0.23 |
ENSDART00000076154
|
hoxb8b
|
homeobox B8b |
| chr8_+_48558100 | 0.23 |
ENSDART00000166165
|
si:ch211-215j19.12
|
si:ch211-215j19.12 |
| chr3_-_18805225 | 0.23 |
ENSDART00000133471
ENSDART00000131758 |
msrb1a
|
methionine sulfoxide reductase B1a |
| chr4_-_20208166 | 0.23 |
ENSDART00000066891
|
gsap
|
gamma-secretase activating protein |
| chr13_+_24842857 | 0.23 |
ENSDART00000123866
|
dusp13a
|
dual specificity phosphatase 13a |
| chr3_+_55131289 | 0.23 |
ENSDART00000111585
|
AL935210.1
|
|
| chr13_+_3252950 | 0.22 |
ENSDART00000020671
|
prph2b
|
peripherin 2b (retinal degeneration, slow) |
| chr5_-_23715861 | 0.22 |
ENSDART00000019992
|
gbgt1l1
|
globoside alpha-1,3-N-acetylgalactosaminyltransferase 1, like 1 |
| chr6_-_8480815 | 0.20 |
ENSDART00000162300
|
rasal3
|
RAS protein activator like 3 |
| chr18_+_33100606 | 0.20 |
ENSDART00000003907
|
si:ch211-229c8.13
|
si:ch211-229c8.13 |
| chr8_+_28467893 | 0.20 |
ENSDART00000189724
|
slc52a3
|
solute carrier family 52 (riboflavin transporter), member 3 |
| chr2_+_54086436 | 0.20 |
ENSDART00000174581
|
CU179656.1
|
|
| chr23_+_21424747 | 0.20 |
ENSDART00000135440
ENSDART00000104214 |
tas1r1
|
taste receptor, type 1, member 1 |
| chr9_-_1984604 | 0.19 |
ENSDART00000082339
|
hoxd12a
|
homeobox D12a |
| chr17_-_51202339 | 0.19 |
ENSDART00000167117
|
si:ch1073-469d17.2
|
si:ch1073-469d17.2 |
| chr9_+_50007556 | 0.19 |
ENSDART00000175587
|
slc38a11
|
solute carrier family 38, member 11 |
| chr21_+_35327589 | 0.19 |
ENSDART00000145191
ENSDART00000132743 |
rars
|
arginyl-tRNA synthetase |
| chr16_-_31644545 | 0.19 |
ENSDART00000181634
|
CR855311.5
|
|
| chr4_-_5831036 | 0.18 |
ENSDART00000166232
|
foxm1
|
forkhead box M1 |
| chr17_-_34952562 | 0.18 |
ENSDART00000021128
|
kidins220a
|
kinase D-interacting substrate 220a |
| chr12_+_18358082 | 0.18 |
ENSDART00000123315
|
si:dkey-7e14.3
|
si:dkey-7e14.3 |
| chr23_+_43834376 | 0.18 |
ENSDART00000148989
|
si:ch211-149b19.4
|
si:ch211-149b19.4 |
| chr3_+_55122662 | 0.18 |
ENSDART00000128380
|
hbbe1.2
|
hemoglobin beta embryonic-1.2 |
| chr22_-_12726556 | 0.18 |
ENSDART00000105776
|
fam207a
|
family with sequence similarity 207, member A |
| chr16_-_30847192 | 0.17 |
ENSDART00000191106
ENSDART00000128158 |
ptk2ab
|
protein tyrosine kinase 2ab |
| chr3_+_55114097 | 0.17 |
ENSDART00000121686
|
hbbe1.1
|
hemoglobin beta embryonic-1.1 |
| chr2_+_51783120 | 0.17 |
ENSDART00000177559
|
crygn1
|
crystallin, gamma N1 |
| chr17_-_4252221 | 0.17 |
ENSDART00000152020
|
gdf3
|
growth differentiation factor 3 |
| chr12_-_17479078 | 0.17 |
ENSDART00000079115
|
papss2b
|
3'-phosphoadenosine 5'-phosphosulfate synthase 2b |
| chr3_+_57991074 | 0.17 |
ENSDART00000076077
|
myadml2
|
myeloid-associated differentiation marker-like 2 |
| chr16_-_4026265 | 0.16 |
ENSDART00000149339
|
si:ch211-175f12.2
|
si:ch211-175f12.2 |
| chr8_-_7078575 | 0.16 |
ENSDART00000137413
|
si:ch73-233m11.2
|
si:ch73-233m11.2 |
| chr23_+_20422661 | 0.16 |
ENSDART00000144047
ENSDART00000104336 |
tnnc2
|
troponin C type 2 (fast) |
| chr1_+_10129099 | 0.16 |
ENSDART00000187740
|
rbm46
|
RNA binding motif protein 46 |
| chr18_+_2585298 | 0.16 |
ENSDART00000166314
ENSDART00000162043 |
p2ry2.2
|
purinergic receptor P2Y2, tandem duplicate 2 |
| chr22_+_22438783 | 0.15 |
ENSDART00000147825
|
kif14
|
kinesin family member 14 |
| chr3_+_56574623 | 0.15 |
ENSDART00000130877
|
rac1b
|
Rac family small GTPase 1b |
| chr23_+_4414343 | 0.14 |
ENSDART00000081821
|
wnt7ab
|
wingless-type MMTV integration site family, member 7Ab |
| chr23_+_4414164 | 0.14 |
ENSDART00000192762
|
wnt7ab
|
wingless-type MMTV integration site family, member 7Ab |
| chr10_-_22854758 | 0.14 |
ENSDART00000079449
|
and3
|
actinodin3 |
| chr19_+_7954679 | 0.14 |
ENSDART00000150909
|
si:dkey-266f7.10
|
si:dkey-266f7.10 |
| chr7_-_15413188 | 0.14 |
ENSDART00000185980
|
si:ch211-106j24.1
|
si:ch211-106j24.1 |
| chr21_+_10739846 | 0.14 |
ENSDART00000084011
|
cplx4a
|
complexin 4a |
| chr14_+_31509922 | 0.14 |
ENSDART00000124499
|
hprt1
|
hypoxanthine phosphoribosyltransferase 1 |
| chr21_+_4320769 | 0.14 |
ENSDART00000168553
|
HTRA2 (1 of many)
|
si:dkey-84o3.4 |
| chr21_+_22985078 | 0.14 |
ENSDART00000156491
|
lpar6b
|
lysophosphatidic acid receptor 6b |
| chr7_-_15413019 | 0.14 |
ENSDART00000172147
|
si:ch211-106j24.1
|
si:ch211-106j24.1 |
| chr12_+_34273240 | 0.14 |
ENSDART00000037904
|
socs3b
|
suppressor of cytokine signaling 3b |
| chr8_-_52859301 | 0.14 |
ENSDART00000162004
|
nr5a1a
|
nuclear receptor subfamily 5, group A, member 1a |
| chr25_+_25085349 | 0.14 |
ENSDART00000192166
|
si:ch73-182e20.4
|
si:ch73-182e20.4 |
| chr11_+_14295011 | 0.14 |
ENSDART00000060226
|
si:ch211-262i1.4
|
si:ch211-262i1.4 |
| chr22_-_20814450 | 0.14 |
ENSDART00000089076
|
dot1l
|
DOT1-like histone H3K79 methyltransferase |
| chr5_-_10012971 | 0.13 |
ENSDART00000179421
|
CR936408.2
|
|
| chr20_-_6131686 | 0.13 |
ENSDART00000145964
ENSDART00000086578 ENSDART00000164090 |
pum2
|
pumilio RNA-binding family member 2 |
| chr13_-_865193 | 0.13 |
ENSDART00000187053
|
AL929536.7
|
|
| chr15_-_47812268 | 0.13 |
ENSDART00000190231
|
tomt
|
transmembrane O-methyltransferase |
| chr20_-_33959815 | 0.13 |
ENSDART00000133081
|
selp
|
selectin P |
| chr25_-_35106751 | 0.12 |
ENSDART00000180682
ENSDART00000073457 |
zgc:194989
|
zgc:194989 |
| chr14_+_34952883 | 0.12 |
ENSDART00000182812
|
il12ba
|
interleukin 12Ba |
| chr13_-_31346392 | 0.12 |
ENSDART00000134343
|
rrp12
|
ribosomal RNA processing 12 homolog |
| chr4_+_51347000 | 0.12 |
ENSDART00000161859
|
si:dkeyp-82b4.3
|
si:dkeyp-82b4.3 |
| chr8_-_49207319 | 0.11 |
ENSDART00000022870
|
fam110a
|
family with sequence similarity 110, member A |
| chr8_-_14484599 | 0.11 |
ENSDART00000057644
|
lhx4
|
LIM homeobox 4 |
| chr23_-_27875140 | 0.11 |
ENSDART00000143662
|
ankrd33aa
|
ankyrin repeat domain 33Aa |
| chr16_+_42667560 | 0.11 |
ENSDART00000023452
|
dpy19l1l
|
dpy-19-like 1, like (H. sapiens) |
| chr5_+_20437124 | 0.11 |
ENSDART00000159001
|
tmem119a
|
transmembrane protein 119a |
| chr13_+_31583034 | 0.10 |
ENSDART00000111763
|
six6a
|
SIX homeobox 6a |
| chr11_+_30314885 | 0.10 |
ENSDART00000187418
ENSDART00000123244 |
ugt1b2
|
UDP glucuronosyltransferase 1 family, polypeptide B2 |
| chr15_-_19128705 | 0.10 |
ENSDART00000152428
|
arhgap32a
|
Rho GTPase activating protein 32a |
| chr5_-_38643188 | 0.10 |
ENSDART00000144972
|
si:ch211-271e10.1
|
si:ch211-271e10.1 |
| chr15_-_38361787 | 0.10 |
ENSDART00000155758
|
trpc2a
|
transient receptor potential cation channel, subfamily C, member 2a |
| chr22_-_10487490 | 0.10 |
ENSDART00000064798
|
aspn
|
asporin (LRR class 1) |
| chr2_+_48638827 | 0.09 |
ENSDART00000163634
|
CR381673.2
|
|
| chr13_-_50624173 | 0.09 |
ENSDART00000184181
|
vox
|
ventral homeobox |
| chr13_-_30150592 | 0.09 |
ENSDART00000143093
|
sar1ab
|
secretion associated, Ras related GTPase 1Ab |
| chr17_+_31580960 | 0.09 |
ENSDART00000188157
|
si:dkey-13p1.4
|
si:dkey-13p1.4 |
| chr4_-_17741513 | 0.08 |
ENSDART00000141680
|
mybpc1
|
myosin binding protein C, slow type |
| chr14_+_46020282 | 0.08 |
ENSDART00000190087
|
c1qtnf2
|
C1q and TNF related 2 |
| chr25_+_30298377 | 0.08 |
ENSDART00000153622
|
C11orf96
|
chromosome 11 open reading frame 96 |
| chr2_+_44528255 | 0.08 |
ENSDART00000193834
|
pask
|
PAS domain containing serine/threonine kinase |
| chr18_-_17485419 | 0.08 |
ENSDART00000018764
|
foxl1
|
forkhead box L1 |
| chr4_+_38937575 | 0.08 |
ENSDART00000150385
|
znf997
|
zinc finger protein 997 |
| chr6_-_15639087 | 0.07 |
ENSDART00000128940
ENSDART00000183092 |
mlpha
|
melanophilin a |
| chr25_+_25085526 | 0.07 |
ENSDART00000171627
|
si:ch73-182e20.4
|
si:ch73-182e20.4 |
| chr20_-_35052823 | 0.07 |
ENSDART00000153033
|
kif26bb
|
kinesin family member 26Bb |
| chr21_+_13389499 | 0.07 |
ENSDART00000151268
|
zgc:113162
|
zgc:113162 |
| chr25_-_3836597 | 0.07 |
ENSDART00000115154
|
si:ch211-247i17.1
|
si:ch211-247i17.1 |
| chr18_+_3022188 | 0.07 |
ENSDART00000170004
|
CU570791.1
|
|
| chr20_+_2669017 | 0.06 |
ENSDART00000058777
|
slc35a1
|
solute carrier family 35 (CMP-sialic acid transporter), member A1 |
| chr17_-_42213822 | 0.06 |
ENSDART00000187904
ENSDART00000180029 |
nkx2.2a
|
NK2 homeobox 2a |
| chr1_-_23370395 | 0.06 |
ENSDART00000143014
ENSDART00000126785 ENSDART00000159138 |
pds5a
|
PDS5 cohesin associated factor A |
| chr24_+_17334682 | 0.06 |
ENSDART00000018868
|
pdia4
|
protein disulfide isomerase family A, member 4 |
| chr24_-_6258869 | 0.06 |
ENSDART00000006636
ENSDART00000178667 |
myo3a
|
myosin IIIA |
| chr14_+_43554490 | 0.06 |
ENSDART00000060351
|
adra1bb
|
adrenoceptor alpha 1Bb |
| chr14_+_30774032 | 0.05 |
ENSDART00000139552
|
atl3
|
atlastin 3 |
| chr23_+_12072980 | 0.05 |
ENSDART00000141646
|
edn3b
|
endothelin 3b |
| chr16_+_20220225 | 0.05 |
ENSDART00000182208
|
colq
|
collagen-like tail subunit (single strand of homotrimer) of asymmetric acetylcholinesterase |
| chr20_-_38610360 | 0.05 |
ENSDART00000168783
ENSDART00000164638 |
slc30a2
|
solute carrier family 30 (zinc transporter), member 2 |
| chr11_+_36355348 | 0.05 |
ENSDART00000145427
|
sypl2a
|
synaptophysin-like 2a |
| chr2_-_30784502 | 0.04 |
ENSDART00000056735
|
rgs20
|
regulator of G protein signaling 20 |
| chr24_+_35827766 | 0.04 |
ENSDART00000144700
|
si:dkeyp-7a3.1
|
si:dkeyp-7a3.1 |
| chr20_-_35052464 | 0.04 |
ENSDART00000037195
|
kif26bb
|
kinesin family member 26Bb |
| chr20_-_7000225 | 0.04 |
ENSDART00000100098
|
adcy1a
|
adenylate cyclase 1a |
| chr1_-_470812 | 0.04 |
ENSDART00000192527
|
zgc:92518
|
zgc:92518 |
| chr17_+_6585333 | 0.04 |
ENSDART00000124293
|
adgrf3a
|
adhesion G protein-coupled receptor F3a |
| chr17_+_6563307 | 0.04 |
ENSDART00000156454
|
adgrf3a
|
adhesion G protein-coupled receptor F3a |
| chr14_+_25464681 | 0.04 |
ENSDART00000067500
ENSDART00000187601 |
si:dkey-280e21.3
|
si:dkey-280e21.3 |
| chr20_+_10498704 | 0.04 |
ENSDART00000192985
|
zgc:100997
|
zgc:100997 |
| chr25_-_29463866 | 0.04 |
ENSDART00000155676
ENSDART00000124645 ENSDART00000181055 ENSDART00000192593 |
ube2q2
|
ubiquitin-conjugating enzyme E2Q family member 2 |
| chr14_-_41091667 | 0.03 |
ENSDART00000170042
|
drp2
|
dystrophin related protein 2 |
| chr1_-_53684005 | 0.03 |
ENSDART00000108906
|
xpo1a
|
exportin 1 (CRM1 homolog, yeast) a |
| chr24_-_25184553 | 0.03 |
ENSDART00000166917
|
plcxd2
|
phosphatidylinositol-specific phospholipase C, X domain containing 2 |
| chr5_+_20599658 | 0.03 |
ENSDART00000143019
|
CMKLR1 (1 of many)
|
si:ch211-240b21.4 |
| chr24_+_7828613 | 0.03 |
ENSDART00000135161
|
zgc:101569
|
zgc:101569 |
| chr4_+_64921810 | 0.03 |
ENSDART00000157654
|
BX928743.1
|
|
| chr19_+_14158075 | 0.02 |
ENSDART00000170335
ENSDART00000168260 |
nudc
|
nudC nuclear distribution protein |
| chr5_-_17876709 | 0.02 |
ENSDART00000141978
|
si:dkey-112e17.1
|
si:dkey-112e17.1 |
| chr24_+_18299175 | 0.02 |
ENSDART00000140994
|
tpk1
|
thiamin pyrophosphokinase 1 |
| chr15_+_16525126 | 0.01 |
ENSDART00000193455
|
galnt17
|
polypeptide N-acetylgalactosaminyltransferase 17 |
| chr9_+_33417969 | 0.01 |
ENSDART00000024795
|
gpr34b
|
G protein-coupled receptor 34b |
| chr6_-_43817761 | 0.01 |
ENSDART00000183945
|
foxp1b
|
forkhead box P1b |
| chr20_-_51186524 | 0.01 |
ENSDART00000027836
ENSDART00000114407 |
rbm25b
|
RNA binding motif protein 25b |
| chr6_+_7123980 | 0.01 |
ENSDART00000179738
ENSDART00000151311 |
si:ch211-237c6.4
|
si:ch211-237c6.4 |
| chr10_-_44482911 | 0.01 |
ENSDART00000085556
|
hip1ra
|
huntingtin interacting protein 1 related a |
| chr12_-_26582607 | 0.00 |
ENSDART00000045186
|
slc16a5a
|
solute carrier family 16 (monocarboxylate transporter), member 5a |
| chr13_-_33317323 | 0.00 |
ENSDART00000110295
ENSDART00000144848 ENSDART00000136701 |
tmem234
|
transmembrane protein 234 |
| chr2_-_57707039 | 0.00 |
ENSDART00000097685
|
CABZ01060891.1
|
|
| chr5_+_68878124 | 0.00 |
ENSDART00000139933
|
spaw
|
southpaw |
Network of associatons between targets according to the STRING database.
First level regulatory network of gli2a+gli3
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 4.0 | GO:0032355 | response to estradiol(GO:0032355) |
| 0.1 | 0.5 | GO:0035434 | copper ion transmembrane transport(GO:0035434) |
| 0.1 | 0.2 | GO:0048785 | hatching gland development(GO:0048785) |
| 0.1 | 0.9 | GO:0006825 | copper ion transport(GO:0006825) |
| 0.1 | 0.2 | GO:0050428 | sulfate assimilation(GO:0000103) purine ribonucleoside bisphosphate metabolic process(GO:0034035) purine ribonucleoside bisphosphate biosynthetic process(GO:0034036) 3'-phosphoadenosine 5'-phosphosulfate metabolic process(GO:0050427) 3'-phosphoadenosine 5'-phosphosulfate biosynthetic process(GO:0050428) |
| 0.0 | 0.1 | GO:0046099 | guanine salvage(GO:0006178) GMP salvage(GO:0032263) guanine biosynthetic process(GO:0046099) |
| 0.0 | 0.7 | GO:0045807 | positive regulation of endocytosis(GO:0045807) |
| 0.0 | 0.2 | GO:0070257 | regulation of mucus secretion(GO:0070255) positive regulation of mucus secretion(GO:0070257) |
| 0.0 | 0.1 | GO:0018406 | protein C-linked glycosylation(GO:0018103) peptidyl-tryptophan modification(GO:0018211) protein C-linked glycosylation via tryptophan(GO:0018317) protein C-linked glycosylation via 2'-alpha-mannosyl-L-tryptophan(GO:0018406) |
| 0.0 | 0.1 | GO:0033690 | positive regulation of osteoblast proliferation(GO:0033690) |
| 0.0 | 0.1 | GO:0034729 | histone H3-K79 methylation(GO:0034729) regulation of transcription regulatory region DNA binding(GO:2000677) |
| 0.0 | 0.2 | GO:0038107 | nodal signaling pathway involved in determination of left/right asymmetry(GO:0038107) regulation of nodal signaling pathway involved in determination of left/right asymmetry(GO:1900145) |
| 0.0 | 0.2 | GO:0032218 | riboflavin transport(GO:0032218) |
| 0.0 | 0.1 | GO:0007638 | mechanosensory behavior(GO:0007638) |
| 0.0 | 0.2 | GO:0034205 | beta-amyloid formation(GO:0034205) amyloid precursor protein catabolic process(GO:0042987) |
| 0.0 | 0.1 | GO:0090113 | regulation of COPII vesicle coating(GO:0003400) regulation of ER to Golgi vesicle-mediated transport by GTP hydrolysis(GO:0090113) |
| 0.0 | 0.1 | GO:0097241 | hematopoietic stem cell migration to bone marrow(GO:0097241) |
| 0.0 | 0.1 | GO:0021530 | spinal cord oligodendrocyte cell fate specification(GO:0021530) |
| 0.0 | 0.1 | GO:0051876 | pigment granule dispersal(GO:0051876) |
| 0.0 | 0.1 | GO:0060585 | regulation of systemic arterial blood pressure by endothelin(GO:0003100) vein smooth muscle contraction(GO:0014826) regulation of prostaglandin-endoperoxide synthase activity(GO:0060584) positive regulation of prostaglandin-endoperoxide synthase activity(GO:0060585) |
| 0.0 | 0.9 | GO:0042738 | drug metabolic process(GO:0017144) drug catabolic process(GO:0042737) exogenous drug catabolic process(GO:0042738) |
| 0.0 | 0.3 | GO:0097178 | ruffle assembly(GO:0097178) |
| 0.0 | 0.2 | GO:0070307 | lens fiber cell development(GO:0070307) lens fiber cell morphogenesis(GO:0070309) |
| 0.0 | 0.3 | GO:0043171 | peptide catabolic process(GO:0043171) |
| 0.0 | 0.2 | GO:0071156 | regulation of cell cycle arrest(GO:0071156) |
| 0.0 | 0.2 | GO:0030259 | lipid glycosylation(GO:0030259) |
| 0.0 | 0.3 | GO:0007032 | endosome organization(GO:0007032) |
| 0.0 | 0.1 | GO:0030325 | adrenal gland development(GO:0030325) |
| 0.0 | 1.0 | GO:0007596 | blood coagulation(GO:0007596) |
| 0.0 | 0.3 | GO:0006884 | cell volume homeostasis(GO:0006884) |
| 0.0 | 0.0 | GO:0061088 | sequestering of zinc ion(GO:0032119) regulation of sequestering of zinc ion(GO:0061088) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | GO:0043514 | interleukin-12 complex(GO:0043514) interleukin-23 complex(GO:0070743) |
| 0.0 | 0.4 | GO:0045277 | respiratory chain complex IV(GO:0045277) |
| 0.0 | 0.2 | GO:0030688 | preribosome, small subunit precursor(GO:0030688) |
| 0.0 | 0.2 | GO:0031838 | haptoglobin-hemoglobin complex(GO:0031838) |
| 0.0 | 0.1 | GO:0001650 | fibrillar center(GO:0001650) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.5 | 3.4 | GO:0045735 | nutrient reservoir activity(GO:0045735) |
| 0.4 | 1.3 | GO:0030267 | hydroxypyruvate reductase activity(GO:0016618) glyoxylate reductase (NADP) activity(GO:0030267) |
| 0.2 | 1.0 | GO:0032052 | bile acid binding(GO:0032052) |
| 0.1 | 0.3 | GO:0047291 | lactosylceramide alpha-2,3-sialyltransferase activity(GO:0047291) |
| 0.1 | 0.3 | GO:0050649 | testosterone 6-beta-hydroxylase activity(GO:0050649) |
| 0.1 | 0.5 | GO:0005375 | copper ion transmembrane transporter activity(GO:0005375) |
| 0.1 | 0.4 | GO:0050508 | glucuronosyl-N-acetylglucosaminyl-proteoglycan 4-alpha-N-acetylglucosaminyltransferase activity(GO:0050508) |
| 0.1 | 0.7 | GO:0000048 | peptidyltransferase activity(GO:0000048) glutathione hydrolase activity(GO:0036374) |
| 0.1 | 1.0 | GO:0004322 | ferroxidase activity(GO:0004322) oxidoreductase activity, oxidizing metal ions, oxygen as acceptor(GO:0016724) |
| 0.1 | 0.2 | GO:0033745 | L-methionine-(R)-S-oxide reductase activity(GO:0033745) |
| 0.1 | 0.2 | GO:0004020 | adenylylsulfate kinase activity(GO:0004020) sulfate adenylyltransferase activity(GO:0004779) sulfate adenylyltransferase (ATP) activity(GO:0004781) |
| 0.0 | 0.1 | GO:0004422 | hypoxanthine phosphoribosyltransferase activity(GO:0004422) |
| 0.0 | 0.1 | GO:0016206 | catechol O-methyltransferase activity(GO:0016206) |
| 0.0 | 0.3 | GO:0004430 | 1-phosphatidylinositol 4-kinase activity(GO:0004430) |
| 0.0 | 0.3 | GO:0008511 | sodium:potassium:chloride symporter activity(GO:0008511) |
| 0.0 | 0.1 | GO:0031151 | histone methyltransferase activity (H3-K79 specific)(GO:0031151) |
| 0.0 | 0.2 | GO:0032217 | riboflavin transporter activity(GO:0032217) |
| 0.0 | 0.1 | GO:0019972 | interleukin-12 binding(GO:0019972) interleukin-12 alpha subunit binding(GO:0042164) |
| 0.0 | 0.4 | GO:0005003 | ephrin receptor activity(GO:0005003) |
| 0.0 | 0.1 | GO:0031708 | endothelin B receptor binding(GO:0031708) |
| 0.0 | 0.2 | GO:0031720 | haptoglobin binding(GO:0031720) |
| 0.0 | 0.9 | GO:0016712 | oxidoreductase activity, acting on paired donors, with incorporation or reduction of molecular oxygen, reduced flavin or flavoprotein as one donor, and incorporation of one atom of oxygen(GO:0016712) |
| 0.0 | 0.4 | GO:0015002 | cytochrome-c oxidase activity(GO:0004129) heme-copper terminal oxidase activity(GO:0015002) oxidoreductase activity, acting on a heme group of donors(GO:0016675) oxidoreductase activity, acting on a heme group of donors, oxygen as acceptor(GO:0016676) |
| 0.0 | 0.2 | GO:0004383 | guanylate cyclase activity(GO:0004383) |
| 0.0 | 0.2 | GO:0045028 | G-protein coupled nucleotide receptor activity(GO:0001608) G-protein coupled purinergic nucleotide receptor activity(GO:0045028) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.9 | PID HIF1 TFPATHWAY | HIF-1-alpha transcription factor network |
| 0.0 | 0.3 | PID WNT CANONICAL PATHWAY | Canonical Wnt signaling pathway |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 1.0 | REACTOME IRON UPTAKE AND TRANSPORT | Genes involved in Iron uptake and transport |
| 0.0 | 0.2 | REACTOME ACTIVATION OF CHAPERONE GENES BY ATF6 ALPHA | Genes involved in Activation of Chaperone Genes by ATF6-alpha |
| 0.0 | 0.2 | REACTOME CLASS C 3 METABOTROPIC GLUTAMATE PHEROMONE RECEPTORS | Genes involved in Class C/3 (Metabotropic glutamate/pheromone receptors) |
| 0.0 | 0.3 | REACTOME SYNTHESIS OF PIPS AT THE EARLY ENDOSOME MEMBRANE | Genes involved in Synthesis of PIPs at the early endosome membrane |
| 0.0 | 0.1 | REACTOME PURINE SALVAGE | Genes involved in Purine salvage |