Project
PRJNA207719: Tissue specific transcriptome profiling
Navigation
Downloads
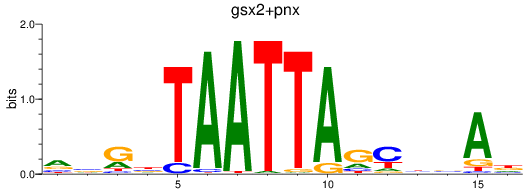
Results for gsx2+pnx
Z-value: 1.09
Transcription factors associated with gsx2+pnx
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
pnx
|
ENSDARG00000025899 | posterior neuron-specific homeobox |
|
gsx2
|
ENSDARG00000043322 | GS homeobox 2 |
|
gsx2
|
ENSDARG00000116417 | GS homeobox 2 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| pnx | dr11_v1_chr10_+_2799285_2799285 | 0.65 | 2.4e-01 | Click! |
| gsx2 | dr11_v1_chr20_-_22484621_22484621 | -0.09 | 8.9e-01 | Click! |
Activity profile of gsx2+pnx motif
Sorted Z-values of gsx2+pnx motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr5_+_51597677 | 0.76 |
ENSDART00000048210
ENSDART00000184797 |
ckmt2b
|
creatine kinase, mitochondrial 2b (sarcomeric) |
| chr25_+_29160102 | 0.73 |
ENSDART00000162854
|
pkmb
|
pyruvate kinase M1/2b |
| chr2_+_55982300 | 0.69 |
ENSDART00000183903
|
nmrk2
|
nicotinamide riboside kinase 2 |
| chr23_+_20110086 | 0.68 |
ENSDART00000054664
|
tnnc1b
|
troponin C type 1b (slow) |
| chr13_+_22479988 | 0.65 |
ENSDART00000188182
ENSDART00000192972 ENSDART00000178372 |
ldb3a
|
LIM domain binding 3a |
| chr6_-_54815886 | 0.63 |
ENSDART00000180793
ENSDART00000007498 |
tnni1b
|
troponin I type 1b (skeletal, slow) |
| chr20_+_26966725 | 0.62 |
ENSDART00000029781
|
ahsa1a
|
AHA1, activator of heat shock protein ATPase homolog 1a |
| chr7_-_71758307 | 0.59 |
ENSDART00000161067
ENSDART00000165253 |
myom1b
|
myomesin 1b |
| chr25_+_35019693 | 0.57 |
ENSDART00000046218
|
flnca
|
filamin C, gamma a (actin binding protein 280) |
| chr12_-_33357655 | 0.55 |
ENSDART00000066233
ENSDART00000148165 |
slc16a3
|
solute carrier family 16 (monocarboxylate transporter), member 3 |
| chr19_-_25119443 | 0.53 |
ENSDART00000148953
|
ptp4a3
|
protein tyrosine phosphatase type IVA, member 3 |
| chr25_+_5035343 | 0.50 |
ENSDART00000011751
|
parvb
|
parvin, beta |
| chr20_-_37813863 | 0.48 |
ENSDART00000147529
|
batf3
|
basic leucine zipper transcription factor, ATF-like 3 |
| chr22_+_16308450 | 0.47 |
ENSDART00000105678
|
lrrc39
|
leucine rich repeat containing 39 |
| chr20_-_9462433 | 0.47 |
ENSDART00000152674
ENSDART00000040557 |
zgc:101840
|
zgc:101840 |
| chr9_-_813511 | 0.44 |
ENSDART00000082368
|
marco
|
macrophage receptor with collagenous structure |
| chr6_+_6924637 | 0.40 |
ENSDART00000065551
ENSDART00000151393 |
zak
|
sterile alpha motif and leucine zipper containing kinase AZK |
| chr6_-_40581376 | 0.40 |
ENSDART00000185412
|
tspo
|
translocator protein |
| chr12_+_22580579 | 0.40 |
ENSDART00000171725
ENSDART00000192290 |
capgb
|
capping protein (actin filament), gelsolin-like b |
| chr19_+_2631565 | 0.40 |
ENSDART00000171487
|
fam126a
|
family with sequence similarity 126, member A |
| chr1_-_14332283 | 0.39 |
ENSDART00000090025
|
wfs1a
|
Wolfram syndrome 1a (wolframin) |
| chr11_+_44135351 | 0.38 |
ENSDART00000182914
|
FO704721.1
|
|
| chr11_-_2131280 | 0.36 |
ENSDART00000008409
|
calcoco1b
|
calcium binding and coiled-coil domain 1b |
| chr4_+_9011448 | 0.36 |
ENSDART00000192357
|
samm50l
|
sorting and assembly machinery component 50 homolog, like |
| chr16_-_42933332 | 0.35 |
ENSDART00000057305
|
thbs3a
|
thrombospondin 3a |
| chr17_-_37395460 | 0.34 |
ENSDART00000148160
ENSDART00000075975 |
crip1
|
cysteine-rich protein 1 |
| chr2_-_2957970 | 0.33 |
ENSDART00000162505
|
si:ch1073-82l19.1
|
si:ch1073-82l19.1 |
| chr4_+_76575585 | 0.33 |
ENSDART00000131588
|
ms4a17a.11
|
membrane-spanning 4-domains, subfamily A, member 17A.11 |
| chr11_-_13107106 | 0.33 |
ENSDART00000184477
|
elovl1b
|
ELOVL fatty acid elongase 1b |
| chr9_+_43799829 | 0.33 |
ENSDART00000186240
|
ube2e3
|
ubiquitin-conjugating enzyme E2E 3 (UBC4/5 homolog, yeast) |
| chr15_-_36533322 | 0.33 |
ENSDART00000156466
ENSDART00000121755 |
si:dkey-262k9.4
|
si:dkey-262k9.4 |
| chr19_-_2861444 | 0.32 |
ENSDART00000169053
|
clec3bb
|
C-type lectin domain family 3, member Bb |
| chr23_-_31512496 | 0.32 |
ENSDART00000158755
ENSDART00000143425 |
eya4
|
EYA transcriptional coactivator and phosphatase 4 |
| chr15_+_31701716 | 0.31 |
ENSDART00000113370
|
b3glcta
|
beta 3-glucosyltransferase a |
| chr12_-_33354409 | 0.31 |
ENSDART00000178515
|
slc16a3
|
solute carrier family 16 (monocarboxylate transporter), member 3 |
| chr5_+_40485503 | 0.31 |
ENSDART00000051055
|
ndufs4
|
NADH dehydrogenase (ubiquinone) Fe-S protein 4, (NADH-coenzyme Q reductase) |
| chr4_+_22480169 | 0.30 |
ENSDART00000146272
ENSDART00000066904 |
ndufb2
|
NADH dehydrogenase (ubiquinone) 1 beta subcomplex, 2 |
| chr25_+_31267268 | 0.30 |
ENSDART00000181239
|
tnni2a.3
|
troponin I type 2a (skeletal, fast), tandem duplicate 3 |
| chr3_+_59784632 | 0.30 |
ENSDART00000084729
|
pecam1
|
platelet/endothelial cell adhesion molecule 1 |
| chr8_+_52619365 | 0.29 |
ENSDART00000162953
|
cyr61l2
|
cysteine-rich, angiogenic inducer, 61 like 2 |
| chr3_-_15734358 | 0.28 |
ENSDART00000137325
|
mvp
|
major vault protein |
| chr3_-_15734530 | 0.27 |
ENSDART00000141142
|
mvp
|
major vault protein |
| chr1_-_51719110 | 0.27 |
ENSDART00000190574
|
rnaseh2a
|
ribonuclease H2, subunit A |
| chr16_-_31754102 | 0.27 |
ENSDART00000185043
|
ptpn6
|
protein tyrosine phosphatase, non-receptor type 6 |
| chr6_-_19271210 | 0.26 |
ENSDART00000163628
ENSDART00000159124 |
zgc:174863
|
zgc:174863 |
| chr25_+_28825657 | 0.26 |
ENSDART00000153625
|
nfybb
|
nuclear transcription factor Y, beta b |
| chr1_+_7517454 | 0.26 |
ENSDART00000016139
|
lancl1
|
LanC antibiotic synthetase component C-like 1 (bacterial) |
| chr24_-_6078222 | 0.26 |
ENSDART00000146830
|
apbb1ip
|
amyloid beta (A4) precursor protein-binding, family B, member 1 interacting protein |
| chr15_-_4528326 | 0.25 |
ENSDART00000158122
ENSDART00000155619 ENSDART00000128602 |
tfdp2
|
transcription factor Dp-2 |
| chr24_+_22485710 | 0.25 |
ENSDART00000146058
|
si:dkey-40h20.1
|
si:dkey-40h20.1 |
| chr20_-_29864390 | 0.25 |
ENSDART00000161834
ENSDART00000132278 |
rnf144ab
|
ring finger protein 144ab |
| chr25_+_31277415 | 0.24 |
ENSDART00000036275
|
tnni2a.4
|
troponin I type 2a (skeletal, fast), tandem duplicate 4 |
| chr25_-_13490744 | 0.23 |
ENSDART00000056721
|
ldhd
|
lactate dehydrogenase D |
| chr8_-_52562672 | 0.23 |
ENSDART00000159333
ENSDART00000159974 |
si:ch73-199g24.2
|
si:ch73-199g24.2 |
| chr17_+_12865746 | 0.23 |
ENSDART00000157083
|
ralgapa1
|
Ral GTPase activating protein, alpha subunit 1 (catalytic) |
| chr24_+_3307857 | 0.23 |
ENSDART00000106527
|
gyg1b
|
glycogenin 1b |
| chr6_-_40778294 | 0.23 |
ENSDART00000019845
|
arpc4
|
actin related protein 2/3 complex, subunit 4 |
| chr10_+_32104305 | 0.22 |
ENSDART00000099880
|
wnt11r
|
wingless-type MMTV integration site family, member 11, related |
| chr15_-_9272328 | 0.22 |
ENSDART00000172114
|
calm2a
|
calmodulin 2a (phosphorylase kinase, delta) |
| chr4_+_77943184 | 0.22 |
ENSDART00000159094
|
pacsin2
|
protein kinase C and casein kinase substrate in neurons 2 |
| chr18_+_48423973 | 0.22 |
ENSDART00000184233
ENSDART00000147074 |
fli1a
|
Fli-1 proto-oncogene, ETS transcription factor a |
| chr13_-_35808904 | 0.22 |
ENSDART00000171667
|
map3k4
|
mitogen-activated protein kinase kinase kinase 4 |
| chr14_+_924876 | 0.22 |
ENSDART00000183908
|
myoz3a
|
myozenin 3a |
| chr14_-_4145594 | 0.21 |
ENSDART00000077348
|
casp3b
|
caspase 3, apoptosis-related cysteine peptidase b |
| chr17_+_21902058 | 0.21 |
ENSDART00000142178
|
plekha1a
|
pleckstrin homology domain containing, family A (phosphoinositide binding specific) member 1a |
| chr5_+_19343880 | 0.21 |
ENSDART00000148130
|
acacb
|
acetyl-CoA carboxylase beta |
| chr16_-_51271962 | 0.20 |
ENSDART00000164021
ENSDART00000046420 |
serpinb1l1
|
serpin peptidase inhibitor, clade B (ovalbumin), member 1, like 1 |
| chr9_-_22834860 | 0.20 |
ENSDART00000146486
|
neb
|
nebulin |
| chr12_+_20352400 | 0.20 |
ENSDART00000066383
|
hbae5
|
hemoglobin, alpha embryonic 5 |
| chr3_+_17933132 | 0.20 |
ENSDART00000104299
ENSDART00000162144 ENSDART00000162242 ENSDART00000166289 ENSDART00000171101 ENSDART00000164853 |
cnp
|
2',3'-cyclic nucleotide 3' phosphodiesterase |
| chr9_-_14683574 | 0.20 |
ENSDART00000144022
|
pard3bb
|
par-3 family cell polarity regulator beta b |
| chr5_-_30074332 | 0.19 |
ENSDART00000147963
|
bco2a
|
beta-carotene oxygenase 2a |
| chr19_-_5669122 | 0.19 |
ENSDART00000112211
|
si:ch211-264f5.2
|
si:ch211-264f5.2 |
| chr19_-_38611814 | 0.19 |
ENSDART00000151958
|
col16a1
|
collagen, type XVI, alpha 1 |
| chr18_-_46258612 | 0.19 |
ENSDART00000153930
|
si:dkey-244a7.1
|
si:dkey-244a7.1 |
| chr1_-_58963395 | 0.19 |
ENSDART00000130415
|
CABZ01115881.1
|
|
| chr24_+_37640626 | 0.18 |
ENSDART00000008047
|
wdr24
|
WD repeat domain 24 |
| chr13_+_35339182 | 0.18 |
ENSDART00000019323
|
jag1b
|
jagged 1b |
| chr23_-_43453975 | 0.18 |
ENSDART00000134142
|
rprd1b
|
regulation of nuclear pre-mRNA domain containing 1B |
| chr4_+_11458078 | 0.18 |
ENSDART00000037600
|
ankrd16
|
ankyrin repeat domain 16 |
| chr16_-_41488023 | 0.17 |
ENSDART00000169312
|
cmtm6
|
CKLF-like MARVEL transmembrane domain containing 6 |
| chr12_+_30265802 | 0.17 |
ENSDART00000182412
|
afap1l2
|
actin filament associated protein 1-like 2 |
| chr13_-_36798204 | 0.17 |
ENSDART00000012357
|
sav1
|
salvador family WW domain containing protein 1 |
| chr6_-_30954268 | 0.17 |
ENSDART00000154523
|
pde4ba
|
phosphodiesterase 4B, cAMP-specific a |
| chr8_-_21372446 | 0.16 |
ENSDART00000061481
ENSDART00000079293 |
ela2l
|
elastase 2 like |
| chr4_-_277081 | 0.16 |
ENSDART00000166174
|
si:ch73-252i11.1
|
si:ch73-252i11.1 |
| chr21_+_25198637 | 0.16 |
ENSDART00000164972
|
si:dkey-183i3.6
|
si:dkey-183i3.6 |
| chr24_-_41320037 | 0.16 |
ENSDART00000129058
|
rheb
|
Ras homolog, mTORC1 binding |
| chr3_+_27798094 | 0.16 |
ENSDART00000075100
ENSDART00000151437 |
carhsp1
|
calcium regulated heat stable protein 1 |
| chr6_+_50381347 | 0.16 |
ENSDART00000055504
|
cyc1
|
cytochrome c-1 |
| chr2_-_37140423 | 0.16 |
ENSDART00000144220
|
tspan37
|
tetraspanin 37 |
| chr10_-_42297889 | 0.16 |
ENSDART00000099262
|
tcf7l1a
|
transcription factor 7 like 1a |
| chr23_+_4741543 | 0.16 |
ENSDART00000144761
|
raf1a
|
Raf-1 proto-oncogene, serine/threonine kinase a |
| chr25_+_31276842 | 0.15 |
ENSDART00000187238
|
tnni2a.4
|
troponin I type 2a (skeletal, fast), tandem duplicate 4 |
| chr23_+_19813677 | 0.15 |
ENSDART00000139192
ENSDART00000142308 |
emd
|
emerin (Emery-Dreifuss muscular dystrophy) |
| chr20_+_26880668 | 0.15 |
ENSDART00000077769
|
serpinb1
|
serpin peptidase inhibitor, clade B (ovalbumin), member 1 |
| chr16_-_51288178 | 0.15 |
ENSDART00000079864
|
zgc:173729
|
zgc:173729 |
| chr13_+_35637048 | 0.15 |
ENSDART00000085037
|
thbs2a
|
thrombospondin 2a |
| chr23_-_1348933 | 0.15 |
ENSDART00000168981
|
CABZ01078120.1
|
|
| chr5_+_69747417 | 0.15 |
ENSDART00000153717
|
si:ch211-275j6.5
|
si:ch211-275j6.5 |
| chr19_-_19379084 | 0.15 |
ENSDART00000165206
|
smarcc1b
|
SWI/SNF related, matrix associated, actin dependent regulator of chromatin, subfamily c, member 1b |
| chr3_-_58798377 | 0.15 |
ENSDART00000161248
|
si:ch73-281f12.4
|
si:ch73-281f12.4 |
| chr7_-_51368681 | 0.15 |
ENSDART00000146385
|
arhgap36
|
Rho GTPase activating protein 36 |
| chr21_+_43328685 | 0.15 |
ENSDART00000109620
ENSDART00000139668 |
sept8a
|
septin 8a |
| chr9_-_3934963 | 0.14 |
ENSDART00000062336
|
ubr3
|
ubiquitin protein ligase E3 component n-recognin 3 |
| chr13_+_35637875 | 0.14 |
ENSDART00000180657
|
thbs2a
|
thrombospondin 2a |
| chr3_-_15210491 | 0.14 |
ENSDART00000037906
|
hirip3
|
HIRA interacting protein 3 |
| chr1_+_513986 | 0.14 |
ENSDART00000109083
ENSDART00000081945 |
txnl4b
|
thioredoxin-like 4B |
| chr3_-_34084387 | 0.14 |
ENSDART00000155365
|
ighv4-3
|
immunoglobulin heavy variable 4-3 |
| chr14_+_30795559 | 0.14 |
ENSDART00000006132
|
cfl1
|
cofilin 1 |
| chr24_+_19415124 | 0.14 |
ENSDART00000186931
|
sulf1
|
sulfatase 1 |
| chr20_+_535458 | 0.13 |
ENSDART00000152453
|
dse
|
dermatan sulfate epimerase |
| chr7_-_30174882 | 0.13 |
ENSDART00000110409
|
frmd5
|
FERM domain containing 5 |
| chr11_+_43043171 | 0.13 |
ENSDART00000180344
|
CABZ01092982.1
|
|
| chr16_-_7828838 | 0.13 |
ENSDART00000191434
ENSDART00000108653 |
tcaim
|
T cell activation inhibitor, mitochondrial |
| chr12_+_3571770 | 0.13 |
ENSDART00000164707
ENSDART00000189819 |
coa3a
|
cytochrome C oxidase assembly factor 3a |
| chr16_+_41826584 | 0.13 |
ENSDART00000147523
|
si:dkey-199f5.6
|
si:dkey-199f5.6 |
| chr23_-_16485190 | 0.13 |
ENSDART00000155038
|
si:dkeyp-100a5.4
|
si:dkeyp-100a5.4 |
| chr12_-_28363111 | 0.13 |
ENSDART00000016283
ENSDART00000164156 |
psmd11b
|
proteasome 26S subunit, non-ATPase 11b |
| chr9_-_950004 | 0.13 |
ENSDART00000168917
|
SCTR
|
secretin receptor |
| chr16_+_13818743 | 0.13 |
ENSDART00000090191
|
flcn
|
folliculin |
| chr22_-_26274177 | 0.13 |
ENSDART00000060978
|
wdr83
|
WD repeat domain containing 83 |
| chr18_+_17827149 | 0.13 |
ENSDART00000190237
ENSDART00000189345 |
ZNF423
|
si:ch211-216l23.1 |
| chr3_+_32832538 | 0.13 |
ENSDART00000139410
|
cd2bp2
|
CD2 (cytoplasmic tail) binding protein 2 |
| chr6_-_55399214 | 0.13 |
ENSDART00000168367
|
ctsa
|
cathepsin A |
| chr9_+_38372216 | 0.13 |
ENSDART00000141895
|
plcd4b
|
phospholipase C, delta 4b |
| chr13_+_22295905 | 0.13 |
ENSDART00000180133
ENSDART00000181125 |
usp54a
|
ubiquitin specific peptidase 54a |
| chr9_-_5263947 | 0.12 |
ENSDART00000088342
|
cytip
|
cytohesin 1 interacting protein |
| chr14_-_7137808 | 0.12 |
ENSDART00000054803
|
trpt1
|
tRNA phosphotransferase 1 |
| chr6_+_24420523 | 0.12 |
ENSDART00000185461
|
tgfbr3
|
transforming growth factor, beta receptor III |
| chr24_-_8777781 | 0.12 |
ENSDART00000082362
ENSDART00000177400 |
mak
|
male germ cell-associated kinase |
| chr18_-_2727764 | 0.12 |
ENSDART00000160841
|
ARHGEF17
|
si:ch211-248g20.5 |
| chr17_+_44697604 | 0.12 |
ENSDART00000156625
|
pgfb
|
placental growth factor b |
| chr21_-_20939488 | 0.12 |
ENSDART00000039043
|
rgs7bpb
|
regulator of G protein signaling 7 binding protein b |
| chr15_+_5088210 | 0.12 |
ENSDART00000183423
|
mxf
|
myxovirus (influenza virus) resistance F |
| chr6_-_33878665 | 0.12 |
ENSDART00000129916
|
tmem69
|
transmembrane protein 69 |
| chr19_-_3724605 | 0.12 |
ENSDART00000123757
|
smim13
|
small integral membrane protein 13 |
| chr16_-_26855936 | 0.11 |
ENSDART00000167320
ENSDART00000078119 |
ino80c
|
INO80 complex subunit C |
| chr13_-_42400647 | 0.11 |
ENSDART00000043069
|
march5
|
membrane-associated ring finger (C3HC4) 5 |
| chr17_+_24722646 | 0.11 |
ENSDART00000138356
|
mtfr1l
|
mitochondrial fission regulator 1-like |
| chr2_-_26720854 | 0.11 |
ENSDART00000148110
|
si:dkey-181m9.8
|
si:dkey-181m9.8 |
| chr18_-_21950751 | 0.11 |
ENSDART00000043452
|
tsnaxip1
|
translin-associated factor X interacting protein 1 |
| chr20_-_14925281 | 0.11 |
ENSDART00000152641
|
dnm3a
|
dynamin 3a |
| chr2_+_30369116 | 0.11 |
ENSDART00000142137
|
pi15b
|
peptidase inhibitor 15b |
| chr24_+_11908833 | 0.11 |
ENSDART00000178622
|
fen1
|
flap structure-specific endonuclease 1 |
| chr11_+_2506516 | 0.11 |
ENSDART00000130886
ENSDART00000189767 |
NABP2
|
si:ch73-190f16.2 |
| chr12_+_26877381 | 0.11 |
ENSDART00000087329
|
znf438
|
zinc finger protein 438 |
| chr9_+_30421489 | 0.11 |
ENSDART00000145025
ENSDART00000132058 |
zgc:113314
|
zgc:113314 |
| chr23_+_29885019 | 0.11 |
ENSDART00000167059
|
aurkaip1
|
aurora kinase A interacting protein 1 |
| chr13_+_15190677 | 0.11 |
ENSDART00000142240
ENSDART00000129045 |
mavs
|
mitochondrial antiviral signaling protein |
| chr24_+_40905100 | 0.10 |
ENSDART00000167854
|
scn12ab
|
sodium channel, voltage gated, type XII, alpha b |
| chr11_+_33818179 | 0.10 |
ENSDART00000109418
|
spoplb
|
speckle-type POZ protein-like b |
| chr19_+_3826782 | 0.10 |
ENSDART00000169222
|
oscp1a
|
organic solute carrier partner 1a |
| chr6_+_9677420 | 0.10 |
ENSDART00000064995
|
sumo1
|
small ubiquitin-like modifier 1 |
| chr20_-_1378514 | 0.10 |
ENSDART00000181830
|
scara5
|
scavenger receptor class A, member 5 (putative) |
| chr23_+_44374041 | 0.10 |
ENSDART00000136056
|
ephb4b
|
eph receptor B4b |
| chr22_+_2254972 | 0.10 |
ENSDART00000144906
|
znf1157
|
zinc finger protein 1157 |
| chr20_+_46513651 | 0.10 |
ENSDART00000152977
|
zc3h14
|
zinc finger CCCH-type containing 14 |
| chr9_-_6372535 | 0.10 |
ENSDART00000149189
|
ecrg4a
|
esophageal cancer related gene 4a |
| chr1_-_23268569 | 0.10 |
ENSDART00000143948
|
rfc1
|
replication factor C (activator 1) 1 |
| chr24_-_36680261 | 0.10 |
ENSDART00000059507
|
ccr10
|
chemokine (C-C motif) receptor 10 |
| chr18_-_46256560 | 0.10 |
ENSDART00000171375
|
si:dkey-244a7.1
|
si:dkey-244a7.1 |
| chr18_-_43884044 | 0.09 |
ENSDART00000087382
|
ddx6
|
DEAD (Asp-Glu-Ala-Asp) box helicase 6 |
| chr24_+_22731228 | 0.09 |
ENSDART00000146733
|
si:dkey-225k4.1
|
si:dkey-225k4.1 |
| chr20_+_53181017 | 0.09 |
ENSDART00000189692
ENSDART00000177109 |
FIG4
|
FIG4 phosphoinositide 5-phosphatase |
| chr15_+_934660 | 0.09 |
ENSDART00000154248
|
si:dkey-77f5.10
|
si:dkey-77f5.10 |
| chr21_+_45757317 | 0.09 |
ENSDART00000163152
|
h2afy
|
H2A histone family, member Y |
| chr22_-_7702958 | 0.09 |
ENSDART00000193046
|
si:ch211-59h6.1
|
si:ch211-59h6.1 |
| chr20_-_9095105 | 0.09 |
ENSDART00000140792
|
oma1
|
OMA1 zinc metallopeptidase |
| chr15_-_35252522 | 0.09 |
ENSDART00000144153
ENSDART00000059195 |
mff
|
mitochondrial fission factor |
| chr6_+_3280939 | 0.09 |
ENSDART00000151359
|
kdm4aa
|
lysine (K)-specific demethylase 4A, genome duplicate a |
| chr8_-_31107537 | 0.09 |
ENSDART00000098925
|
vgll4l
|
vestigial like 4 like |
| chr7_-_24046999 | 0.09 |
ENSDART00000144616
ENSDART00000124653 ENSDART00000127813 |
dhrs4
|
dehydrogenase/reductase (SDR family) member 4 |
| chr19_-_44064842 | 0.09 |
ENSDART00000151541
|
mterf3
|
mitochondrial transcription termination factor 3 |
| chr8_+_20140321 | 0.09 |
ENSDART00000012120
|
acsbg2
|
acyl-CoA synthetase bubblegum family member 2 |
| chr24_+_11908480 | 0.09 |
ENSDART00000024224
|
fen1
|
flap structure-specific endonuclease 1 |
| chr14_+_23717165 | 0.09 |
ENSDART00000006373
|
ndfip1
|
Nedd4 family interacting protein 1 |
| chr11_-_44979281 | 0.09 |
ENSDART00000190972
|
ldb1b
|
LIM-domain binding 1b |
| chr15_+_47492838 | 0.09 |
ENSDART00000122372
|
sik3
|
SIK family kinase 3 |
| chr4_-_3353595 | 0.08 |
ENSDART00000009076
|
pik3cg
|
phosphatidylinositol-4,5-bisphosphate 3-kinase, catalytic subunit gamma |
| chr11_+_25129013 | 0.08 |
ENSDART00000132879
|
ndrg3a
|
ndrg family member 3a |
| chr25_-_36827491 | 0.08 |
ENSDART00000170941
|
CU929365.1
|
|
| chr22_-_30770751 | 0.08 |
ENSDART00000172115
|
AL831726.2
|
|
| chr20_-_14924858 | 0.08 |
ENSDART00000047039
|
dnm3a
|
dynamin 3a |
| chr5_-_38063585 | 0.08 |
ENSDART00000164947
|
si:dkey-111e8.5
|
si:dkey-111e8.5 |
| chr5_-_52010122 | 0.08 |
ENSDART00000073627
ENSDART00000163898 ENSDART00000051003 |
cdk7
|
cyclin-dependent kinase 7 |
| chr20_+_25225112 | 0.08 |
ENSDART00000153088
ENSDART00000127291 ENSDART00000130494 |
moxd1
|
monooxygenase, DBH-like 1 |
| chr11_+_11303458 | 0.08 |
ENSDART00000162486
ENSDART00000160703 |
si:dkey-23f9.4
|
si:dkey-23f9.4 |
| chr15_-_2493771 | 0.08 |
ENSDART00000184906
|
neu4
|
sialidase 4 |
| chr12_+_5977777 | 0.08 |
ENSDART00000152302
|
si:ch211-131k2.2
|
si:ch211-131k2.2 |
| chr3_+_29445473 | 0.07 |
ENSDART00000020381
|
grap2a
|
GRB2-related adaptor protein 2a |
| chr8_-_49908978 | 0.07 |
ENSDART00000172642
|
agtpbp1
|
ATP/GTP binding protein 1 |
| chr4_-_45100253 | 0.07 |
ENSDART00000163870
|
si:dkey-51d8.3
|
si:dkey-51d8.3 |
| chr7_+_8456999 | 0.07 |
ENSDART00000172880
|
jac4
|
jacalin 4 |
| chr10_+_28160265 | 0.07 |
ENSDART00000022484
|
rnft1
|
ring finger protein, transmembrane 1 |
| chr16_+_54209504 | 0.07 |
ENSDART00000020033
|
xrcc1
|
X-ray repair complementing defective repair in Chinese hamster cells 1 |
| chr2_+_30368800 | 0.07 |
ENSDART00000179564
|
pi15b
|
peptidase inhibitor 15b |
| chr1_-_57629639 | 0.07 |
ENSDART00000158984
|
zmp:0000001289
|
zmp:0000001289 |
| chr19_-_31402429 | 0.07 |
ENSDART00000137292
|
tmem106bb
|
transmembrane protein 106Bb |
| chr21_-_26490186 | 0.07 |
ENSDART00000009889
|
zgc:110540
|
zgc:110540 |
Network of associatons between targets according to the STRING database.
First level regulatory network of gsx2+pnx
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.4 | GO:0007008 | outer mitochondrial membrane organization(GO:0007008) protein import into mitochondrial outer membrane(GO:0045040) |
| 0.1 | 0.3 | GO:0097237 | cellular response to antibiotic(GO:0071236) cellular response to toxic substance(GO:0097237) |
| 0.1 | 0.5 | GO:0043137 | DNA replication, removal of RNA primer(GO:0043137) |
| 0.1 | 0.2 | GO:0006089 | lactate metabolic process(GO:0006089) |
| 0.1 | 0.4 | GO:0060231 | mesenchymal to epithelial transition(GO:0060231) |
| 0.1 | 0.6 | GO:0046716 | muscle cell cellular homeostasis(GO:0046716) |
| 0.1 | 0.8 | GO:0042396 | phosphagen metabolic process(GO:0006599) phosphocreatine metabolic process(GO:0006603) phosphagen biosynthetic process(GO:0042396) phosphocreatine biosynthetic process(GO:0046314) |
| 0.1 | 0.2 | GO:0032637 | interleukin-8 production(GO:0032637) regulation of interleukin-8 production(GO:0032677) positive regulation of interleukin-8 production(GO:0032757) |
| 0.1 | 0.6 | GO:0032781 | positive regulation of ATPase activity(GO:0032781) |
| 0.1 | 0.2 | GO:2001295 | malonyl-CoA biosynthetic process(GO:2001295) |
| 0.0 | 0.2 | GO:0051660 | establishment of centrosome localization(GO:0051660) |
| 0.0 | 0.1 | GO:0030043 | actin filament fragmentation(GO:0030043) |
| 0.0 | 0.2 | GO:0060829 | negative regulation of canonical Wnt signaling pathway involved in neural plate anterior/posterior pattern formation(GO:0060829) |
| 0.0 | 2.0 | GO:0003009 | skeletal muscle contraction(GO:0003009) |
| 0.0 | 0.3 | GO:0016576 | histone dephosphorylation(GO:0016576) negative regulation of signal transduction in absence of ligand(GO:1901099) regulation of extrinsic apoptotic signaling pathway in absence of ligand(GO:2001239) negative regulation of extrinsic apoptotic signaling pathway in absence of ligand(GO:2001240) |
| 0.0 | 0.1 | GO:1900364 | negative regulation of mRNA 3'-end processing(GO:0031441) negative regulation of mRNA polyadenylation(GO:1900364) |
| 0.0 | 0.2 | GO:0071691 | cardiac muscle thin filament assembly(GO:0071691) |
| 0.0 | 0.1 | GO:0070314 | G1 to G0 transition(GO:0070314) |
| 0.0 | 0.1 | GO:0070131 | regulation of mitochondrial translation(GO:0070129) positive regulation of mitochondrial translation(GO:0070131) |
| 0.0 | 0.3 | GO:0070979 | protein K11-linked ubiquitination(GO:0070979) |
| 0.0 | 0.1 | GO:0071596 | ubiquitin-dependent protein catabolic process via the N-end rule pathway(GO:0071596) |
| 0.0 | 0.1 | GO:0030205 | dermatan sulfate metabolic process(GO:0030205) |
| 0.0 | 0.1 | GO:0046333 | octopamine biosynthetic process(GO:0006589) dopamine catabolic process(GO:0042420) norepinephrine biosynthetic process(GO:0042421) octopamine metabolic process(GO:0046333) |
| 0.0 | 0.1 | GO:0071276 | cellular response to cadmium ion(GO:0071276) |
| 0.0 | 0.2 | GO:1903846 | positive regulation of transforming growth factor beta receptor signaling pathway(GO:0030511) positive regulation of cellular response to transforming growth factor beta stimulus(GO:1903846) |
| 0.0 | 0.2 | GO:0044857 | membrane raft assembly(GO:0001765) plasma membrane raft assembly(GO:0044854) plasma membrane raft organization(GO:0044857) caveola assembly(GO:0070836) |
| 0.0 | 0.1 | GO:0061668 | mitochondrial ribosome assembly(GO:0061668) |
| 0.0 | 0.1 | GO:0098586 | cytoplasmic pattern recognition receptor signaling pathway in response to virus(GO:0039528) cellular response to virus(GO:0098586) |
| 0.0 | 0.5 | GO:0030032 | lamellipodium assembly(GO:0030032) |
| 0.0 | 0.1 | GO:0035610 | protein side chain deglutamylation(GO:0035610) |
| 0.0 | 0.1 | GO:0048340 | paraxial mesoderm morphogenesis(GO:0048340) paraxial mesoderm formation(GO:0048341) |
| 0.0 | 0.1 | GO:0021910 | smoothened signaling pathway involved in ventral spinal cord patterning(GO:0021910) regulation of myotome development(GO:2000290) |
| 0.0 | 0.3 | GO:0098884 | postsynaptic neurotransmitter receptor internalization(GO:0098884) |
| 0.0 | 0.4 | GO:0032008 | positive regulation of TOR signaling(GO:0032008) |
| 0.0 | 0.2 | GO:0006122 | mitochondrial electron transport, ubiquinol to cytochrome c(GO:0006122) |
| 0.0 | 0.5 | GO:0055008 | cardiac muscle tissue morphogenesis(GO:0055008) |
| 0.0 | 0.3 | GO:0034625 | fatty acid elongation, saturated fatty acid(GO:0019367) fatty acid elongation, unsaturated fatty acid(GO:0019368) fatty acid elongation, monounsaturated fatty acid(GO:0034625) fatty acid elongation, polyunsaturated fatty acid(GO:0034626) |
| 0.0 | 0.7 | GO:0031102 | neuron projection regeneration(GO:0031102) |
| 0.0 | 0.1 | GO:0007217 | tachykinin receptor signaling pathway(GO:0007217) |
| 0.0 | 0.1 | GO:0034755 | iron ion transmembrane transport(GO:0034755) protein homotrimerization(GO:0070207) |
| 0.0 | 0.1 | GO:1904292 | regulation of ERAD pathway(GO:1904292) |
| 0.0 | 0.1 | GO:0000012 | single strand break repair(GO:0000012) |
| 0.0 | 0.2 | GO:0030216 | keratinocyte differentiation(GO:0030216) |
| 0.0 | 0.1 | GO:0006388 | tRNA splicing, via endonucleolytic cleavage and ligation(GO:0006388) |
| 0.0 | 0.3 | GO:0051014 | actin filament severing(GO:0051014) |
| 0.0 | 0.0 | GO:0015919 | peroxisomal membrane transport(GO:0015919) protein import into peroxisome membrane(GO:0045046) |
| 0.0 | 0.2 | GO:0060325 | face morphogenesis(GO:0060325) |
| 0.0 | 0.1 | GO:0034063 | stress granule assembly(GO:0034063) |
| 0.0 | 0.1 | GO:1990845 | adaptive thermogenesis(GO:1990845) |
| 0.0 | 0.0 | GO:0038093 | Fc receptor signaling pathway(GO:0038093) |
| 0.0 | 0.0 | GO:0030579 | ubiquitin-dependent SMAD protein catabolic process(GO:0030579) |
| 0.0 | 0.1 | GO:0060753 | mast cell chemotaxis(GO:0002551) regulation of mast cell chemotaxis(GO:0060753) positive regulation of mast cell chemotaxis(GO:0060754) mast cell migration(GO:0097531) |
| 0.0 | 0.2 | GO:0000186 | activation of MAPKK activity(GO:0000186) |
| 0.0 | 0.1 | GO:1904105 | positive regulation of convergent extension involved in gastrulation(GO:1904105) |
| 0.0 | 0.1 | GO:0070301 | cellular response to hydrogen peroxide(GO:0070301) |
| 0.0 | 0.9 | GO:0015718 | monocarboxylic acid transport(GO:0015718) |
| 0.0 | 0.2 | GO:0015671 | oxygen transport(GO:0015671) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.3 | GO:0032299 | ribonuclease H2 complex(GO:0032299) |
| 0.1 | 1.1 | GO:0031430 | M band(GO:0031430) |
| 0.0 | 2.0 | GO:0005861 | troponin complex(GO:0005861) |
| 0.0 | 0.4 | GO:0001401 | mitochondrial sorting and assembly machinery complex(GO:0001401) |
| 0.0 | 0.3 | GO:0016602 | CCAAT-binding factor complex(GO:0016602) |
| 0.0 | 0.3 | GO:0031462 | Cul2-RING ubiquitin ligase complex(GO:0031462) |
| 0.0 | 0.2 | GO:0031838 | haptoglobin-hemoglobin complex(GO:0031838) |
| 0.0 | 0.3 | GO:0098844 | postsynaptic endocytic zone membrane(GO:0098844) |
| 0.0 | 0.2 | GO:0045275 | mitochondrial respiratory chain complex III(GO:0005750) respiratory chain complex III(GO:0045275) |
| 0.0 | 0.3 | GO:0005682 | U5 snRNP(GO:0005682) |
| 0.0 | 1.6 | GO:0030018 | Z disc(GO:0030018) |
| 0.0 | 0.6 | GO:0005747 | mitochondrial respiratory chain complex I(GO:0005747) NADH dehydrogenase complex(GO:0030964) respiratory chain complex I(GO:0045271) |
| 0.0 | 0.1 | GO:0071797 | LUBAC complex(GO:0071797) |
| 0.0 | 0.2 | GO:0005885 | Arp2/3 protein complex(GO:0005885) |
| 0.0 | 0.1 | GO:0032584 | growth cone membrane(GO:0032584) |
| 0.0 | 0.1 | GO:0008541 | proteasome regulatory particle, lid subcomplex(GO:0008541) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.7 | GO:0004743 | pyruvate kinase activity(GO:0004743) |
| 0.1 | 0.7 | GO:0050262 | ribosylnicotinamide kinase activity(GO:0050262) |
| 0.1 | 0.9 | GO:0015129 | lactate transmembrane transporter activity(GO:0015129) |
| 0.1 | 0.5 | GO:0004523 | RNA-DNA hybrid ribonuclease activity(GO:0004523) |
| 0.1 | 0.8 | GO:0004111 | creatine kinase activity(GO:0004111) phosphotransferase activity, nitrogenous group as acceptor(GO:0016775) |
| 0.1 | 0.2 | GO:0003989 | acetyl-CoA carboxylase activity(GO:0003989) |
| 0.0 | 0.7 | GO:0051879 | Hsp90 protein binding(GO:0051879) |
| 0.0 | 0.2 | GO:0043295 | glutathione binding(GO:0043295) |
| 0.0 | 0.6 | GO:0051393 | muscle alpha-actinin binding(GO:0051371) alpha-actinin binding(GO:0051393) |
| 0.0 | 0.2 | GO:0004457 | lactate dehydrogenase activity(GO:0004457) |
| 0.0 | 0.2 | GO:0042169 | SH2 domain binding(GO:0042169) |
| 0.0 | 0.1 | GO:0047757 | chondroitin-glucuronate 5-epimerase activity(GO:0047757) |
| 0.0 | 0.1 | GO:0004500 | dopamine beta-monooxygenase activity(GO:0004500) |
| 0.0 | 0.1 | GO:0004185 | serine-type carboxypeptidase activity(GO:0004185) |
| 0.0 | 0.1 | GO:0031834 | neurokinin receptor binding(GO:0031834) substance P receptor binding(GO:0031835) |
| 0.0 | 0.7 | GO:0048306 | calcium-dependent protein binding(GO:0048306) |
| 0.0 | 0.2 | GO:0031433 | telethonin binding(GO:0031433) FATZ binding(GO:0051373) |
| 0.0 | 0.1 | GO:0050431 | transforming growth factor beta binding(GO:0050431) |
| 0.0 | 0.2 | GO:0031720 | haptoglobin binding(GO:0031720) |
| 0.0 | 0.1 | GO:0001223 | transcription coactivator binding(GO:0001223) |
| 0.0 | 0.1 | GO:0050699 | WW domain binding(GO:0050699) |
| 0.0 | 0.1 | GO:0043621 | protein self-association(GO:0043621) |
| 0.0 | 0.3 | GO:0102338 | fatty acid elongase activity(GO:0009922) 3-oxo-arachidoyl-CoA synthase activity(GO:0102336) 3-oxo-cerotoyl-CoA synthase activity(GO:0102337) 3-oxo-lignoceronyl-CoA synthase activity(GO:0102338) |
| 0.0 | 0.1 | GO:0003747 | translation release factor activity(GO:0003747) translation termination factor activity(GO:0008079) |
| 0.0 | 0.1 | GO:0097108 | hedgehog family protein binding(GO:0097108) |
| 0.0 | 0.1 | GO:0030274 | LIM domain binding(GO:0030274) |
| 0.0 | 0.1 | GO:0005172 | vascular endothelial growth factor receptor binding(GO:0005172) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.5 | PID TCR CALCIUM PATHWAY | Calcium signaling in the CD4+ TCR pathway |
| 0.0 | 0.4 | PID P38 GAMMA DELTA PATHWAY | Signaling mediated by p38-gamma and p38-delta |
| 0.0 | 0.1 | SA FAS SIGNALING | The TNF-type receptor Fas induces apoptosis on ligand binding. |
| 0.0 | 0.5 | PID PRL SIGNALING EVENTS PATHWAY | Signaling events mediated by PRL |
| 0.0 | 0.6 | ST WNT BETA CATENIN PATHWAY | Wnt/beta-catenin Pathway |
| 0.0 | 0.5 | PID ILK PATHWAY | Integrin-linked kinase signaling |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.3 | REACTOME PD1 SIGNALING | Genes involved in PD-1 signaling |
| 0.1 | 0.9 | REACTOME BILE SALT AND ORGANIC ANION SLC TRANSPORTERS | Genes involved in Bile salt and organic anion SLC transporters |
| 0.0 | 0.3 | REACTOME PECAM1 INTERACTIONS | Genes involved in PECAM1 interactions |
| 0.0 | 0.5 | REACTOME CELL EXTRACELLULAR MATRIX INTERACTIONS | Genes involved in Cell-extracellular matrix interactions |
| 0.0 | 0.2 | REACTOME REGULATION OF RHEB GTPASE ACTIVITY BY AMPK | Genes involved in Regulation of Rheb GTPase activity by AMPK |
| 0.0 | 0.2 | REACTOME EARLY PHASE OF HIV LIFE CYCLE | Genes involved in Early Phase of HIV Life Cycle |
| 0.0 | 0.1 | REACTOME ENDOSOMAL VACUOLAR PATHWAY | Genes involved in Endosomal/Vacuolar pathway |
| 0.0 | 0.1 | REACTOME REGULATION OF IFNG SIGNALING | Genes involved in Regulation of IFNG signaling |
| 0.0 | 0.3 | REACTOME P130CAS LINKAGE TO MAPK SIGNALING FOR INTEGRINS | Genes involved in p130Cas linkage to MAPK signaling for integrins |
| 0.0 | 0.1 | REACTOME CHEMOKINE RECEPTORS BIND CHEMOKINES | Genes involved in Chemokine receptors bind chemokines |
| 0.0 | 0.2 | REACTOME ACTIVATED AMPK STIMULATES FATTY ACID OXIDATION IN MUSCLE | Genes involved in Activated AMPK stimulates fatty-acid oxidation in muscle |
| 0.0 | 0.1 | REACTOME G BETA GAMMA SIGNALLING THROUGH PI3KGAMMA | Genes involved in G beta:gamma signalling through PI3Kgamma |
| 0.0 | 0.1 | REACTOME SEMA3A PAK DEPENDENT AXON REPULSION | Genes involved in Sema3A PAK dependent Axon repulsion |
| 0.0 | 0.8 | REACTOME RESPIRATORY ELECTRON TRANSPORT | Genes involved in Respiratory electron transport |