Project
PRJNA207719: Tissue specific transcriptome profiling
Navigation
Downloads
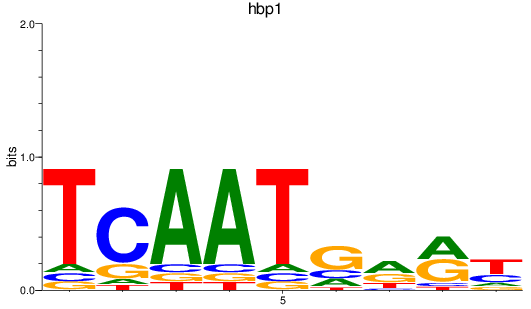
Results for hbp1
Z-value: 0.84
Transcription factors associated with hbp1
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
hbp1
|
ENSDARG00000028517 | HMG-box transcription factor 1 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| hbp1 | dr11_v1_chr25_-_3469576_3469576 | 0.38 | 5.3e-01 | Click! |
Activity profile of hbp1 motif
Sorted Z-values of hbp1 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr18_+_13792490 | 1.27 |
ENSDART00000136754
|
cdh13
|
cadherin 13, H-cadherin (heart) |
| chr12_-_11457625 | 0.91 |
ENSDART00000012318
|
htra1b
|
HtrA serine peptidase 1b |
| chr17_+_15297398 | 0.76 |
ENSDART00000156574
|
si:ch211-270g19.5
|
si:ch211-270g19.5 |
| chr8_+_48613040 | 0.67 |
ENSDART00000121432
|
nppa
|
natriuretic peptide A |
| chr19_-_32487469 | 0.57 |
ENSDART00000050130
|
gmpr
|
guanosine monophosphate reductase |
| chr11_+_36243774 | 0.55 |
ENSDART00000023323
|
zgc:172270
|
zgc:172270 |
| chr21_+_45626136 | 0.49 |
ENSDART00000158742
|
irf1b
|
interferon regulatory factor 1b |
| chr3_-_34528306 | 0.49 |
ENSDART00000023039
|
sept9a
|
septin 9a |
| chr23_-_24343363 | 0.40 |
ENSDART00000166392
|
fam131c
|
family with sequence similarity 131, member C |
| chr4_+_77948970 | 0.40 |
ENSDART00000149636
|
pacsin2
|
protein kinase C and casein kinase substrate in neurons 2 |
| chr6_-_40722200 | 0.40 |
ENSDART00000035101
|
kbtbd12
|
kelch repeat and BTB (POZ) domain containing 12 |
| chr20_-_49729446 | 0.38 |
ENSDART00000111089
|
filip1b
|
filamin A interacting protein 1b |
| chr22_+_10646928 | 0.38 |
ENSDART00000038465
|
rassf1
|
Ras association (RalGDS/AF-6) domain family 1 |
| chr11_-_25418856 | 0.34 |
ENSDART00000013714
|
gata1a
|
GATA binding protein 1a |
| chr1_-_33380011 | 0.28 |
ENSDART00000141347
ENSDART00000136383 |
cd99
|
CD99 molecule |
| chr12_+_23812530 | 0.28 |
ENSDART00000066331
|
svila
|
supervillin a |
| chr14_-_30945515 | 0.27 |
ENSDART00000161540
|
si:zfos-80g12.1
|
si:zfos-80g12.1 |
| chr1_-_33380340 | 0.27 |
ENSDART00000181515
|
cd99
|
CD99 molecule |
| chr13_-_28672474 | 0.26 |
ENSDART00000164508
|
nt5c2a
|
5'-nucleotidase, cytosolic IIa |
| chr20_-_44496245 | 0.25 |
ENSDART00000012229
|
fkbp1b
|
FK506 binding protein 1b |
| chr20_-_53996193 | 0.25 |
ENSDART00000004756
|
hsp90aa1.1
|
heat shock protein 90, alpha (cytosolic), class A member 1, tandem duplicate 1 |
| chr22_+_35068046 | 0.24 |
ENSDART00000161660
ENSDART00000169573 |
si:ch73-173h19.3
|
si:ch73-173h19.3 |
| chr3_+_11101585 | 0.24 |
ENSDART00000172103
|
sstr5
|
somatostatin receptor 5 |
| chr5_-_4230370 | 0.22 |
ENSDART00000133187
|
si:ch211-283g2.2
|
si:ch211-283g2.2 |
| chr6_+_29402997 | 0.22 |
ENSDART00000104298
|
ndufb5
|
NADH dehydrogenase (ubiquinone) 1 beta subcomplex 5 |
| chr11_+_41064822 | 0.22 |
ENSDART00000167495
|
svbp
|
small vasohibin binding protein |
| chr14_+_6159162 | 0.20 |
ENSDART00000128638
|
bscl2l
|
Bernardinelli-Seip congenital lipodystrophy 2, like |
| chr16_+_46453176 | 0.18 |
ENSDART00000132793
|
rpz3
|
rapunzel 3 |
| chr4_-_73190246 | 0.18 |
ENSDART00000170842
|
LO018260.1
|
|
| chr7_-_25697285 | 0.17 |
ENSDART00000082620
|
dysf
|
dysferlin, limb girdle muscular dystrophy 2B (autosomal recessive) |
| chr21_-_42831033 | 0.17 |
ENSDART00000160998
|
stk10
|
serine/threonine kinase 10 |
| chr14_+_6159356 | 0.15 |
ENSDART00000157730
|
bscl2l
|
Bernardinelli-Seip congenital lipodystrophy 2, like |
| chr17_-_26911852 | 0.14 |
ENSDART00000045842
|
rcan3
|
regulator of calcineurin 3 |
| chr12_+_4260779 | 0.13 |
ENSDART00000081382
|
mmp25b
|
matrix metallopeptidase 25b |
| chr18_+_22138924 | 0.13 |
ENSDART00000183961
|
ripor1
|
RHO family interacting cell polarization regulator 1 |
| chr23_-_38054 | 0.13 |
ENSDART00000170393
|
CABZ01074076.1
|
|
| chr11_+_45219558 | 0.12 |
ENSDART00000167828
|
tmc6b
|
transmembrane channel-like 6b |
| chr16_+_7242610 | 0.12 |
ENSDART00000081477
|
sri
|
sorcin |
| chr25_+_23777469 | 0.12 |
ENSDART00000067179
|
mrpl23
|
mitochondrial ribosomal protein L23 |
| chr8_+_27555314 | 0.11 |
ENSDART00000135568
ENSDART00000016696 |
rhocb
|
ras homolog family member Cb |
| chr5_+_30518036 | 0.11 |
ENSDART00000161836
|
hmbsa
|
hydroxymethylbilane synthase a |
| chr16_-_41439659 | 0.11 |
ENSDART00000191624
|
cpne4a
|
copine IVa |
| chr16_+_43344475 | 0.11 |
ENSDART00000085282
|
mrpl32
|
mitochondrial ribosomal protein L32 |
| chr6_-_24103666 | 0.10 |
ENSDART00000164915
|
scinla
|
scinderin like a |
| chr24_-_31194847 | 0.10 |
ENSDART00000158808
|
cnn3a
|
calponin 3, acidic a |
| chr17_-_26537928 | 0.10 |
ENSDART00000155692
ENSDART00000122366 |
ccser2a
|
coiled-coil serine-rich protein 2a |
| chr15_+_37936458 | 0.09 |
ENSDART00000154491
|
si:ch73-380l3.2
|
si:ch73-380l3.2 |
| chr18_+_16986903 | 0.09 |
ENSDART00000142088
|
si:ch211-218c6.8
|
si:ch211-218c6.8 |
| chr12_+_20412293 | 0.09 |
ENSDART00000115015
ENSDART00000177573 |
arhgap17a
|
Rho GTPase activating protein 17a |
| chr16_-_9423735 | 0.09 |
ENSDART00000185645
|
ccr8.1
|
chemokine (C-C motif) receptor 8.1 |
| chr23_-_32092443 | 0.09 |
ENSDART00000133688
|
letmd1
|
LETM1 domain containing 1 |
| chr6_+_23887314 | 0.08 |
ENSDART00000163188
|
znf648
|
zinc finger protein 648 |
| chr4_-_9909371 | 0.08 |
ENSDART00000102656
|
si:dkey-22l11.6
|
si:dkey-22l11.6 |
| chr12_-_28548789 | 0.08 |
ENSDART00000153062
|
si:ch73-180n10.1
|
si:ch73-180n10.1 |
| chr16_-_47427016 | 0.08 |
ENSDART00000074575
|
sept7b
|
septin 7b |
| chr20_+_48116476 | 0.08 |
ENSDART00000043938
|
tram2
|
translocation associated membrane protein 2 |
| chr24_+_38301080 | 0.07 |
ENSDART00000105672
|
mybpc2b
|
myosin binding protein C, fast type b |
| chr4_-_55641422 | 0.07 |
ENSDART00000165178
|
znf1074
|
zinc finger protein 1074 |
| chr17_+_21514777 | 0.07 |
ENSDART00000154633
|
chst15
|
carbohydrate (N-acetylgalactosamine 4-sulfate 6-O) sulfotransferase 15 |
| chr8_-_4010887 | 0.07 |
ENSDART00000163678
|
mtmr3
|
myotubularin related protein 3 |
| chr23_-_40895168 | 0.07 |
ENSDART00000061037
|
si:dkey-194e6.1
|
si:dkey-194e6.1 |
| chr9_+_30577925 | 0.06 |
ENSDART00000139811
|
commd6
|
COMM domain containing 6 |
| chr1_+_40263671 | 0.06 |
ENSDART00000045697
|
zgc:56493
|
zgc:56493 |
| chr12_+_9183626 | 0.06 |
ENSDART00000020192
|
tubgcp2
|
tubulin, gamma complex associated protein 2 |
| chr10_-_24659822 | 0.06 |
ENSDART00000171842
ENSDART00000163102 |
proser1
|
proline and serine rich 1 |
| chr7_+_21272833 | 0.06 |
ENSDART00000052942
|
serpinh2
|
serine (or cysteine) peptidase inhibitor, clade H, member 2 |
| chr17_+_49096222 | 0.05 |
ENSDART00000185900
|
tiam2a
|
T cell lymphoma invasion and metastasis 2a |
| chr2_-_11027258 | 0.05 |
ENSDART00000081072
ENSDART00000193824 ENSDART00000187036 ENSDART00000097741 |
ssbp3a
|
single stranded DNA binding protein 3a |
| chr1_-_633356 | 0.05 |
ENSDART00000171019
|
appa
|
amyloid beta (A4) precursor protein a |
| chr15_-_28107502 | 0.05 |
ENSDART00000152270
ENSDART00000167129 |
cryba1a
|
crystallin, beta A1a |
| chr20_+_1088658 | 0.05 |
ENSDART00000162991
|
BX537249.1
|
|
| chr4_+_32649955 | 0.05 |
ENSDART00000135750
|
znf1042
|
zinc finger protein 1042 |
| chr5_-_19444930 | 0.05 |
ENSDART00000136259
ENSDART00000188499 |
kctd10
|
potassium channel tetramerization domain containing 10 |
| chr14_-_34512859 | 0.05 |
ENSDART00000140368
|
si:ch211-232m8.3
|
si:ch211-232m8.3 |
| chr22_+_2504524 | 0.05 |
ENSDART00000136686
|
znf1170
|
zinc finger protein 1170 |
| chr24_+_13735616 | 0.05 |
ENSDART00000184267
|
msc
|
musculin (activated B-cell factor-1) |
| chr20_+_47491247 | 0.05 |
ENSDART00000113412
|
lin28b
|
lin-28 homolog B (C. elegans) |
| chr10_-_15405564 | 0.05 |
ENSDART00000020665
|
sgtb
|
small glutamine-rich tetratricopeptide repeat (TPR)-containing, beta |
| chr5_+_9434288 | 0.04 |
ENSDART00000162089
|
ugt2a7
|
UDP glucuronosyltransferase 2 family, polypeptide A7 |
| chr15_-_27972474 | 0.04 |
ENSDART00000162753
|
CR391930.1
|
|
| chr18_+_9362455 | 0.04 |
ENSDART00000187025
|
sema3ab
|
sema domain, immunoglobulin domain (Ig), short basic domain, secreted, (semaphorin) 3Ab |
| chr15_-_2497568 | 0.04 |
ENSDART00000080398
|
neu4
|
sialidase 4 |
| chr4_-_43388943 | 0.04 |
ENSDART00000150796
|
si:dkey-29j8.2
|
si:dkey-29j8.2 |
| chr16_+_21918503 | 0.04 |
ENSDART00000167919
|
znf687a
|
zinc finger protein 687a |
| chr15_+_45643787 | 0.04 |
ENSDART00000055995
ENSDART00000157750 |
sagb
|
S-antigen; retina and pineal gland (arrestin) b |
| chr22_-_26865181 | 0.04 |
ENSDART00000138311
|
hmox2a
|
heme oxygenase 2a |
| chr1_-_45341760 | 0.04 |
ENSDART00000149183
ENSDART00000148289 ENSDART00000110390 |
zgc:101679
|
zgc:101679 |
| chr10_+_24660017 | 0.04 |
ENSDART00000079597
|
vps36
|
vacuolar protein sorting 36 homolog (S. cerevisiae) |
| chr4_+_31328329 | 0.04 |
ENSDART00000189539
|
si:rp71-5o12.3
|
si:rp71-5o12.3 |
| chr21_+_38855551 | 0.03 |
ENSDART00000171977
|
ddx52
|
DEAD (Asp-Glu-Ala-Asp) box polypeptide 52 |
| chr6_+_38880166 | 0.03 |
ENSDART00000019939
ENSDART00000144286 |
bin2b
|
bridging integrator 2b |
| chr13_-_22843562 | 0.03 |
ENSDART00000142738
|
pbld
|
phenazine biosynthesis like protein domain containing |
| chr8_-_22157301 | 0.03 |
ENSDART00000158383
|
nphp4
|
nephronophthisis 4 |
| chr25_-_35182347 | 0.03 |
ENSDART00000115210
|
ano9a
|
anoctamin 9a |
| chr16_+_21917673 | 0.03 |
ENSDART00000170604
|
znf687a
|
zinc finger protein 687a |
| chr10_+_24660225 | 0.03 |
ENSDART00000190695
|
vps36
|
vacuolar protein sorting 36 homolog (S. cerevisiae) |
| chr22_+_7923713 | 0.03 |
ENSDART00000167210
|
CABZ01034691.1
|
|
| chr5_+_20112032 | 0.03 |
ENSDART00000130554
|
isg15
|
ISG15 ubiquitin-like modifier |
| chr15_+_31816835 | 0.03 |
ENSDART00000189658
ENSDART00000186634 ENSDART00000193032 ENSDART00000180401 |
frya
|
furry homolog a (Drosophila) |
| chr3_+_45365098 | 0.02 |
ENSDART00000052746
ENSDART00000156555 |
ube2ia
|
ubiquitin-conjugating enzyme E2Ia |
| chr15_+_1199407 | 0.02 |
ENSDART00000163827
|
mfsd1
|
major facilitator superfamily domain containing 1 |
| chr1_+_44911405 | 0.02 |
ENSDART00000182465
|
wu:fc21g02
|
wu:fc21g02 |
| chr4_+_77920666 | 0.02 |
ENSDART00000129523
|
ttll1
|
tubulin tyrosine ligase-like family, member 1 |
| chr4_-_76027176 | 0.02 |
ENSDART00000165718
|
si:dkey-71l4.1
|
si:dkey-71l4.1 |
| chr1_+_7414318 | 0.02 |
ENSDART00000127426
|
si:ch73-383l1.1
|
si:ch73-383l1.1 |
| chr20_-_27086143 | 0.02 |
ENSDART00000008590
|
itpk1a
|
inositol-tetrakisphosphate 1-kinase a |
| chr21_+_21263988 | 0.02 |
ENSDART00000089651
ENSDART00000108978 |
ccdc61
|
coiled-coil domain containing 61 |
| chr1_-_40735381 | 0.01 |
ENSDART00000093269
|
zgc:153642
|
zgc:153642 |
| chr23_+_5490854 | 0.01 |
ENSDART00000175403
|
tulp1a
|
tubby like protein 1a |
| chr4_-_59159690 | 0.01 |
ENSDART00000164706
|
znf1149
|
zinc finger protein 1149 |
| chr22_-_7778265 | 0.01 |
ENSDART00000097276
|
si:ch73-44m9.3
|
si:ch73-44m9.3 |
| chr22_+_1821718 | 0.01 |
ENSDART00000132220
|
znf1002
|
zinc finger protein 1002 |
| chr16_-_51299061 | 0.01 |
ENSDART00000148677
|
serpinb1l4
|
serpin peptidase inhibitor, clade B (ovalbumin), member 1, like 4 |
| chr2_+_9061885 | 0.01 |
ENSDART00000028906
|
pigk
|
phosphatidylinositol glycan anchor biosynthesis, class K |
| chr13_+_18515126 | 0.01 |
ENSDART00000044697
|
tlr4ba
|
toll-like receptor 4b, duplicate a |
| chr7_+_51816321 | 0.01 |
ENSDART00000073846
|
si:ch211-122f10.4
|
si:ch211-122f10.4 |
| chr10_-_20524387 | 0.01 |
ENSDART00000159060
|
ddhd2
|
DDHD domain containing 2 |
| chr23_-_11128601 | 0.01 |
ENSDART00000131232
|
cntn3a.2
|
contactin 3a, tandem duplicate 2 |
| chr21_+_21612214 | 0.01 |
ENSDART00000008099
|
b9d2
|
B9 domain containing 2 |
| chr7_-_20836625 | 0.01 |
ENSDART00000192566
|
cldn15a
|
claudin 15a |
| chr4_-_68868229 | 0.01 |
ENSDART00000159219
|
si:dkey-264f17.5
|
si:dkey-264f17.5 |
| chr9_+_21819082 | 0.00 |
ENSDART00000136902
ENSDART00000101991 |
txndc9
|
thioredoxin domain containing 9 |
| chr19_+_46240171 | 0.00 |
ENSDART00000162785
|
mapk15
|
mitogen-activated protein kinase 15 |
| chr9_-_50000144 | 0.00 |
ENSDART00000123416
|
scn1a
|
sodium channel, voltage-gated, type I, alpha |
| chr10_-_20524592 | 0.00 |
ENSDART00000185048
|
ddhd2
|
DDHD domain containing 2 |
| chr1_-_17689858 | 0.00 |
ENSDART00000181078
|
cfap97
|
cilia and flagella associated protein 97 |
Network of associatons between targets according to the STRING database.
First level regulatory network of hbp1
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.7 | GO:1905072 | cardiac jelly development(GO:1905072) |
| 0.2 | 1.3 | GO:1902262 | apoptotic process involved in patterning of blood vessels(GO:1902262) |
| 0.1 | 0.5 | GO:2000389 | neutrophil extravasation(GO:0072672) regulation of neutrophil extravasation(GO:2000389) positive regulation of neutrophil extravasation(GO:2000391) |
| 0.1 | 0.9 | GO:0044854 | membrane raft assembly(GO:0001765) plasma membrane raft assembly(GO:0044854) plasma membrane raft organization(GO:0044857) caveola assembly(GO:0070836) |
| 0.1 | 0.3 | GO:0030224 | monocyte differentiation(GO:0030224) |
| 0.1 | 0.2 | GO:1905048 | regulation of metallopeptidase activity(GO:1905048) |
| 0.1 | 0.2 | GO:0045429 | regulation of nitric oxide biosynthetic process(GO:0045428) positive regulation of nitric oxide biosynthetic process(GO:0045429) positive regulation of reactive oxygen species biosynthetic process(GO:1903428) positive regulation of nitric oxide metabolic process(GO:1904407) |
| 0.1 | 0.8 | GO:0043462 | regulation of ATPase activity(GO:0043462) |
| 0.0 | 0.5 | GO:0050688 | regulation of defense response to virus(GO:0050688) |
| 0.0 | 0.3 | GO:0034389 | lipid particle organization(GO:0034389) |
| 0.0 | 0.2 | GO:0001778 | plasma membrane repair(GO:0001778) monocyte activation(GO:0042117) |
| 0.0 | 0.3 | GO:0071233 | response to leucine(GO:0043201) cellular response to leucine(GO:0071233) regulation of response to reactive oxygen species(GO:1901031) |
| 0.0 | 0.6 | GO:0006144 | purine nucleobase metabolic process(GO:0006144) |
| 0.0 | 0.1 | GO:0018160 | peptidyl-pyrromethane cofactor linkage(GO:0018160) |
| 0.0 | 0.1 | GO:1902766 | skeletal muscle satellite cell migration(GO:1902766) |
| 0.0 | 0.0 | GO:1903646 | regulation of protein folding(GO:1903332) positive regulation of protein folding(GO:1903334) regulation of chaperone-mediated protein folding(GO:1903644) positive regulation of chaperone-mediated protein folding(GO:1903646) |
| 0.0 | 0.1 | GO:0097374 | sensory neuron axon guidance(GO:0097374) |
| 0.0 | 0.2 | GO:0071385 | cellular response to corticosteroid stimulus(GO:0071384) cellular response to glucocorticoid stimulus(GO:0071385) |
| 0.0 | 0.0 | GO:0030213 | hyaluronan biosynthetic process(GO:0030213) |
| 0.0 | 0.4 | GO:0051014 | actin filament severing(GO:0051014) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.6 | GO:1902560 | GMP reductase complex(GO:1902560) |
| 0.0 | 0.3 | GO:0061700 | GATOR2 complex(GO:0061700) |
| 0.0 | 0.8 | GO:0033017 | sarcoplasmic reticulum membrane(GO:0033017) |
| 0.0 | 1.3 | GO:0016342 | catenin complex(GO:0016342) |
| 0.0 | 0.9 | GO:0044853 | caveola(GO:0005901) plasma membrane raft(GO:0044853) |
| 0.0 | 0.1 | GO:0008275 | gamma-tubulin small complex(GO:0008275) |
| 0.0 | 0.0 | GO:0072380 | TRC complex(GO:0072380) |
| 0.0 | 0.6 | GO:0031105 | septin ring(GO:0005940) septin complex(GO:0031105) septin cytoskeleton(GO:0032156) |
| 0.0 | 0.2 | GO:0043209 | myelin sheath(GO:0043209) |
| 0.0 | 0.1 | GO:0000814 | ESCRT II complex(GO:0000814) |
| 0.0 | 0.2 | GO:0030315 | T-tubule(GO:0030315) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.6 | GO:0003920 | GMP reductase activity(GO:0003920) oxidoreductase activity, acting on NAD(P)H, nitrogenous group as acceptor(GO:0016657) |
| 0.2 | 0.8 | GO:0042030 | ATPase inhibitor activity(GO:0042030) |
| 0.1 | 0.2 | GO:0005006 | epidermal growth factor-activated receptor activity(GO:0005006) |
| 0.0 | 0.3 | GO:0070728 | leucine binding(GO:0070728) |
| 0.0 | 0.9 | GO:0005520 | insulin-like growth factor binding(GO:0005520) |
| 0.0 | 0.1 | GO:0008597 | calcium-dependent protein serine/threonine phosphatase regulator activity(GO:0008597) |
| 0.0 | 0.2 | GO:0004994 | somatostatin receptor activity(GO:0004994) |
| 0.0 | 0.1 | GO:0004418 | hydroxymethylbilane synthase activity(GO:0004418) |
| 0.0 | 0.1 | GO:0051059 | NF-kappaB binding(GO:0051059) |
| 0.0 | 1.3 | GO:0045296 | cadherin binding(GO:0045296) |
| 0.0 | 0.3 | GO:0008253 | 5'-nucleotidase activity(GO:0008253) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.7 | PID AP1 PATHWAY | AP-1 transcription factor network |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.6 | REACTOME PURINE SALVAGE | Genes involved in Purine salvage |
| 0.1 | 1.3 | REACTOME ADHERENS JUNCTIONS INTERACTIONS | Genes involved in Adherens junctions interactions |
| 0.0 | 0.7 | REACTOME YAP1 AND WWTR1 TAZ STIMULATED GENE EXPRESSION | Genes involved in YAP1- and WWTR1 (TAZ)-stimulated gene expression |