Project
PRJNA207719: Tissue specific transcriptome profiling
Navigation
Downloads
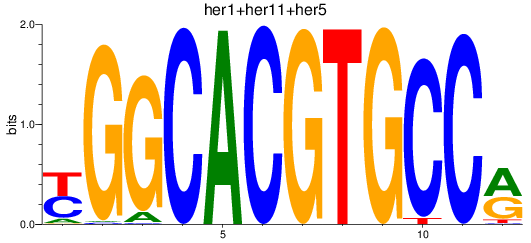
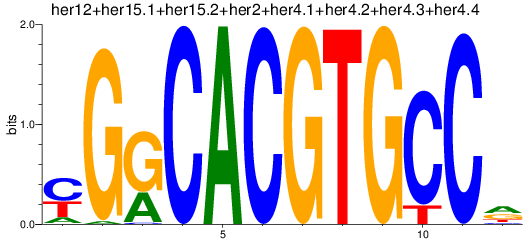
Results for her1+her11+her5_her12+her15.1+her15.2+her2+her4.1+her4.2+her4.3+her4.4
Z-value: 0.60
Transcription factors associated with her1+her11+her5_her12+her15.1+her15.2+her2+her4.1+her4.2+her4.3+her4.4
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
her11
|
ENSDARG00000002707 | hairy-related 11 |
|
her5
|
ENSDARG00000008796 | hairy-related 5 |
|
her1
|
ENSDARG00000014722 | hairy-related 1 |
|
her4.4
|
ENSDARG00000009822 | hairy-related 4, tandem duplicate 4 |
|
her12
|
ENSDARG00000032963 | hairy-related 12 |
|
her2
|
ENSDARG00000038205 | hairy-related 2 |
|
her15.2
|
ENSDARG00000054560 | hairy and enhancer of split-related 15, tandem duplicate 2 |
|
her15.1
|
ENSDARG00000054562 | hairy and enhancer of split-related 15, tandem duplicate 1 |
|
her4.2
|
ENSDARG00000056729 | hairy-related 4, tandem duplicate 2 |
|
her4.1
|
ENSDARG00000056732 | hairy-related 4, tandem duplicate 1 |
|
her4.3
|
ENSDARG00000070770 | hairy-related 4, tandem duplicate 3 |
|
her4.2
|
ENSDARG00000094426 | hairy-related 4, tandem duplicate 2 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| her4.3 | dr11_v1_chr23_+_21459263_21459263 | 0.96 | 1.0e-02 | Click! |
| her4.2 | dr11_v1_chr23_-_21471022_21471022 | 0.96 | 1.1e-02 | Click! |
| her4.4 | dr11_v1_chr23_-_21463788_21463788 | 0.95 | 1.1e-02 | Click! |
| her4.1 | dr11_v1_chr23_-_21453614_21453614 | 0.94 | 1.6e-02 | Click! |
| her12 | dr11_v1_chr23_-_21446985_21446985 | 0.82 | 8.7e-02 | Click! |
| her2 | dr11_v1_chr11_-_41966854_41966854 | 0.82 | 9.2e-02 | Click! |
| her15.1 | dr11_v1_chr11_+_41981959_41981959 | 0.64 | 2.5e-01 | Click! |
| her15.2 | dr11_v1_chr11_-_41996957_41996957 | 0.62 | 2.7e-01 | Click! |
| her11 | dr11_v1_chr14_-_30540967_30540967 | -0.59 | 3.0e-01 | Click! |
| her1 | dr11_v1_chr5_-_68795063_68795063 | 0.00 | 1.0e+00 | Click! |
Activity profile of her1+her11+her5_her12+her15.1+her15.2+her2+her4.1+her4.2+her4.3+her4.4 motif
Sorted Z-values of her1+her11+her5_her12+her15.1+her15.2+her2+her4.1+her4.2+her4.3+her4.4 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr6_-_60147517 | 1.82 |
ENSDART00000083453
|
slc32a1
|
solute carrier family 32 (GABA vesicular transporter), member 1 |
| chr7_-_40993456 | 1.00 |
ENSDART00000031700
|
en2a
|
engrailed homeobox 2a |
| chr9_-_44295071 | 0.93 |
ENSDART00000011837
|
neurod1
|
neuronal differentiation 1 |
| chr23_-_21463788 | 0.67 |
ENSDART00000079265
|
her4.4
|
hairy-related 4, tandem duplicate 4 |
| chr2_+_29976419 | 0.61 |
ENSDART00000056748
|
en2b
|
engrailed homeobox 2b |
| chr6_+_39184236 | 0.59 |
ENSDART00000156187
|
tac3b
|
tachykinin 3b |
| chr18_+_6126506 | 0.57 |
ENSDART00000125725
|
si:ch1073-390k14.1
|
si:ch1073-390k14.1 |
| chr16_-_45069882 | 0.46 |
ENSDART00000058384
|
gapdhs
|
glyceraldehyde-3-phosphate dehydrogenase, spermatogenic |
| chr8_+_21146262 | 0.45 |
ENSDART00000045684
|
porcn
|
porcupine O-acyltransferase |
| chr23_+_21459263 | 0.44 |
ENSDART00000104209
|
her4.3
|
hairy-related 4, tandem duplicate 3 |
| chr23_+_22819971 | 0.36 |
ENSDART00000111345
|
rerea
|
arginine-glutamic acid dipeptide (RE) repeats a |
| chr10_-_27566481 | 0.33 |
ENSDART00000078920
|
auts2a
|
autism susceptibility candidate 2a |
| chr15_-_14469704 | 0.32 |
ENSDART00000185077
|
numbl
|
numb homolog (Drosophila)-like |
| chr5_-_45894802 | 0.32 |
ENSDART00000097648
|
crfb6
|
cytokine receptor family member b6 |
| chr20_-_27733683 | 0.32 |
ENSDART00000103317
ENSDART00000138139 |
zgc:153157
|
zgc:153157 |
| chr12_+_33038757 | 0.27 |
ENSDART00000153146
|
rbfox3a
|
RNA binding fox-1 homolog 3a |
| chr19_+_42469058 | 0.26 |
ENSDART00000076915
|
si:dkey-166k12.1
|
si:dkey-166k12.1 |
| chr11_-_3959889 | 0.25 |
ENSDART00000159683
|
pbrm1
|
polybromo 1 |
| chr15_-_20731297 | 0.24 |
ENSDART00000114464
|
tpst1
|
tyrosylprotein sulfotransferase 1 |
| chr5_-_38342992 | 0.23 |
ENSDART00000140337
|
mink1
|
misshapen-like kinase 1 |
| chr7_+_32021669 | 0.23 |
ENSDART00000173976
|
mettl15
|
methyltransferase like 15 |
| chr20_+_19212962 | 0.20 |
ENSDART00000063706
|
fndc4a
|
fibronectin type III domain containing 4a |
| chr11_-_3959477 | 0.16 |
ENSDART00000045971
|
pbrm1
|
polybromo 1 |
| chr5_-_18897482 | 0.16 |
ENSDART00000010101
ENSDART00000099434 |
aacs
|
acetoacetyl-CoA synthetase |
| chr18_-_25855263 | 0.16 |
ENSDART00000042074
|
sema4ba
|
sema domain, immunoglobulin domain (Ig), transmembrane domain (TM) and short cytoplasmic domain, (semaphorin) 4Ba |
| chr5_-_46329880 | 0.15 |
ENSDART00000156577
|
si:ch211-130m23.5
|
si:ch211-130m23.5 |
| chr4_-_1818315 | 0.15 |
ENSDART00000067433
|
ube2nb
|
ubiquitin-conjugating enzyme E2Nb |
| chr8_+_39760258 | 0.12 |
ENSDART00000037914
|
cox6a1
|
cytochrome c oxidase subunit VIa polypeptide 1 |
| chr23_-_31969786 | 0.11 |
ENSDART00000134550
|
ormdl2
|
ORMDL sphingolipid biosynthesis regulator 2 |
| chr17_-_37195354 | 0.10 |
ENSDART00000190963
|
asxl2
|
additional sex combs like transcriptional regulator 2 |
| chr21_-_38730557 | 0.09 |
ENSDART00000150984
ENSDART00000111885 |
taf9
|
TAF9 RNA polymerase II, TATA box binding protein (TBP)-associated factor |
| chr17_-_37195163 | 0.09 |
ENSDART00000108514
|
asxl2
|
additional sex combs like transcriptional regulator 2 |
| chr9_+_45789887 | 0.09 |
ENSDART00000135202
|
si:dkey-34f9.3
|
si:dkey-34f9.3 |
| chr12_+_30046320 | 0.09 |
ENSDART00000179904
ENSDART00000153394 |
ablim1b
|
actin binding LIM protein 1b |
| chr3_+_32425202 | 0.08 |
ENSDART00000156464
|
prr12b
|
proline rich 12b |
| chr21_+_1586379 | 0.08 |
ENSDART00000150961
|
wdr91
|
WD repeat domain 91 |
| chr23_+_24989387 | 0.06 |
ENSDART00000172299
ENSDART00000145307 |
arhgap4a
|
Rho GTPase activating protein 4a |
| chr18_+_30370559 | 0.06 |
ENSDART00000165450
|
gse1
|
Gse1 coiled-coil protein |
| chr18_+_30370339 | 0.06 |
ENSDART00000158871
|
gse1
|
Gse1 coiled-coil protein |
| chr25_-_8625601 | 0.05 |
ENSDART00000155280
|
GDPGP1
|
zgc:153343 |
| chr1_+_5576151 | 0.03 |
ENSDART00000109756
|
cpo
|
carboxypeptidase O |
| chr4_-_8093753 | 0.02 |
ENSDART00000133434
|
wnk1b
|
WNK lysine deficient protein kinase 1b |
| chr19_-_35596207 | 0.01 |
ENSDART00000136811
|
col8a2
|
collagen, type VIII, alpha 2 |
| chr22_-_38951128 | 0.01 |
ENSDART00000085685
ENSDART00000104422 |
rbbp9
|
retinoblastoma binding protein 9 |
Network of associatons between targets according to the STRING database.
First level regulatory network of her1+her11+her5_her12+her15.1+her15.2+her2+her4.1+her4.2+her4.3+her4.4
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 1.8 | GO:0098700 | neurotransmitter loading into synaptic vesicle(GO:0098700) |
| 0.2 | 0.5 | GO:0061355 | Wnt protein secretion(GO:0061355) |
| 0.1 | 0.6 | GO:0007217 | tachykinin receptor signaling pathway(GO:0007217) |
| 0.1 | 1.0 | GO:0060385 | axonogenesis involved in innervation(GO:0060385) |
| 0.0 | 1.1 | GO:0048936 | peripheral nervous system neuron axonogenesis(GO:0048936) |
| 0.0 | 0.4 | GO:0031063 | regulation of histone deacetylation(GO:0031063) |
| 0.0 | 1.3 | GO:0045746 | negative regulation of Notch signaling pathway(GO:0045746) |
| 0.0 | 0.2 | GO:0006477 | protein sulfation(GO:0006477) peptidyl-tyrosine sulfation(GO:0006478) |
| 0.0 | 0.1 | GO:2000303 | regulation of sphingolipid biosynthetic process(GO:0090153) negative regulation of sphingolipid biosynthetic process(GO:0090155) negative regulation of ceramide biosynthetic process(GO:1900060) regulation of membrane lipid metabolic process(GO:1905038) regulation of ceramide biosynthetic process(GO:2000303) |
| 0.0 | 0.5 | GO:0043153 | entrainment of circadian clock by photoperiod(GO:0043153) |
| 0.0 | 0.1 | GO:0042769 | DNA damage response, detection of DNA damage(GO:0042769) |
| 0.0 | 0.2 | GO:0032024 | positive regulation of insulin secretion(GO:0032024) |
| 0.0 | 0.2 | GO:0070475 | rRNA base methylation(GO:0070475) |
| 0.0 | 0.3 | GO:0021884 | forebrain neuron development(GO:0021884) |
| 0.0 | 0.6 | GO:0030901 | midbrain development(GO:0030901) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 1.8 | GO:0030285 | integral component of synaptic vesicle membrane(GO:0030285) intrinsic component of synaptic vesicle membrane(GO:0098563) |
| 0.1 | 0.4 | GO:0016586 | RSC complex(GO:0016586) |
| 0.0 | 0.2 | GO:0035517 | PR-DUB complex(GO:0035517) |
| 0.0 | 0.1 | GO:0035339 | SPOTS complex(GO:0035339) |
| 0.0 | 0.1 | GO:0031371 | ubiquitin conjugating enzyme complex(GO:0031371) |
| 0.0 | 0.1 | GO:0031313 | extrinsic component of endosome membrane(GO:0031313) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 1.8 | GO:0015295 | solute:proton symporter activity(GO:0015295) |
| 0.1 | 0.5 | GO:0004365 | glyceraldehyde-3-phosphate dehydrogenase (NAD+) (phosphorylating) activity(GO:0004365) glyceraldehyde-3-phosphate dehydrogenase (NAD(P)+) (phosphorylating) activity(GO:0043891) |
| 0.1 | 0.2 | GO:0016434 | rRNA (cytosine) methyltransferase activity(GO:0016434) |
| 0.0 | 0.2 | GO:0042975 | peroxisome proliferator activated receptor binding(GO:0042975) |
| 0.0 | 0.2 | GO:0008476 | protein-tyrosine sulfotransferase activity(GO:0008476) |
| 0.0 | 0.5 | GO:0017147 | Wnt-protein binding(GO:0017147) |
| 0.0 | 0.3 | GO:0005246 | calcium channel regulator activity(GO:0005246) |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 1.8 | REACTOME GABA SYNTHESIS RELEASE REUPTAKE AND DEGRADATION | Genes involved in GABA synthesis, release, reuptake and degradation |
| 0.0 | 0.9 | REACTOME REGULATION OF GENE EXPRESSION IN BETA CELLS | Genes involved in Regulation of gene expression in beta cells |
| 0.0 | 0.5 | REACTOME GLYCOLYSIS | Genes involved in Glycolysis |