Project
PRJNA207719: Tissue specific transcriptome profiling
Navigation
Downloads
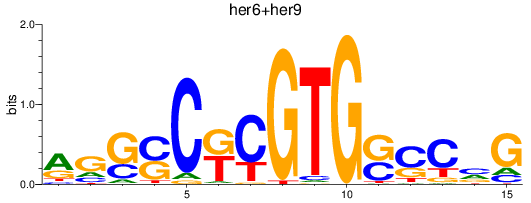
Results for her6+her9
Z-value: 0.36
Transcription factors associated with her6+her9
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
her6
|
ENSDARG00000006514 | hairy-related 6 |
|
her9
|
ENSDARG00000056438 | hairy-related 9 |
|
her6
|
ENSDARG00000111099 | hairy-related 6 |
|
her9
|
ENSDARG00000115775 | hairy-related 9 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| her6 | dr11_v1_chr6_-_36552844_36552844 | -0.38 | 5.3e-01 | Click! |
| her9 | dr11_v1_chr23_-_23401305_23401305 | -0.23 | 7.1e-01 | Click! |
Activity profile of her6+her9 motif
Sorted Z-values of her6+her9 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr2_+_68789 | 0.15 |
ENSDART00000058569
|
cldn1
|
claudin 1 |
| chr5_-_12560569 | 0.14 |
ENSDART00000133587
|
wsb2
|
WD repeat and SOCS box containing 2 |
| chr1_+_31942961 | 0.12 |
ENSDART00000007522
|
anos1a
|
anosmin 1a |
| chr5_-_29122834 | 0.11 |
ENSDART00000087197
|
whrnb
|
whirlin b |
| chr5_-_29122615 | 0.10 |
ENSDART00000144802
|
whrnb
|
whirlin b |
| chr18_+_3169579 | 0.09 |
ENSDART00000164724
ENSDART00000186340 ENSDART00000181247 ENSDART00000168056 |
pak1
|
p21 protein (Cdc42/Rac)-activated kinase 1 |
| chr4_-_77551860 | 0.08 |
ENSDART00000188176
|
AL935186.6
|
|
| chr17_-_3986236 | 0.08 |
ENSDART00000188794
ENSDART00000160830 |
PLCB1
|
si:ch1073-140o9.2 |
| chr13_+_7241170 | 0.07 |
ENSDART00000109434
|
aifm2
|
apoptosis-inducing factor, mitochondrion-associated, 2 |
| chr19_-_35450661 | 0.07 |
ENSDART00000113574
ENSDART00000136895 |
anln
|
anillin, actin binding protein |
| chr8_+_36509885 | 0.07 |
ENSDART00000109530
|
slc7a4
|
solute carrier family 7, member 4 |
| chr7_+_24528430 | 0.07 |
ENSDART00000133022
|
si:dkeyp-75h12.2
|
si:dkeyp-75h12.2 |
| chr15_-_12319065 | 0.07 |
ENSDART00000162973
ENSDART00000170543 |
fxyd6
|
FXYD domain containing ion transport regulator 6 |
| chr25_+_19954576 | 0.07 |
ENSDART00000149335
|
kcna1a
|
potassium voltage-gated channel, shaker-related subfamily, member 1a |
| chr7_+_529522 | 0.07 |
ENSDART00000190811
|
nrxn2b
|
neurexin 2b |
| chr21_-_44731865 | 0.07 |
ENSDART00000013814
|
ammecr1
|
Alport syndrome, mental retardation, midface hypoplasia and elliptocytosis chromosomal region gene 1 homolog (human) |
| chr8_-_39977026 | 0.06 |
ENSDART00000141707
|
asphd2
|
aspartate beta-hydroxylase domain containing 2 |
| chr19_-_15397946 | 0.06 |
ENSDART00000143480
|
hivep3a
|
human immunodeficiency virus type I enhancer binding protein 3a |
| chr24_-_33291784 | 0.06 |
ENSDART00000124938
|
si:ch1073-406l10.2
|
si:ch1073-406l10.2 |
| chr4_-_77563411 | 0.06 |
ENSDART00000186841
|
AL935186.8
|
|
| chr4_+_9669717 | 0.06 |
ENSDART00000004604
|
si:dkey-153k10.9
|
si:dkey-153k10.9 |
| chr13_+_35925490 | 0.06 |
ENSDART00000046115
|
mfsd2aa
|
major facilitator superfamily domain containing 2aa |
| chr15_-_1765098 | 0.06 |
ENSDART00000149980
ENSDART00000093074 |
bud23
|
BUD23, rRNA methyltransferase and ribosome maturation factor |
| chr9_+_500052 | 0.06 |
ENSDART00000166707
|
CU984600.1
|
|
| chr19_-_6385594 | 0.06 |
ENSDART00000104950
|
atp1a3a
|
ATPase Na+/K+ transporting subunit alpha 3a |
| chr10_-_2942900 | 0.05 |
ENSDART00000002622
|
oclna
|
occludin a |
| chr14_-_2189889 | 0.05 |
ENSDART00000181557
ENSDART00000106707 |
pcdh2ab9
pcdh2ab11
|
protocadherin 2 alpha b 9 protocadherin 2 alpha b 11 |
| chr2_+_58841181 | 0.05 |
ENSDART00000164102
|
cirbpa
|
cold inducible RNA binding protein a |
| chr20_-_8304271 | 0.05 |
ENSDART00000179057
|
dab1a
|
Dab, reelin signal transducer, homolog 1a (Drosophila) |
| chr3_-_29977495 | 0.05 |
ENSDART00000077111
|
hsd17b14
|
hydroxysteroid (17-beta) dehydrogenase 14 |
| chr14_-_30387894 | 0.05 |
ENSDART00000176136
|
slc7a2
|
solute carrier family 7 (cationic amino acid transporter, y+ system), member 2 |
| chr20_-_16498991 | 0.04 |
ENSDART00000104137
|
ches1
|
checkpoint suppressor 1 |
| chr18_+_41772474 | 0.04 |
ENSDART00000097547
|
trpc6b
|
transient receptor potential cation channel, subfamily C, member 6b |
| chr5_-_32890807 | 0.04 |
ENSDART00000007512
|
pole3
|
polymerase (DNA directed), epsilon 3 (p17 subunit) |
| chr5_-_54395488 | 0.04 |
ENSDART00000160781
|
zmynd19
|
zinc finger, MYND-type containing 19 |
| chr9_-_6624663 | 0.04 |
ENSDART00000092537
|
GPR45
|
si:dkeyp-118h3.5 |
| chr3_+_431208 | 0.04 |
ENSDART00000154296
ENSDART00000048733 |
si:ch73-308m11.1
si:dkey-167k11.5
|
si:ch73-308m11.1 si:dkey-167k11.5 |
| chr14_-_32520295 | 0.04 |
ENSDART00000165463
|
atp11c
|
ATPase phospholipid transporting 11C |
| chr11_-_12800945 | 0.04 |
ENSDART00000191178
|
txlng
|
taxilin gamma |
| chr3_-_11828206 | 0.03 |
ENSDART00000018159
|
si:ch211-262e15.1
|
si:ch211-262e15.1 |
| chr11_-_12801157 | 0.03 |
ENSDART00000103449
|
txlng
|
taxilin gamma |
| chr23_+_44732863 | 0.03 |
ENSDART00000160044
ENSDART00000172268 |
atp1b2a
|
ATPase Na+/K+ transporting subunit beta 2a |
| chr3_-_58823762 | 0.03 |
ENSDART00000101253
|
luc7l3
|
LUC7-like 3 pre-mRNA splicing factor |
| chr25_+_388258 | 0.03 |
ENSDART00000166834
|
rfx7b
|
regulatory factor X7b |
| chr2_-_58257624 | 0.03 |
ENSDART00000098940
|
foxl2b
|
forkhead box L2b |
| chr7_+_17938128 | 0.03 |
ENSDART00000141044
|
mta2
|
metastasis associated 1 family, member 2 |
| chr1_-_22756898 | 0.03 |
ENSDART00000158915
|
prom1b
|
prominin 1 b |
| chr1_+_49955869 | 0.03 |
ENSDART00000150517
|
gstcd
|
glutathione S-transferase, C-terminal domain containing |
| chr5_-_36916790 | 0.03 |
ENSDART00000143827
|
kptn
|
kaptin (actin binding protein) |
| chr20_-_6196989 | 0.02 |
ENSDART00000013343
|
b4galt6
|
UDP-Gal:betaGlcNAc beta 1,4- galactosyltransferase, polypeptide 6 |
| chr4_+_68562464 | 0.02 |
ENSDART00000192954
|
BX548011.4
|
|
| chr3_+_14641962 | 0.02 |
ENSDART00000091070
|
zgc:158403
|
zgc:158403 |
| chr13_-_45022301 | 0.02 |
ENSDART00000183589
ENSDART00000125633 ENSDART00000074787 |
khdrbs1a
|
KH domain containing, RNA binding, signal transduction associated 1a |
| chr1_+_37195919 | 0.02 |
ENSDART00000159684
ENSDART00000172742 ENSDART00000158395 |
dclk2a
|
doublecortin-like kinase 2a |
| chr25_-_21280584 | 0.02 |
ENSDART00000143644
|
tmem60
|
transmembrane protein 60 |
| chr17_-_20666616 | 0.02 |
ENSDART00000020167
|
slc16a9a
|
solute carrier family 16, member 9a |
| chr21_+_1647990 | 0.02 |
ENSDART00000148540
|
fech
|
ferrochelatase |
| chr13_-_45022527 | 0.02 |
ENSDART00000159021
|
khdrbs1a
|
KH domain containing, RNA binding, signal transduction associated 1a |
| chr1_-_52461322 | 0.02 |
ENSDART00000083836
|
si:ch211-217k17.7
|
si:ch211-217k17.7 |
| chr1_+_37196106 | 0.02 |
ENSDART00000008756
ENSDART00000157503 ENSDART00000162971 ENSDART00000191004 ENSDART00000078206 ENSDART00000045111 |
dclk2a
|
doublecortin-like kinase 2a |
| chr24_+_39537565 | 0.02 |
ENSDART00000140868
ENSDART00000145358 |
si:dkey-161j23.5
|
si:dkey-161j23.5 |
| chr14_+_12109535 | 0.01 |
ENSDART00000054620
ENSDART00000121913 |
kctd12b
|
potassium channel tetramerisation domain containing 12b |
| chr3_-_8765165 | 0.01 |
ENSDART00000191131
|
CABZ01058333.1
|
|
| chr6_-_40195510 | 0.01 |
ENSDART00000156156
|
col7a1
|
collagen, type VII, alpha 1 |
| chr20_-_5328637 | 0.01 |
ENSDART00000075974
|
ism2b
|
isthmin 2b |
| chr23_-_6765653 | 0.01 |
ENSDART00000192310
|
FP102169.1
|
|
| chr16_+_11029762 | 0.01 |
ENSDART00000091183
|
erfl3
|
Ets2 repressor factor like 3 |
| chr4_+_62576422 | 0.01 |
ENSDART00000180679
|
si:ch211-79g12.2
|
si:ch211-79g12.2 |
| chr9_-_43538328 | 0.01 |
ENSDART00000140526
|
znf385b
|
zinc finger protein 385B |
| chr7_-_6438544 | 0.00 |
ENSDART00000173180
|
zgc:165555
|
zgc:165555 |
| chr14_-_41075262 | 0.00 |
ENSDART00000180518
|
drp2
|
dystrophin related protein 2 |
| chr14_+_15064870 | 0.00 |
ENSDART00000167075
|
nkx1.2lb
|
NK1 transcription factor related 2-like,b |
| chr20_-_40766387 | 0.00 |
ENSDART00000061173
|
hsdl1
|
hydroxysteroid dehydrogenase like 1 |
| chr14_+_12110020 | 0.00 |
ENSDART00000192462
|
kctd12b
|
potassium channel tetramerisation domain containing 12b |
Network of associatons between targets according to the STRING database.
First level regulatory network of her6+her9
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | GO:0008356 | asymmetric cell division(GO:0008356) |
| 0.0 | 0.1 | GO:0003232 | bulbus arteriosus development(GO:0003232) |
| 0.0 | 0.1 | GO:0035633 | maintenance of blood-brain barrier(GO:0035633) |
| 0.0 | 0.1 | GO:0036265 | RNA (guanine-N7)-methylation(GO:0036265) |
| 0.0 | 0.2 | GO:0060088 | auditory receptor cell stereocilium organization(GO:0060088) |
| 0.0 | 0.0 | GO:0048496 | maintenance of organ identity(GO:0048496) |
| 0.0 | 0.0 | GO:0006272 | leading strand elongation(GO:0006272) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.2 | GO:0002141 | stereocilia coupling link(GO:0002139) stereocilia ankle link(GO:0002141) stereocilia ankle link complex(GO:0002142) stereocilium tip(GO:0032426) |
| 0.0 | 0.0 | GO:0008622 | epsilon DNA polymerase complex(GO:0008622) |
| 0.0 | 0.1 | GO:0005826 | actomyosin contractile ring(GO:0005826) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | GO:0016649 | electron-transferring-flavoprotein dehydrogenase activity(GO:0004174) oxidoreductase activity, acting on the CH-NH group of donors, quinone or similar compound as acceptor(GO:0016649) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | PID EPHA2 FWD PATHWAY | EPHA2 forward signaling |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | REACTOME TIGHT JUNCTION INTERACTIONS | Genes involved in Tight junction interactions |
| 0.0 | 0.1 | REACTOME CD28 DEPENDENT VAV1 PATHWAY | Genes involved in CD28 dependent Vav1 pathway |