Project
PRJNA207719: Tissue specific transcriptome profiling
Navigation
Downloads
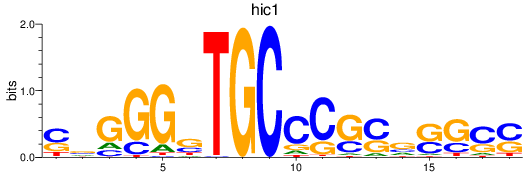
Results for hic1
Z-value: 0.85
Transcription factors associated with hic1
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
hic1
|
ENSDARG00000055493 | hypermethylated in cancer 1 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| hic1 | dr11_v1_chr15_+_25683069_25683069 | -0.91 | 3.1e-02 | Click! |
Activity profile of hic1 motif
Sorted Z-values of hic1 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr4_-_77557279 | 1.35 |
ENSDART00000180113
|
AL935186.10
|
|
| chr4_-_55728559 | 1.28 |
ENSDART00000186201
|
CT583728.14
|
|
| chr4_-_77561679 | 1.28 |
ENSDART00000180809
|
AL935186.9
|
|
| chr4_+_55778679 | 1.20 |
ENSDART00000183009
|
CT583728.19
|
|
| chr4_+_55794876 | 1.20 |
ENSDART00000189043
|
CT583728.17
|
|
| chr4_+_55810436 | 1.20 |
ENSDART00000182875
|
CT583728.16
|
|
| chr4_-_68568233 | 1.20 |
ENSDART00000184284
|
BX548011.1
|
|
| chr1_+_135903 | 0.85 |
ENSDART00000124837
|
f10
|
coagulation factor X |
| chr14_-_21064199 | 0.72 |
ENSDART00000172099
|
si:dkey-74k8.3
|
si:dkey-74k8.3 |
| chr14_-_21063977 | 0.68 |
ENSDART00000164373
|
si:dkey-74k8.3
|
si:dkey-74k8.3 |
| chr17_-_49412313 | 0.31 |
ENSDART00000152100
|
mthfd1b
|
methylenetetrahydrofolate dehydrogenase (NADP+ dependent) 1b |
| chr3_+_46762703 | 0.27 |
ENSDART00000133283
|
prkcsh
|
protein kinase C substrate 80K-H |
| chr10_+_23088 | 0.25 |
ENSDART00000022840
|
riox2
|
ribosomal oxygenase 2 |
| chr23_+_45845423 | 0.24 |
ENSDART00000183404
|
lmnl3
|
lamin L3 |
| chr5_+_62723233 | 0.22 |
ENSDART00000183718
|
nanos2
|
nanos homolog 2 |
| chr6_+_28203 | 0.22 |
ENSDART00000191561
|
CZQB01141835.1
|
|
| chr16_+_25285998 | 0.18 |
ENSDART00000154112
|
si:dkey-29h14.10
|
si:dkey-29h14.10 |
| chr9_+_7358749 | 0.17 |
ENSDART00000081660
|
ihha
|
Indian hedgehog homolog a |
| chr1_-_59571758 | 0.17 |
ENSDART00000193546
ENSDART00000167087 |
wfikkn1
|
WAP, follistatin/kazal, immunoglobulin, kunitz and netrin domain containing 1 |
| chr11_-_6420917 | 0.17 |
ENSDART00000193717
|
FO681393.2
|
|
| chr23_+_45845159 | 0.16 |
ENSDART00000023944
|
lmnl3
|
lamin L3 |
| chr23_-_31060350 | 0.16 |
ENSDART00000145598
ENSDART00000191491 |
si:ch211-197l9.5
|
si:ch211-197l9.5 |
| chr1_+_18863060 | 0.15 |
ENSDART00000139241
|
rnf38
|
ring finger protein 38 |
| chr2_-_49370042 | 0.15 |
ENSDART00000180515
|
CT583662.1
|
|
| chr10_-_21484065 | 0.14 |
ENSDART00000064545
|
fgf18b
|
fibroblast growth factor 18b |
| chr16_+_2565433 | 0.13 |
ENSDART00000188014
ENSDART00000171378 |
preb
|
prolactin regulatory element binding |
| chr3_+_23721808 | 0.11 |
ENSDART00000012470
|
hoxb4a
|
homeobox B4a |
| chr7_-_24995631 | 0.11 |
ENSDART00000173955
ENSDART00000173791 |
rcor2
|
REST corepressor 2 |
| chr10_-_9192450 | 0.10 |
ENSDART00000139783
|
si:dkeyp-41f9.4
|
si:dkeyp-41f9.4 |
| chr19_-_12967986 | 0.08 |
ENSDART00000151064
|
slc25a32a
|
solute carrier family 25 (mitochondrial folate carrier), member 32a |
| chr2_+_21063660 | 0.08 |
ENSDART00000022765
|
riok1
|
RIO kinase 1 (yeast) |
| chr19_+_17642356 | 0.08 |
ENSDART00000176431
|
CR382334.1
|
|
| chr19_-_22488952 | 0.05 |
ENSDART00000179856
ENSDART00000141503 |
pleca
|
plectin a |
| chr13_-_37519774 | 0.05 |
ENSDART00000141420
ENSDART00000185478 |
sgpp1
|
sphingosine-1-phosphate phosphatase 1 |
| chr4_-_68831753 | 0.04 |
ENSDART00000189170
|
si:dkey-264f17.1
|
si:dkey-264f17.1 |
| chr21_+_13389499 | 0.04 |
ENSDART00000151268
|
zgc:113162
|
zgc:113162 |
| chr8_-_13678415 | 0.04 |
ENSDART00000134153
ENSDART00000143331 |
si:dkey-258f14.3
|
si:dkey-258f14.3 |
| chr23_+_14590767 | 0.03 |
ENSDART00000143675
|
si:rp71-79p20.2
|
si:rp71-79p20.2 |
| chr3_-_61592417 | 0.03 |
ENSDART00000155082
|
nptx2a
|
neuronal pentraxin 2a |
| chr17_+_132555 | 0.01 |
ENSDART00000158159
|
zgc:77287
|
zgc:77287 |
| chr19_-_3574060 | 0.01 |
ENSDART00000105120
|
tmem170b
|
transmembrane protein 170B |
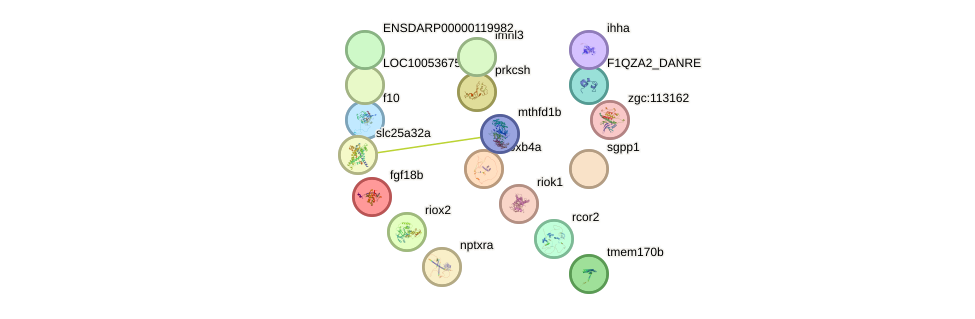
Network of associatons between targets according to the STRING database.
First level regulatory network of hic1
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 1.4 | GO:0071480 | response to gamma radiation(GO:0010332) cellular response to gamma radiation(GO:0071480) |
| 0.1 | 0.3 | GO:0070544 | histone H3-K36 demethylation(GO:0070544) |
| 0.0 | 0.1 | GO:0090113 | regulation of COPII vesicle coating(GO:0003400) regulation of ER to Golgi vesicle-mediated transport by GTP hydrolysis(GO:0090113) |
| 0.0 | 0.2 | GO:0048795 | swim bladder morphogenesis(GO:0048795) |
| 0.0 | 0.3 | GO:0035999 | tetrahydrofolate interconversion(GO:0035999) |
| 0.0 | 0.3 | GO:0006491 | N-glycan processing(GO:0006491) |
| 0.0 | 0.8 | GO:0007596 | blood coagulation(GO:0007596) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.3 | GO:0017177 | glucosidase II complex(GO:0017177) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.3 | GO:0004329 | formate-tetrahydrofolate ligase activity(GO:0004329) |
| 0.0 | 0.1 | GO:0008517 | folic acid transporter activity(GO:0008517) |
| 0.0 | 0.2 | GO:0005113 | patched binding(GO:0005113) |
| 0.0 | 0.3 | GO:0051864 | histone demethylase activity (H3-K36 specific)(GO:0051864) |
| 0.0 | 0.1 | GO:0005105 | type 1 fibroblast growth factor receptor binding(GO:0005105) |
| 0.0 | 0.2 | GO:0008191 | metalloendopeptidase inhibitor activity(GO:0008191) |
| 0.0 | 0.0 | GO:0042392 | sphingosine-1-phosphate phosphatase activity(GO:0042392) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.8 | PID INTEGRIN2 PATHWAY | Beta2 integrin cell surface interactions |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.8 | REACTOME GAMMA CARBOXYLATION TRANSPORT AND AMINO TERMINAL CLEAVAGE OF PROTEINS | Genes involved in Gamma-carboxylation, transport, and amino-terminal cleavage of proteins |
| 0.0 | 0.3 | REACTOME ADVANCED GLYCOSYLATION ENDPRODUCT RECEPTOR SIGNALING | Genes involved in Advanced glycosylation endproduct receptor signaling |