Project
PRJNA207719: Tissue specific transcriptome profiling
Navigation
Downloads
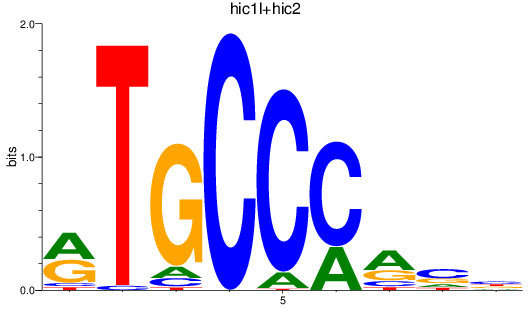
Results for hic1l+hic2
Z-value: 0.63
Transcription factors associated with hic1l+hic2
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
hic1l
|
ENSDARG00000045660 | hypermethylated in cancer 1 like |
|
hic2
|
ENSDARG00000100497 | hypermethylated in cancer 2 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| hic2 | dr11_v1_chr10_+_3153973_3153973 | -0.76 | 1.4e-01 | Click! |
| hic1l | dr11_v1_chr25_-_25575241_25575241 | -0.59 | 2.9e-01 | Click! |
Activity profile of hic1l+hic2 motif
Sorted Z-values of hic1l+hic2 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr22_-_25612680 | 0.78 |
ENSDART00000114167
|
si:ch211-12h2.8
|
si:ch211-12h2.8 |
| chr14_-_21064199 | 0.52 |
ENSDART00000172099
|
si:dkey-74k8.3
|
si:dkey-74k8.3 |
| chr19_+_14109348 | 0.43 |
ENSDART00000159015
|
zgc:175136
|
zgc:175136 |
| chr12_-_22524388 | 0.42 |
ENSDART00000020942
|
shbg
|
sex hormone-binding globulin |
| chr7_+_29952719 | 0.42 |
ENSDART00000173737
|
tpma
|
alpha-tropomyosin |
| chr7_+_29952169 | 0.41 |
ENSDART00000173540
ENSDART00000173940 ENSDART00000173906 ENSDART00000173772 ENSDART00000173506 ENSDART00000039657 |
tpma
|
alpha-tropomyosin |
| chr21_+_25765734 | 0.40 |
ENSDART00000021664
|
cldnb
|
claudin b |
| chr2_-_37883256 | 0.32 |
ENSDART00000035685
|
hbl4
|
hexose-binding lectin 4 |
| chr17_+_53294228 | 0.31 |
ENSDART00000158172
|
si:ch1073-416d2.3
|
si:ch1073-416d2.3 |
| chr14_-_25444774 | 0.29 |
ENSDART00000183448
|
slc26a2
|
solute carrier family 26 (anion exchanger), member 2 |
| chr2_-_41942666 | 0.28 |
ENSDART00000075673
|
ebi3
|
Epstein-Barr virus induced 3 |
| chr25_-_31423493 | 0.28 |
ENSDART00000027661
|
myod1
|
myogenic differentiation 1 |
| chr4_-_77551860 | 0.26 |
ENSDART00000188176
|
AL935186.6
|
|
| chr2_+_24936766 | 0.26 |
ENSDART00000025962
|
gyg1a
|
glycogenin 1a |
| chr10_+_15608326 | 0.25 |
ENSDART00000188770
|
zfand5b
|
zinc finger, AN1-type domain 5b |
| chr2_-_44255537 | 0.25 |
ENSDART00000011188
ENSDART00000093298 |
atp1a2a
|
ATPase Na+/K+ transporting subunit alpha 2 |
| chr8_+_15254564 | 0.24 |
ENSDART00000024433
|
slc5a9
|
solute carrier family 5 (sodium/sugar cotransporter), member 9 |
| chr24_-_9989634 | 0.23 |
ENSDART00000115275
|
zgc:152652
|
zgc:152652 |
| chr16_+_35594670 | 0.22 |
ENSDART00000163275
|
si:ch211-1i11.3
|
si:ch211-1i11.3 |
| chr1_-_55810730 | 0.20 |
ENSDART00000100551
|
zgc:136908
|
zgc:136908 |
| chr14_-_21063977 | 0.20 |
ENSDART00000164373
|
si:dkey-74k8.3
|
si:dkey-74k8.3 |
| chr24_-_29822913 | 0.19 |
ENSDART00000160929
|
aglb
|
amylo-alpha-1, 6-glucosidase, 4-alpha-glucanotransferase b |
| chr15_+_25158104 | 0.17 |
ENSDART00000128267
|
slc35f2l
|
info solute carrier family 35, member F2, like |
| chr25_+_16915974 | 0.17 |
ENSDART00000188923
|
zgc:77158
|
zgc:77158 |
| chr3_+_23692462 | 0.16 |
ENSDART00000145934
|
hoxb7a
|
homeobox B7a |
| chr19_-_13962469 | 0.16 |
ENSDART00000159062
|
paqr7a
|
progestin and adipoQ receptor family member VII, a |
| chr6_+_40723554 | 0.16 |
ENSDART00000103833
|
slc26a6l
|
solute carrier family 26, member 6, like |
| chr24_+_34806822 | 0.16 |
ENSDART00000148407
ENSDART00000188328 |
mchr2
|
melanin-concentrating hormone receptor 2 |
| chr21_+_5257018 | 0.16 |
ENSDART00000183100
ENSDART00000191525 |
loxhd1a
|
lipoxygenase homology domains 1a |
| chr2_-_14798295 | 0.16 |
ENSDART00000143430
ENSDART00000145869 |
si:ch73-366i20.1
|
si:ch73-366i20.1 |
| chr8_-_38616712 | 0.16 |
ENSDART00000141827
|
si:ch211-198d23.1
|
si:ch211-198d23.1 |
| chr15_-_29348212 | 0.15 |
ENSDART00000133117
|
tsku
|
tsukushi small leucine rich proteoglycan homolog (Xenopus laevis) |
| chr11_+_12879635 | 0.15 |
ENSDART00000182515
ENSDART00000081296 |
si:dkey-11m19.5
|
si:dkey-11m19.5 |
| chr6_+_8630355 | 0.15 |
ENSDART00000161749
ENSDART00000193976 |
tspeara
|
thrombospondin-type laminin G domain and EAR repeats a |
| chr18_-_16123222 | 0.14 |
ENSDART00000061189
|
sspn
|
sarcospan (Kras oncogene-associated gene) |
| chr13_-_7573670 | 0.14 |
ENSDART00000102538
|
pitx3
|
paired-like homeodomain 3 |
| chr5_-_57641257 | 0.14 |
ENSDART00000149282
|
hspb2
|
heat shock protein, alpha-crystallin-related, b2 |
| chr6_-_49634787 | 0.14 |
ENSDART00000188538
|
BX323459.1
|
|
| chr16_+_12836143 | 0.14 |
ENSDART00000067741
|
cacng6b
|
calcium channel, voltage-dependent, gamma subunit 6b |
| chr23_-_27875140 | 0.14 |
ENSDART00000143662
|
ankrd33aa
|
ankyrin repeat domain 33Aa |
| chr4_-_25485404 | 0.13 |
ENSDART00000041402
|
pfkfb3
|
6-phosphofructo-2-kinase/fructose-2,6-biphosphatase 3 |
| chr20_+_51466046 | 0.13 |
ENSDART00000178065
|
tlr5b
|
toll-like receptor 5b |
| chr23_-_1144425 | 0.13 |
ENSDART00000098271
|
psmb11b
|
proteasome subunit beta11b |
| chr7_+_35268880 | 0.13 |
ENSDART00000182231
|
dpep2
|
dipeptidase 2 |
| chr18_+_45666489 | 0.13 |
ENSDART00000180147
ENSDART00000151351 |
prrg4
|
proline rich Gla (G-carboxyglutamic acid) 4 (transmembrane) |
| chr5_-_63438717 | 0.12 |
ENSDART00000186899
|
cntrl
|
centriolin |
| chr16_+_35595312 | 0.12 |
ENSDART00000170438
|
si:ch211-1i11.3
|
si:ch211-1i11.3 |
| chr6_-_27108844 | 0.12 |
ENSDART00000073883
|
dtymk
|
deoxythymidylate kinase (thymidylate kinase) |
| chr23_-_1587955 | 0.12 |
ENSDART00000136037
|
fndc7b
|
fibronectin type III domain containing 7b |
| chr23_-_2037566 | 0.12 |
ENSDART00000191312
ENSDART00000127443 |
prdm5
|
PR domain containing 5 |
| chr22_-_24285432 | 0.12 |
ENSDART00000164083
|
si:ch211-117l17.4
|
si:ch211-117l17.4 |
| chr25_+_11110380 | 0.12 |
ENSDART00000164218
|
AKAP13
|
si:dkey-187e18.1 |
| chr10_+_41199660 | 0.12 |
ENSDART00000125314
|
adrb3b
|
adrenoceptor beta 3b |
| chr19_-_24958393 | 0.12 |
ENSDART00000098592
|
si:ch211-195b13.6
|
si:ch211-195b13.6 |
| chr2_-_23600821 | 0.11 |
ENSDART00000146217
|
BX677666.1
|
|
| chr23_+_4324625 | 0.11 |
ENSDART00000146302
ENSDART00000136792 ENSDART00000135027 ENSDART00000179819 |
sgk2a
|
serum/glucocorticoid regulated kinase 2a |
| chr16_-_25606889 | 0.11 |
ENSDART00000077447
ENSDART00000131528 |
zgc:110410
|
zgc:110410 |
| chr12_-_44010532 | 0.11 |
ENSDART00000183875
|
si:ch211-182p11.1
|
si:ch211-182p11.1 |
| chr2_+_17181777 | 0.11 |
ENSDART00000112063
|
ptger4c
|
prostaglandin E receptor 4 (subtype EP4) c |
| chr8_+_38417461 | 0.11 |
ENSDART00000132718
|
nkx6.3
|
NK6 homeobox 3 |
| chr13_-_9061944 | 0.11 |
ENSDART00000164186
ENSDART00000102051 |
si:dkey-112g5.12
|
si:dkey-112g5.12 |
| chr3_-_61592417 | 0.11 |
ENSDART00000155082
|
nptx2a
|
neuronal pentraxin 2a |
| chr14_+_6159162 | 0.11 |
ENSDART00000128638
|
bscl2l
|
Bernardinelli-Seip congenital lipodystrophy 2, like |
| chr11_-_16021424 | 0.11 |
ENSDART00000193291
ENSDART00000170731 ENSDART00000104107 |
zgc:173544
|
zgc:173544 |
| chr23_-_31060350 | 0.11 |
ENSDART00000145598
ENSDART00000191491 |
si:ch211-197l9.5
|
si:ch211-197l9.5 |
| chr7_+_22981909 | 0.10 |
ENSDART00000122449
|
ccnb3
|
cyclin B3 |
| chr3_-_32170850 | 0.10 |
ENSDART00000055307
ENSDART00000157366 |
tnnt1
|
troponin T type 1 (skeletal, slow) |
| chr16_-_25606235 | 0.10 |
ENSDART00000192741
|
zgc:110410
|
zgc:110410 |
| chr22_+_19262161 | 0.10 |
ENSDART00000163654
|
si:dkey-21e2.12
|
si:dkey-21e2.12 |
| chr4_-_8030583 | 0.10 |
ENSDART00000113628
|
si:ch211-240l19.8
|
si:ch211-240l19.8 |
| chr18_+_20677090 | 0.10 |
ENSDART00000060243
|
cftr
|
cystic fibrosis transmembrane conductance regulator (ATP-binding cassette sub-family C, member 7) |
| chr23_+_27740788 | 0.10 |
ENSDART00000053871
|
dhh
|
desert hedgehog |
| chr7_+_22982201 | 0.09 |
ENSDART00000134116
|
ccnb3
|
cyclin B3 |
| chr12_-_4243268 | 0.09 |
ENSDART00000131275
|
zgc:92313
|
zgc:92313 |
| chr22_+_29009541 | 0.09 |
ENSDART00000169449
|
pimr97
|
Pim proto-oncogene, serine/threonine kinase, related 97 |
| chr2_-_14793343 | 0.09 |
ENSDART00000132264
|
si:ch73-366i20.1
|
si:ch73-366i20.1 |
| chr19_+_23298037 | 0.09 |
ENSDART00000183948
|
irgf1
|
immunity-related GTPase family, f1 |
| chr22_-_5006119 | 0.09 |
ENSDART00000187998
|
rx1
|
retinal homeobox gene 1 |
| chr15_-_37779978 | 0.09 |
ENSDART00000157202
|
si:dkey-42l23.3
|
si:dkey-42l23.3 |
| chr16_-_41840668 | 0.09 |
ENSDART00000146150
|
si:dkey-199f5.4
|
si:dkey-199f5.4 |
| chr1_+_44992207 | 0.09 |
ENSDART00000172357
|
tmem132a
|
transmembrane protein 132A |
| chr8_+_46536893 | 0.09 |
ENSDART00000124023
|
pimr187
|
Pim proto-oncogene, serine/threonine kinase, related 187 |
| chr2_+_30547018 | 0.09 |
ENSDART00000193747
|
ankrd33bb
|
ankyrin repeat domain 33Bb |
| chr22_+_7742211 | 0.09 |
ENSDART00000140896
|
CELA1 (1 of many)
|
zgc:92511 |
| chr14_-_897874 | 0.09 |
ENSDART00000167395
|
rgs14a
|
regulator of G protein signaling 14a |
| chr5_-_30984271 | 0.09 |
ENSDART00000051392
|
spns3
|
spinster homolog 3 (Drosophila) |
| chr1_+_30723380 | 0.08 |
ENSDART00000127943
ENSDART00000062628 ENSDART00000127670 |
bora
|
bora, aurora kinase A activator |
| chr15_-_37425468 | 0.08 |
ENSDART00000059630
|
si:ch211-113j13.2
|
si:ch211-113j13.2 |
| chr5_+_32815745 | 0.08 |
ENSDART00000181535
|
crata
|
carnitine O-acetyltransferase a |
| chr9_-_41817712 | 0.08 |
ENSDART00000182657
|
si:dkeyp-30e7.2
|
si:dkeyp-30e7.2 |
| chr1_+_32791920 | 0.08 |
ENSDART00000109369
|
zgc:174320
|
zgc:174320 |
| chr1_+_30723677 | 0.08 |
ENSDART00000177900
|
bora
|
bora, aurora kinase A activator |
| chr18_+_33100606 | 0.07 |
ENSDART00000003907
|
si:ch211-229c8.13
|
si:ch211-229c8.13 |
| chr21_+_18877130 | 0.07 |
ENSDART00000136893
|
si:dkey-65l23.2
|
si:dkey-65l23.2 |
| chr23_-_32404022 | 0.07 |
ENSDART00000156387
ENSDART00000155508 |
si:ch211-66i15.4
|
si:ch211-66i15.4 |
| chr19_+_17642356 | 0.07 |
ENSDART00000176431
|
CR382334.1
|
|
| chr22_+_7497319 | 0.07 |
ENSDART00000034564
|
CELA1 (1 of many)
|
zgc:92511 |
| chr4_+_74396786 | 0.07 |
ENSDART00000127501
ENSDART00000174347 |
zmp:0000001020
|
zmp:0000001020 |
| chr21_-_22681534 | 0.07 |
ENSDART00000159233
|
gig2f
|
grass carp reovirus (GCRV)-induced gene 2f |
| chr3_+_23221047 | 0.07 |
ENSDART00000009393
|
col1a1a
|
collagen, type I, alpha 1a |
| chr7_+_26172396 | 0.07 |
ENSDART00000173975
|
si:ch211-196f2.7
|
si:ch211-196f2.7 |
| chr5_-_26181863 | 0.07 |
ENSDART00000098500
|
ccdc125
|
coiled-coil domain containing 125 |
| chr21_-_36619599 | 0.07 |
ENSDART00000065208
|
nop16
|
NOP16 nucleolar protein homolog (yeast) |
| chr24_-_39858710 | 0.07 |
ENSDART00000134251
|
slc12a7b
|
solute carrier family 12 (potassium/chloride transporter), member 7b |
| chr21_+_25375172 | 0.07 |
ENSDART00000078748
|
or132-5
|
odorant receptor, family H, subfamily 132, member 5 |
| chr2_+_11206317 | 0.07 |
ENSDART00000144982
|
lhx8a
|
LIM homeobox 8a |
| chr12_+_33894665 | 0.07 |
ENSDART00000004769
|
mfsd13a
|
major facilitator superfamily domain containing 13A |
| chr14_-_5677979 | 0.06 |
ENSDART00000182156
|
tlx2
|
T cell leukemia homeobox 2 |
| chr23_-_6765653 | 0.06 |
ENSDART00000192310
|
FP102169.1
|
|
| chr12_+_33894396 | 0.06 |
ENSDART00000130853
ENSDART00000152988 |
mfsd13a
|
major facilitator superfamily domain containing 13A |
| chr8_-_53548145 | 0.06 |
ENSDART00000180665
|
parapinopsina
|
parapinopsin a |
| chr25_+_31222069 | 0.06 |
ENSDART00000159373
|
tnni2a.1
|
troponin I type 2a (skeletal, fast), tandem duplicate 1 |
| chr4_+_54519511 | 0.06 |
ENSDART00000161653
|
znf974
|
zinc finger protein 974 |
| chr3_-_1387292 | 0.06 |
ENSDART00000163535
|
ddx47
|
DEAD (Asp-Glu-Ala-Asp) box polypeptide 47 |
| chr7_+_16963091 | 0.06 |
ENSDART00000173770
|
nav2a
|
neuron navigator 2a |
| chr4_-_8014463 | 0.06 |
ENSDART00000036153
|
ccdc3a
|
coiled-coil domain containing 3a |
| chr8_-_7475917 | 0.06 |
ENSDART00000082157
|
gata1b
|
GATA binding protein 1b |
| chr8_-_14144707 | 0.06 |
ENSDART00000148061
|
si:dkey-6n6.1
|
si:dkey-6n6.1 |
| chr21_-_31210749 | 0.06 |
ENSDART00000185356
|
zgc:152891
|
zgc:152891 |
| chr5_-_25733745 | 0.06 |
ENSDART00000051566
|
zgc:101016
|
zgc:101016 |
| chr5_-_67145505 | 0.06 |
ENSDART00000011295
|
rom1a
|
retinal outer segment membrane protein 1a |
| chr7_-_38689562 | 0.06 |
ENSDART00000167209
|
aplnr2
|
apelin receptor 2 |
| chr17_+_14886828 | 0.06 |
ENSDART00000010507
ENSDART00000131052 |
ptger2a
|
prostaglandin E receptor 2a (subtype EP2) |
| chr8_+_18101608 | 0.06 |
ENSDART00000140941
|
glis1b
|
GLIS family zinc finger 1b |
| chr5_+_39537195 | 0.05 |
ENSDART00000051256
|
fgf5
|
fibroblast growth factor 5 |
| chr9_+_38737924 | 0.05 |
ENSDART00000147652
|
kansl1l
|
KAT8 regulatory NSL complex subunit 1-like |
| chr22_-_9618867 | 0.05 |
ENSDART00000131246
|
si:ch73-286h23.4
|
si:ch73-286h23.4 |
| chr23_+_11285662 | 0.05 |
ENSDART00000111028
|
chl1a
|
cell adhesion molecule L1-like a |
| chr5_-_54148271 | 0.05 |
ENSDART00000163034
|
arhgef15
|
Rho guanine nucleotide exchange factor (GEF) 15 |
| chr6_+_18544791 | 0.05 |
ENSDART00000167463
ENSDART00000169599 |
atad5b
|
ATPase family, AAA domain containing 5b |
| chr10_-_12650259 | 0.05 |
ENSDART00000191567
|
CT990561.2
|
|
| chr3_-_46403778 | 0.05 |
ENSDART00000074422
|
cdip1
|
cell death-inducing p53 target 1 |
| chr15_+_31471808 | 0.05 |
ENSDART00000110078
|
or102-3
|
odorant receptor, family C, subfamily 102, member 3 |
| chr7_+_26172682 | 0.05 |
ENSDART00000173631
|
si:ch211-196f2.7
|
si:ch211-196f2.7 |
| chr25_+_36292465 | 0.05 |
ENSDART00000152649
|
bmb
|
brambleberry |
| chr21_+_13244450 | 0.05 |
ENSDART00000146062
|
specc1lb
|
sperm antigen with calponin homology and coiled-coil domains 1-like b |
| chr9_+_29994439 | 0.05 |
ENSDART00000012447
|
tmem30c
|
transmembrane protein 30C |
| chr17_+_34215886 | 0.05 |
ENSDART00000186775
|
mpp5a
|
membrane protein, palmitoylated 5a (MAGUK p55 subfamily member 5) |
| chr22_+_5123479 | 0.05 |
ENSDART00000111822
ENSDART00000081910 |
loxl5a
|
lysyl oxidase-like 5a |
| chr12_-_44151296 | 0.05 |
ENSDART00000168734
|
si:ch73-329n5.3
|
si:ch73-329n5.3 |
| chr16_-_49846359 | 0.05 |
ENSDART00000184246
|
znf385d
|
zinc finger protein 385D |
| chr1_-_34685007 | 0.05 |
ENSDART00000157471
|
klf5a
|
Kruppel-like factor 5a |
| chr8_-_1219815 | 0.05 |
ENSDART00000016800
ENSDART00000149969 |
znf367
|
zinc finger protein 367 |
| chr13_+_39297802 | 0.04 |
ENSDART00000133636
|
si:dkey-85a20.4
|
si:dkey-85a20.4 |
| chr15_-_38202630 | 0.04 |
ENSDART00000183772
|
rhoga
|
ras homolog family member Ga |
| chr11_+_20057182 | 0.04 |
ENSDART00000184979
|
fezf2
|
FEZ family zinc finger 2 |
| chr2_-_37889111 | 0.04 |
ENSDART00000168939
ENSDART00000098529 |
mbl2
|
mannose binding lectin 2 |
| chr16_-_41807397 | 0.04 |
ENSDART00000138577
|
si:dkey-199f5.7
|
si:dkey-199f5.7 |
| chr25_-_27541621 | 0.04 |
ENSDART00000130678
|
spam1
|
sperm adhesion molecule 1 |
| chr18_+_41560822 | 0.04 |
ENSDART00000158503
|
baz1b
|
bromodomain adjacent to zinc finger domain, 1B |
| chr12_+_5251647 | 0.04 |
ENSDART00000124097
|
plce1
|
phospholipase C, epsilon 1 |
| chr14_+_50807558 | 0.04 |
ENSDART00000183649
|
si:ch211-59p23.1
|
si:ch211-59p23.1 |
| chr1_-_54425791 | 0.04 |
ENSDART00000039911
|
pkd1a
|
polycystic kidney disease 1a |
| chr3_+_24207243 | 0.03 |
ENSDART00000023454
ENSDART00000136400 |
adsl
|
adenylosuccinate lyase |
| chr3_+_13830599 | 0.03 |
ENSDART00000159282
|
ptger1b
|
prostaglandin E receptor 1b (subtype EP1) |
| chr5_-_10236599 | 0.03 |
ENSDART00000099834
|
si:ch73-42k18.1
|
si:ch73-42k18.1 |
| chr6_-_426041 | 0.03 |
ENSDART00000162789
|
fam83fb
|
family with sequence similarity 83, member Fb |
| chr12_-_20362041 | 0.03 |
ENSDART00000184145
ENSDART00000105952 |
aqp8a.2
|
aquaporin 8a, tandem duplicate 2 |
| chr17_+_32360673 | 0.03 |
ENSDART00000155519
|
si:ch211-139d20.3
|
si:ch211-139d20.3 |
| chr2_+_30531726 | 0.03 |
ENSDART00000146518
|
ankrd33bb
|
ankyrin repeat domain 33Bb |
| chr15_-_28082310 | 0.03 |
ENSDART00000152620
|
dhrs13a.3
|
dehydrogenase/reductase (SDR family) member 13a, duplicate 3 |
| chr3_-_11828206 | 0.03 |
ENSDART00000018159
|
si:ch211-262e15.1
|
si:ch211-262e15.1 |
| chr20_+_48543492 | 0.03 |
ENSDART00000175480
|
LO018254.1
|
|
| chr10_-_22828302 | 0.03 |
ENSDART00000079459
ENSDART00000100468 |
per1a
|
period circadian clock 1a |
| chr13_+_21674445 | 0.03 |
ENSDART00000100931
ENSDART00000078567 |
mtg1
|
mitochondrial ribosome-associated GTPase 1 |
| chr13_-_14926318 | 0.03 |
ENSDART00000142785
|
cdc25b
|
cell division cycle 25B |
| chr23_+_2669 | 0.03 |
ENSDART00000011146
|
twist3
|
twist3 |
| chr18_-_50845804 | 0.03 |
ENSDART00000158517
|
PDPR
|
si:cabz01113374.3 |
| chr7_-_22981796 | 0.03 |
ENSDART00000167565
|
si:dkey-171c9.3
|
si:dkey-171c9.3 |
| chr15_-_17138640 | 0.03 |
ENSDART00000080777
|
mrpl28
|
mitochondrial ribosomal protein L28 |
| chr23_+_22293682 | 0.03 |
ENSDART00000187304
|
CR354612.1
|
|
| chr16_-_25663846 | 0.03 |
ENSDART00000031304
|
derl1
|
derlin 1 |
| chr21_-_8370500 | 0.02 |
ENSDART00000055328
|
nek6
|
NIMA-related kinase 6 |
| chr19_-_10810006 | 0.02 |
ENSDART00000151157
|
si:dkey-3n22.9
|
si:dkey-3n22.9 |
| chr13_-_33207367 | 0.02 |
ENSDART00000146138
ENSDART00000109667 ENSDART00000182741 |
trip11
|
thyroid hormone receptor interactor 11 |
| chr3_-_49163683 | 0.02 |
ENSDART00000166146
|
dnajb1a
|
DnaJ (Hsp40) homolog, subfamily B, member 1a |
| chr25_-_3087556 | 0.02 |
ENSDART00000193249
|
best1
|
bestrophin 1 |
| chr7_-_65236649 | 0.02 |
ENSDART00000185070
|
pkd1l2a
|
polycystic kidney disease 1 like 2a |
| chr9_-_34885902 | 0.02 |
ENSDART00000137504
|
p2ry8
|
P2Y receptor family member 8 |
| chr22_+_12477996 | 0.02 |
ENSDART00000177704
|
CR847870.3
|
|
| chr6_-_29402578 | 0.02 |
ENSDART00000115031
|
mrpl47
|
mitochondrial ribosomal protein L47 |
| chr15_-_669476 | 0.02 |
ENSDART00000153687
ENSDART00000030603 |
si:ch211-210b2.2
|
si:ch211-210b2.2 |
| chr7_-_65191937 | 0.02 |
ENSDART00000173234
|
pkd1l2a
|
polycystic kidney disease 1 like 2a |
| chr15_-_7337148 | 0.02 |
ENSDART00000182568
|
SLC7A1 (1 of many)
|
high affinity cationic amino acid transporter 1 |
| chr14_-_14643190 | 0.02 |
ENSDART00000167119
|
znf185
|
zinc finger protein 185 with LIM domain |
| chr20_-_25481306 | 0.02 |
ENSDART00000182495
|
si:dkey-183n20.15
|
si:dkey-183n20.15 |
| chr17_-_24890843 | 0.01 |
ENSDART00000184984
ENSDART00000135569 ENSDART00000193661 |
gale
|
UDP-galactose-4-epimerase |
| chr15_+_33851280 | 0.01 |
ENSDART00000162308
|
inpp5ka
|
inositol polyphosphate-5-phosphatase Ka |
| chr25_+_19681662 | 0.01 |
ENSDART00000104347
|
si:dkeyp-110c12.3
|
si:dkeyp-110c12.3 |
| chr20_+_15600167 | 0.01 |
ENSDART00000171991
|
faslg
|
Fas ligand (TNF superfamily, member 6) |
| chr25_-_19574146 | 0.01 |
ENSDART00000156811
|
si:ch211-59o9.10
|
si:ch211-59o9.10 |
| chr5_-_6508250 | 0.01 |
ENSDART00000060535
|
crybb3
|
crystallin, beta B3 |
| chr7_+_34786591 | 0.01 |
ENSDART00000173700
|
si:dkey-148a17.5
|
si:dkey-148a17.5 |
| chr16_-_53800047 | 0.01 |
ENSDART00000158047
|
znrf2b
|
zinc and ring finger 2b |
| chr5_+_50912729 | 0.01 |
ENSDART00000190837
|
col4a3bpa
|
collagen, type IV, alpha 3 (Goodpasture antigen) binding protein a |
| chr5_-_65037371 | 0.01 |
ENSDART00000170560
|
anxa1b
|
annexin A1b |
| chr25_+_30238021 | 0.01 |
ENSDART00000123220
|
alkbh3
|
alkB homolog 3, alpha-ketoglutarate-dependent dioxygenase |
| chr5_+_57641554 | 0.00 |
ENSDART00000171309
ENSDART00000157992 ENSDART00000164742 |
cryabb
|
crystallin, alpha B, b |
| chr11_-_41220794 | 0.00 |
ENSDART00000192895
|
mrps16
|
mitochondrial ribosomal protein S16 |
| chr23_-_21797517 | 0.00 |
ENSDART00000110041
|
lrrc38a
|
leucine rich repeat containing 38a |
Network of associatons between targets according to the STRING database.
First level regulatory network of hic1l+hic2
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.7 | GO:0010332 | response to gamma radiation(GO:0010332) cellular response to gamma radiation(GO:0071480) |
| 0.1 | 0.3 | GO:0044320 | leptin-mediated signaling pathway(GO:0033210) cellular response to leptin stimulus(GO:0044320) response to leptin(GO:0044321) |
| 0.1 | 0.3 | GO:0002074 | extraocular skeletal muscle development(GO:0002074) |
| 0.1 | 0.2 | GO:0065001 | specification of axis polarity(GO:0065001) |
| 0.1 | 0.2 | GO:0051228 | mitotic spindle disassembly(GO:0051228) spindle disassembly(GO:0051230) |
| 0.0 | 0.1 | GO:0009211 | pyrimidine deoxyribonucleoside triphosphate metabolic process(GO:0009211) |
| 0.0 | 0.2 | GO:0003413 | chondrocyte differentiation involved in endochondral bone morphogenesis(GO:0003413) growth plate cartilage chondrocyte differentiation(GO:0003418) |
| 0.0 | 0.2 | GO:0034695 | response to prostaglandin E(GO:0034695) cellular response to prostaglandin E stimulus(GO:0071380) |
| 0.0 | 0.1 | GO:0034146 | toll-like receptor 5 signaling pathway(GO:0034146) |
| 0.0 | 0.2 | GO:0045852 | pH elevation(GO:0045852) intracellular pH elevation(GO:0051454) |
| 0.0 | 0.1 | GO:0045730 | respiratory burst(GO:0045730) |
| 0.0 | 0.1 | GO:0002025 | vasodilation by norepinephrine-epinephrine involved in regulation of systemic arterial blood pressure(GO:0002025) |
| 0.0 | 0.3 | GO:0000186 | activation of MAPKK activity(GO:0000186) |
| 0.0 | 0.1 | GO:0035630 | bone mineralization involved in bone maturation(GO:0035630) |
| 0.0 | 0.0 | GO:0097095 | frontonasal suture morphogenesis(GO:0097095) |
| 0.0 | 0.1 | GO:0019254 | carnitine metabolic process, CoA-linked(GO:0019254) |
| 0.0 | 0.1 | GO:0034389 | lipid particle organization(GO:0034389) |
| 0.0 | 0.1 | GO:0031444 | slow-twitch skeletal muscle fiber contraction(GO:0031444) |
| 0.0 | 0.3 | GO:0008272 | sulfate transport(GO:0008272) |
| 0.0 | 0.1 | GO:0061577 | calcium ion transmembrane transport via high voltage-gated calcium channel(GO:0061577) regulation of calcium ion transmembrane transport via high voltage-gated calcium channel(GO:1902514) |
| 0.0 | 0.2 | GO:0044247 | polysaccharide catabolic process(GO:0000272) glycogen catabolic process(GO:0005980) glucan catabolic process(GO:0009251) cellular polysaccharide catabolic process(GO:0044247) |
| 0.0 | 0.1 | GO:0016539 | intein-mediated protein splicing(GO:0016539) protein splicing(GO:0030908) |
| 0.0 | 0.1 | GO:0016037 | light absorption(GO:0016037) |
| 0.0 | 0.0 | GO:0016332 | establishment or maintenance of polarity of embryonic epithelium(GO:0016332) |
| 0.0 | 0.2 | GO:0060236 | regulation of mitotic spindle organization(GO:0060236) |
| 0.0 | 0.1 | GO:0070782 | phosphatidylserine exposure on apoptotic cell surface(GO:0070782) |
| 0.0 | 0.1 | GO:0000741 | karyogamy(GO:0000741) pronuclear fusion(GO:0007344) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.2 | GO:0034098 | VCP-NPL4-UFD1 AAA ATPase complex(GO:0034098) |
| 0.0 | 0.2 | GO:0036156 | inner dynein arm(GO:0036156) |
| 0.0 | 0.3 | GO:0005771 | multivesicular body(GO:0005771) |
| 0.0 | 0.1 | GO:1990454 | L-type voltage-gated calcium channel complex(GO:1990454) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.2 | GO:0004135 | glycogen debranching enzyme activity(GO:0004133) 4-alpha-glucanotransferase activity(GO:0004134) amylo-alpha-1,6-glucosidase activity(GO:0004135) |
| 0.0 | 0.2 | GO:0005412 | glucose:sodium symporter activity(GO:0005412) |
| 0.0 | 0.1 | GO:0009041 | uridylate kinase activity(GO:0009041) |
| 0.0 | 0.3 | GO:0070888 | E-box binding(GO:0070888) |
| 0.0 | 0.1 | GO:0051380 | norepinephrine binding(GO:0051380) |
| 0.0 | 0.1 | GO:0070573 | metallodipeptidase activity(GO:0070573) |
| 0.0 | 0.5 | GO:0019531 | oxalate transmembrane transporter activity(GO:0019531) |
| 0.0 | 0.2 | GO:0005391 | sodium:potassium-exchanging ATPase activity(GO:0005391) |
| 0.0 | 0.2 | GO:0004096 | catalase activity(GO:0004096) |
| 0.0 | 0.2 | GO:0008506 | sugar:proton symporter activity(GO:0005351) cation:sugar symporter activity(GO:0005402) sucrose:proton symporter activity(GO:0008506) sucrose transmembrane transporter activity(GO:0008515) disaccharide transmembrane transporter activity(GO:0015154) oligosaccharide transmembrane transporter activity(GO:0015157) |
| 0.0 | 0.2 | GO:0004957 | prostaglandin E receptor activity(GO:0004957) |
| 0.0 | 0.1 | GO:0004092 | carnitine O-acetyltransferase activity(GO:0004092) |
| 0.0 | 0.1 | GO:0016165 | linoleate 13S-lipoxygenase activity(GO:0016165) |
| 0.0 | 0.1 | GO:0070052 | collagen V binding(GO:0070052) |
| 0.0 | 0.3 | GO:0004709 | MAP kinase kinase kinase activity(GO:0004709) |
| 0.0 | 0.1 | GO:0060182 | apelin receptor activity(GO:0060182) |
| 0.0 | 0.7 | GO:0042562 | hormone binding(GO:0042562) |
| 0.0 | 0.1 | GO:0005113 | patched binding(GO:0005113) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.3 | PID IL27 PATHWAY | IL27-mediated signaling events |
| 0.0 | 0.4 | PID AR NONGENOMIC PATHWAY | Nongenotropic Androgen signaling |
| 0.0 | 0.3 | PID HDAC CLASSIII PATHWAY | Signaling events mediated by HDAC Class III |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | REACTOME SIGNALING BY ACTIVATED POINT MUTANTS OF FGFR1 | Genes involved in Signaling by activated point mutants of FGFR1 |