Project
PRJNA207719: Tissue specific transcriptome profiling
Navigation
Downloads
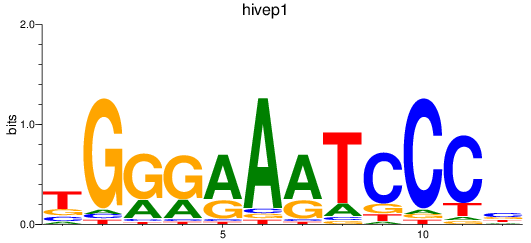
Results for hivep1
Z-value: 0.97
Transcription factors associated with hivep1
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
hivep1
|
ENSDARG00000103658 | HIVEP zinc finger 1 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| hivep1 | dr11_v1_chr19_-_3488860_3488860 | -0.94 | 1.9e-02 | Click! |
Activity profile of hivep1 motif
Sorted Z-values of hivep1 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr14_-_1454045 | 0.73 |
ENSDART00000161460
|
pmt
|
phosphoethanolamine methyltransferase |
| chr16_-_27749172 | 0.70 |
ENSDART00000145198
|
steap4
|
STEAP family member 4 |
| chr5_+_28830643 | 0.67 |
ENSDART00000051448
|
serpina7
|
serpin peptidase inhibitor, clade A (alpha-1 antiproteinase, antitrypsin), member 7 |
| chr6_+_54576520 | 0.66 |
ENSDART00000093199
ENSDART00000127519 ENSDART00000157142 |
tead3b
|
TEA domain family member 3 b |
| chr10_+_19596214 | 0.62 |
ENSDART00000183110
|
CABZ01059626.1
|
|
| chr19_-_24555935 | 0.60 |
ENSDART00000132660
ENSDART00000162801 |
polr3gla
|
polymerase (RNA) III (DNA directed) polypeptide G like a |
| chr1_-_52447364 | 0.59 |
ENSDART00000140740
|
si:ch211-217k17.10
|
si:ch211-217k17.10 |
| chr20_-_40754794 | 0.58 |
ENSDART00000187251
|
cx32.3
|
connexin 32.3 |
| chr3_-_36750068 | 0.58 |
ENSDART00000173388
|
abcc6b.1
|
ATP-binding cassette, sub-family C (CFTR/MRP), member 6b, tandem duplicate 1 |
| chr16_+_46294337 | 0.57 |
ENSDART00000040769
|
nr2f5
|
nuclear receptor subfamily 2, group F, member 5 |
| chr10_-_8079737 | 0.54 |
ENSDART00000059014
ENSDART00000179549 |
zgc:173443
si:ch211-251f6.6
|
zgc:173443 si:ch211-251f6.6 |
| chr5_+_9246458 | 0.52 |
ENSDART00000081772
|
susd1
|
sushi domain containing 1 |
| chr15_-_36365840 | 0.52 |
ENSDART00000192926
|
si:dkey-23k10.3
|
si:dkey-23k10.3 |
| chr9_-_43082945 | 0.52 |
ENSDART00000142257
|
ccdc141
|
coiled-coil domain containing 141 |
| chr24_-_21689146 | 0.51 |
ENSDART00000105917
|
urad
|
ureidoimidazoline (2-oxo-4-hydroxy-4-carboxy-5-) decarboxylase |
| chr13_+_30692669 | 0.49 |
ENSDART00000187818
|
CR762483.1
|
|
| chr16_+_23403602 | 0.49 |
ENSDART00000159848
|
s100w
|
S100 calcium binding protein W |
| chr16_-_25608453 | 0.48 |
ENSDART00000140140
|
zgc:110410
|
zgc:110410 |
| chr7_-_60831082 | 0.48 |
ENSDART00000073654
ENSDART00000136999 |
pcxb
|
pyruvate carboxylase b |
| chr4_+_18806251 | 0.46 |
ENSDART00000138662
|
slc26a3.2
|
solute carrier family 26 (anion exchanger), member 3, tandem duplicate 2 |
| chr2_-_37465517 | 0.45 |
ENSDART00000139983
|
si:dkey-57k2.6
|
si:dkey-57k2.6 |
| chr16_-_25739331 | 0.44 |
ENSDART00000189455
|
bcl3
|
B cell CLL/lymphoma 3 |
| chr7_+_21275152 | 0.43 |
ENSDART00000173612
|
serpinh2
|
serine (or cysteine) peptidase inhibitor, clade H, member 2 |
| chr19_-_12404590 | 0.42 |
ENSDART00000103703
|
ftr56
|
finTRIM family, member 56 |
| chr19_-_24555623 | 0.42 |
ENSDART00000176022
|
polr3gla
|
polymerase (RNA) III (DNA directed) polypeptide G like a |
| chr16_+_38394371 | 0.41 |
ENSDART00000137954
|
cd83
|
CD83 molecule |
| chr5_+_28830388 | 0.40 |
ENSDART00000149150
|
serpina7
|
serpin peptidase inhibitor, clade A (alpha-1 antiproteinase, antitrypsin), member 7 |
| chr7_-_73717082 | 0.38 |
ENSDART00000164301
ENSDART00000082625 |
BX664721.2
|
|
| chr24_+_26006730 | 0.38 |
ENSDART00000140384
ENSDART00000139184 |
ccl20b
|
chemokine (C-C motif) ligand 20b |
| chr2_-_2642476 | 0.38 |
ENSDART00000124032
|
serbp1b
|
SERPINE1 mRNA binding protein 1b |
| chr17_+_53356403 | 0.37 |
ENSDART00000186502
ENSDART00000002562 |
CABZ01068209.1
zmp:0000000527
|
zmp:0000000527 |
| chr18_-_5527050 | 0.37 |
ENSDART00000145400
ENSDART00000132498 ENSDART00000146209 |
zgc:153317
|
zgc:153317 |
| chr3_+_22273123 | 0.37 |
ENSDART00000044157
|
scn4ab
|
sodium channel, voltage-gated, type IV, alpha, b |
| chr5_+_9246018 | 0.37 |
ENSDART00000081769
ENSDART00000183189 |
susd1
|
sushi domain containing 1 |
| chr8_-_20230559 | 0.36 |
ENSDART00000193677
|
mllt1a
|
MLLT1, super elongation complex subunit a |
| chr6_-_54815886 | 0.36 |
ENSDART00000180793
ENSDART00000007498 |
tnni1b
|
troponin I type 1b (skeletal, slow) |
| chr15_-_3736149 | 0.35 |
ENSDART00000182986
|
lpar6a
|
lysophosphatidic acid receptor 6a |
| chr22_+_3184500 | 0.35 |
ENSDART00000176409
ENSDART00000160604 |
ftsj3
|
FtsJ RNA methyltransferase homolog 3 |
| chr14_+_1014109 | 0.35 |
ENSDART00000157945
|
f8
|
coagulation factor VIII, procoagulant component |
| chr2_-_879800 | 0.34 |
ENSDART00000019733
|
irf4a
|
interferon regulatory factor 4a |
| chr8_-_25247284 | 0.34 |
ENSDART00000132697
|
gnat2
|
guanine nucleotide binding protein (G protein), alpha transducing activity polypeptide 2 |
| chr12_-_4835378 | 0.33 |
ENSDART00000172093
|
si:ch211-93e11.8
|
si:ch211-93e11.8 |
| chr25_+_34641536 | 0.33 |
ENSDART00000167033
|
CABZ01079011.1
|
|
| chr22_+_28969071 | 0.33 |
ENSDART00000163427
|
pimr95
|
Pim proto-oncogene, serine/threonine kinase, related 95 |
| chr8_-_18667693 | 0.32 |
ENSDART00000100516
|
stap2b
|
signal transducing adaptor family member 2b |
| chr20_+_43691208 | 0.32 |
ENSDART00000152976
ENSDART00000045185 |
lin9
|
lin-9 DREAM MuvB core complex component |
| chr23_+_36115541 | 0.31 |
ENSDART00000130090
|
hoxc6a
|
homeobox C6a |
| chr6_-_39270851 | 0.31 |
ENSDART00000148839
|
arhgef25b
|
Rho guanine nucleotide exchange factor (GEF) 25b |
| chr12_-_4206869 | 0.30 |
ENSDART00000106572
|
si:dkey-32n7.9
|
si:dkey-32n7.9 |
| chr14_-_24081929 | 0.29 |
ENSDART00000158576
|
msx2a
|
muscle segment homeobox 2a |
| chr23_-_31974060 | 0.29 |
ENSDART00000168087
|
BX927210.1
|
|
| chr13_+_21601149 | 0.29 |
ENSDART00000179369
|
sh2d4ba
|
SH2 domain containing 4Ba |
| chr7_-_34262080 | 0.28 |
ENSDART00000183246
|
si:ch211-98n17.5
|
si:ch211-98n17.5 |
| chr5_-_30984271 | 0.28 |
ENSDART00000051392
|
spns3
|
spinster homolog 3 (Drosophila) |
| chr9_-_51563575 | 0.28 |
ENSDART00000167034
ENSDART00000148918 |
tank
|
TRAF family member-associated NFKB activator |
| chr20_-_30376433 | 0.28 |
ENSDART00000190737
|
rps7
|
ribosomal protein S7 |
| chr13_+_21600946 | 0.28 |
ENSDART00000144045
|
sh2d4ba
|
SH2 domain containing 4Ba |
| chr16_-_25741225 | 0.28 |
ENSDART00000130641
|
bcl3
|
B cell CLL/lymphoma 3 |
| chr9_-_813693 | 0.28 |
ENSDART00000174793
|
marco
|
macrophage receptor with collagenous structure |
| chr25_-_27843066 | 0.27 |
ENSDART00000179684
ENSDART00000186000 ENSDART00000190065 |
asb15a
|
ankyrin repeat and SOCS box containing 15a |
| chr5_+_22579975 | 0.27 |
ENSDART00000080877
|
tnfsf10l4
|
tumor necrosis factor (ligand) superfamily, member 10 like 4 |
| chr19_-_7144548 | 0.27 |
ENSDART00000147177
ENSDART00000134850 |
psmb8a
psmb13a
|
proteasome subunit beta 8A proteasome subunit beta 13a |
| chr25_-_14637660 | 0.27 |
ENSDART00000143666
|
nav2b
|
neuron navigator 2b |
| chr5_-_42904329 | 0.27 |
ENSDART00000112807
|
cxcl20
|
chemokine (C-X-C motif) ligand 20 |
| chr10_+_15088534 | 0.27 |
ENSDART00000142865
|
si:ch211-95j8.3
|
si:ch211-95j8.3 |
| chr23_-_33738570 | 0.26 |
ENSDART00000131680
|
si:ch211-210c8.7
|
si:ch211-210c8.7 |
| chr15_-_18200358 | 0.26 |
ENSDART00000158569
|
si:ch211-247l8.8
|
si:ch211-247l8.8 |
| chr19_+_7717449 | 0.26 |
ENSDART00000104719
ENSDART00000146747 |
tuft1b
|
tuftelin 1b |
| chr3_+_6325895 | 0.26 |
ENSDART00000167386
ENSDART00000165000 |
zgc:136767
|
zgc:136767 |
| chr3_-_27061637 | 0.26 |
ENSDART00000157126
|
atf7ip2
|
activating transcription factor 7 interacting protein 2 |
| chr10_+_35491216 | 0.26 |
ENSDART00000109705
|
phldb2a
|
pleckstrin homology-like domain, family B, member 2a |
| chr2_-_36674005 | 0.25 |
ENSDART00000004899
|
ccnl1b
|
cyclin L1b |
| chr8_-_48704050 | 0.25 |
ENSDART00000163916
|
pimr182
|
Pim proto-oncogene, serine/threonine kinase, related 182 |
| chr6_-_436658 | 0.25 |
ENSDART00000191515
|
grap2b
|
GRB2-related adaptor protein 2b |
| chr7_+_22585447 | 0.24 |
ENSDART00000149144
|
chrnb1l
|
cholinergic receptor, nicotinic, beta 1 (muscle) like |
| chr18_+_7073130 | 0.24 |
ENSDART00000101216
ENSDART00000148947 |
si:dkey-88e18.2
|
si:dkey-88e18.2 |
| chr20_+_16881883 | 0.24 |
ENSDART00000130107
|
nfkbiaa
|
nuclear factor of kappa light polypeptide gene enhancer in B-cells inhibitor, alpha a |
| chr23_+_26733232 | 0.24 |
ENSDART00000035080
|
zgc:158263
|
zgc:158263 |
| chr9_-_23894392 | 0.24 |
ENSDART00000133417
|
asb18
|
ankyrin repeat and SOCS box containing 18 |
| chr3_+_31662126 | 0.24 |
ENSDART00000113441
|
mylk5
|
myosin, light chain kinase 5 |
| chr20_+_23501535 | 0.23 |
ENSDART00000177922
ENSDART00000058532 |
palld
|
palladin, cytoskeletal associated protein |
| chr13_+_40692804 | 0.23 |
ENSDART00000109822
|
hps1
|
Hermansky-Pudlak syndrome 1 |
| chr23_+_43849190 | 0.23 |
ENSDART00000017375
|
cnga1
|
cyclic nucleotide gated channel alpha 1 |
| chr3_-_31619463 | 0.23 |
ENSDART00000124559
|
moto
|
minamoto |
| chr18_+_40572294 | 0.23 |
ENSDART00000147003
|
si:ch211-132b12.2
|
si:ch211-132b12.2 |
| chr9_-_49978335 | 0.23 |
ENSDART00000170814
|
scn1a
|
sodium channel, voltage-gated, type I, alpha |
| chr4_+_38550788 | 0.22 |
ENSDART00000157412
|
si:ch211-209n20.1
|
si:ch211-209n20.1 |
| chr20_+_13883131 | 0.22 |
ENSDART00000003248
ENSDART00000152611 |
nek2
|
NIMA-related kinase 2 |
| chr15_+_17251191 | 0.22 |
ENSDART00000156587
|
si:ch73-223p23.2
|
si:ch73-223p23.2 |
| chr8_-_12432604 | 0.22 |
ENSDART00000133350
ENSDART00000140699 ENSDART00000101174 |
traf1
|
TNF receptor-associated factor 1 |
| chr5_-_16218777 | 0.21 |
ENSDART00000141698
|
kremen1
|
kringle containing transmembrane protein 1 |
| chr2_-_43583896 | 0.21 |
ENSDART00000161711
|
itgb1b
|
integrin, beta 1b |
| chr9_-_32663496 | 0.21 |
ENSDART00000078384
|
gzm3.2
|
granzyme 3, tandem duplicate 2 |
| chr8_-_20230802 | 0.21 |
ENSDART00000063400
|
mllt1a
|
MLLT1, super elongation complex subunit a |
| chr20_-_51307815 | 0.21 |
ENSDART00000098833
|
nfkbie
|
nuclear factor of kappa light polypeptide gene enhancer in B-cells inhibitor, epsilon |
| chr22_+_9922301 | 0.21 |
ENSDART00000105924
|
blf
|
bloody fingers |
| chr6_-_442163 | 0.21 |
ENSDART00000163564
ENSDART00000189134 ENSDART00000169789 |
grap2b
|
GRB2-related adaptor protein 2b |
| chr2_-_24398324 | 0.21 |
ENSDART00000165226
|
zgc:154006
|
zgc:154006 |
| chr24_+_15670013 | 0.21 |
ENSDART00000185826
|
CU929414.1
|
|
| chr12_+_22576404 | 0.20 |
ENSDART00000172053
|
capgb
|
capping protein (actin filament), gelsolin-like b |
| chr18_+_30567945 | 0.20 |
ENSDART00000078894
|
irf8
|
interferon regulatory factor 8 |
| chr16_+_35535171 | 0.20 |
ENSDART00000167001
|
cited4b
|
Cbp/p300-interacting transactivator, with Glu/Asp-rich carboxy-terminal domain, 4b |
| chr11_-_8320868 | 0.20 |
ENSDART00000160570
|
si:ch211-247n2.1
|
si:ch211-247n2.1 |
| chr1_-_59571758 | 0.20 |
ENSDART00000193546
ENSDART00000167087 |
wfikkn1
|
WAP, follistatin/kazal, immunoglobulin, kunitz and netrin domain containing 1 |
| chr15_-_16076399 | 0.20 |
ENSDART00000135658
ENSDART00000133755 ENSDART00000080413 |
srsf1a
|
serine/arginine-rich splicing factor 1a |
| chr13_-_25196758 | 0.20 |
ENSDART00000184722
|
adka
|
adenosine kinase a |
| chr7_-_7398350 | 0.19 |
ENSDART00000012637
|
zgc:101810
|
zgc:101810 |
| chr1_+_17892944 | 0.19 |
ENSDART00000013021
|
tlr3
|
toll-like receptor 3 |
| chr3_+_56574623 | 0.19 |
ENSDART00000130877
|
rac1b
|
Rac family small GTPase 1b |
| chr3_-_58733718 | 0.19 |
ENSDART00000154603
|
si:ch73-281f12.4
|
si:ch73-281f12.4 |
| chr9_+_35014728 | 0.19 |
ENSDART00000100700
|
gabpa
|
GA binding protein transcription factor, alpha subunit |
| chr17_+_51627209 | 0.19 |
ENSDART00000056886
|
zgc:113142
|
zgc:113142 |
| chr14_+_14806692 | 0.19 |
ENSDART00000193050
|
fhdc2
|
FH2 domain containing 2 |
| chr7_+_4384863 | 0.19 |
ENSDART00000042955
ENSDART00000134653 |
slc12a10.3
|
slc12a10.3 solute carrier family 12 (sodium/potassium/chloride transporters), member 10, tandem duplicate 3 |
| chr2_+_30547018 | 0.19 |
ENSDART00000193747
|
ankrd33bb
|
ankyrin repeat domain 33Bb |
| chr23_+_3594171 | 0.19 |
ENSDART00000159609
|
si:dkey-9l20.3
|
si:dkey-9l20.3 |
| chr16_+_11683548 | 0.19 |
ENSDART00000138000
|
si:dkey-250k15.7
|
si:dkey-250k15.7 |
| chr15_-_37779978 | 0.18 |
ENSDART00000157202
|
si:dkey-42l23.3
|
si:dkey-42l23.3 |
| chr10_+_37927100 | 0.18 |
ENSDART00000172548
|
bhlha9
|
basic helix-loop-helix family, member a9 |
| chr13_-_4664403 | 0.18 |
ENSDART00000023803
ENSDART00000177957 |
c1d
|
C1D nuclear receptor corepressor |
| chr8_+_24745041 | 0.18 |
ENSDART00000148872
|
slc16a4
|
solute carrier family 16, member 4 |
| chr24_-_31422574 | 0.18 |
ENSDART00000158567
|
cngb3.1
|
cyclic nucleotide gated channel beta 3, tandem duplicate 1 |
| chr19_+_41173386 | 0.18 |
ENSDART00000142773
|
asb4
|
ankyrin repeat and SOCS box containing 4 |
| chr1_-_18585046 | 0.18 |
ENSDART00000147228
|
fam114a1
|
family with sequence similarity 114, member A1 |
| chr16_-_40426837 | 0.18 |
ENSDART00000193690
|
plekhf2
|
pleckstrin homology domain containing, family F (with FYVE domain) member 2 |
| chr8_+_32722842 | 0.18 |
ENSDART00000147594
|
hmcn2
|
hemicentin 2 |
| chr7_+_29293452 | 0.17 |
ENSDART00000127358
|
si:ch211-112g6.4
|
si:ch211-112g6.4 |
| chr22_+_8313513 | 0.17 |
ENSDART00000181169
ENSDART00000103911 |
CABZ01077217.1
|
|
| chr3_+_8826905 | 0.17 |
ENSDART00000167474
ENSDART00000168088 |
si:dkeyp-30d6.2
|
si:dkeyp-30d6.2 |
| chr4_+_8016457 | 0.17 |
ENSDART00000014036
|
optn
|
optineurin |
| chr6_-_46589726 | 0.17 |
ENSDART00000084334
|
ptgis
|
prostaglandin I2 (prostacyclin) synthase |
| chr6_+_9175886 | 0.17 |
ENSDART00000165333
|
si:ch211-207l14.1
|
si:ch211-207l14.1 |
| chr19_-_3261019 | 0.17 |
ENSDART00000134407
|
si:ch211-133n4.12
|
si:ch211-133n4.12 |
| chr19_+_43392446 | 0.17 |
ENSDART00000147290
|
yrk
|
Yes-related kinase |
| chr20_+_98179 | 0.17 |
ENSDART00000022725
|
si:ch1073-155h21.1
|
si:ch1073-155h21.1 |
| chr20_-_41992878 | 0.17 |
ENSDART00000100967
|
si:dkeyp-114g9.1
|
si:dkeyp-114g9.1 |
| chr22_+_38767780 | 0.17 |
ENSDART00000149499
|
alpi.1
|
alkaline phosphatase, intestinal, tandem duplicate 1 |
| chr24_+_9003998 | 0.16 |
ENSDART00000179656
ENSDART00000191314 |
CR318624.2
|
|
| chr7_+_32021982 | 0.16 |
ENSDART00000173848
|
mettl15
|
methyltransferase like 15 |
| chr7_-_19003504 | 0.16 |
ENSDART00000157618
|
HACD4
|
si:ch73-41h24.1 |
| chr16_+_35535375 | 0.16 |
ENSDART00000171675
|
cited4b
|
Cbp/p300-interacting transactivator, with Glu/Asp-rich carboxy-terminal domain, 4b |
| chr25_-_6557854 | 0.16 |
ENSDART00000181740
|
cspg4
|
chondroitin sulfate proteoglycan 4 |
| chr10_-_41980797 | 0.16 |
ENSDART00000076575
|
rhof
|
ras homolog family member F |
| chr16_-_22729119 | 0.16 |
ENSDART00000132944
|
tlr19
|
toll-like receptor 19 |
| chr22_+_24378064 | 0.15 |
ENSDART00000180592
|
BX649608.2
|
|
| chr20_-_49657134 | 0.15 |
ENSDART00000151248
|
col12a1b
|
collagen, type XII, alpha 1b |
| chr8_+_26410539 | 0.15 |
ENSDART00000168780
|
ifrd2
|
interferon-related developmental regulator 2 |
| chr10_-_43844537 | 0.15 |
ENSDART00000114208
|
tlr8b
|
toll-like receptor 8b |
| chr5_+_37032038 | 0.15 |
ENSDART00000045036
|
nfkbib
|
nuclear factor of kappa light polypeptide gene enhancer in B-cells inhibitor, beta |
| chr22_-_651719 | 0.15 |
ENSDART00000148692
|
apobec2a
|
apolipoprotein B mRNA editing enzyme, catalytic polypeptide-like 2a |
| chr2_+_111919 | 0.15 |
ENSDART00000149391
|
fggy
|
FGGY carbohydrate kinase domain containing |
| chr11_-_36450770 | 0.15 |
ENSDART00000128889
|
zbtb40
|
zinc finger and BTB domain containing 40 |
| chr5_+_67390115 | 0.15 |
ENSDART00000193255
|
ebf2
|
early B cell factor 2 |
| chr1_+_292545 | 0.14 |
ENSDART00000148261
|
cenpe
|
centromere protein E |
| chr9_+_32859967 | 0.14 |
ENSDART00000168992
|
si:dkey-145p14.5
|
si:dkey-145p14.5 |
| chr13_+_22717939 | 0.14 |
ENSDART00000188288
|
nfkb2
|
nuclear factor of kappa light polypeptide gene enhancer in B-cells 2 (p49/p100) |
| chr3_+_57268363 | 0.14 |
ENSDART00000180725
|
TMEM235 (1 of many)
|
transmembrane protein 235 |
| chr20_-_44055095 | 0.14 |
ENSDART00000125898
ENSDART00000082265 |
runx2b
|
runt-related transcription factor 2b |
| chr21_+_5257316 | 0.14 |
ENSDART00000184441
|
loxhd1a
|
lipoxygenase homology domains 1a |
| chr7_-_17591007 | 0.14 |
ENSDART00000171023
|
CU672228.1
|
|
| chr18_-_16924221 | 0.13 |
ENSDART00000122102
|
wee1
|
WEE1 G2 checkpoint kinase |
| chr15_-_28677725 | 0.13 |
ENSDART00000060255
|
blmh
|
bleomycin hydrolase |
| chr14_-_35883019 | 0.13 |
ENSDART00000112379
|
fam198b
|
family with sequence similarity 198, member B |
| chr11_+_40544269 | 0.13 |
ENSDART00000175424
ENSDART00000184673 |
mmel1
|
membrane metallo-endopeptidase-like 1 |
| chr23_+_14590767 | 0.13 |
ENSDART00000143675
|
si:rp71-79p20.2
|
si:rp71-79p20.2 |
| chr9_+_35014513 | 0.12 |
ENSDART00000100701
|
gabpa
|
GA binding protein transcription factor, alpha subunit |
| chr24_-_21131607 | 0.12 |
ENSDART00000010126
|
zdhhc23b
|
zinc finger, DHHC-type containing 23b |
| chr15_-_38202630 | 0.12 |
ENSDART00000183772
|
rhoga
|
ras homolog family member Ga |
| chr9_-_12624622 | 0.12 |
ENSDART00000146535
|
tspearb
|
thrombospondin-type laminin G domain and EAR repeats b |
| chr19_+_43504480 | 0.12 |
ENSDART00000159421
|
BX649384.3
|
|
| chr3_-_32079916 | 0.12 |
ENSDART00000040900
|
baxb
|
BCL2 associated X, apoptosis regulator b |
| chr2_-_53592532 | 0.12 |
ENSDART00000184066
|
ccl25a
|
chemokine (C-C motif) ligand 25a |
| chr5_-_10007897 | 0.11 |
ENSDART00000109052
|
CR936408.1
|
Danio rerio uncharacterized LOC799523 (LOC799523), mRNA. |
| chr17_+_33313566 | 0.11 |
ENSDART00000045040
|
si:ch211-132f19.7
|
si:ch211-132f19.7 |
| chr11_-_21304452 | 0.11 |
ENSDART00000163008
|
RASSF5
|
si:dkey-85p17.3 |
| chr14_-_38828057 | 0.11 |
ENSDART00000186088
|
spdl1
|
spindle apparatus coiled-coil protein 1 |
| chr12_+_17754859 | 0.11 |
ENSDART00000112119
|
bhlha15
|
basic helix-loop-helix family, member a15 |
| chr22_-_16180467 | 0.11 |
ENSDART00000171331
ENSDART00000185607 |
vcam1b
|
vascular cell adhesion molecule 1b |
| chr14_+_21007957 | 0.10 |
ENSDART00000162316
|
pimr194
|
Pim proto-oncogene, serine/threonine kinase, related 194 |
| chr17_-_53329704 | 0.10 |
ENSDART00000193895
|
exd1
|
exonuclease 3'-5' domain containing 1 |
| chr7_-_28696556 | 0.10 |
ENSDART00000148822
|
adgrg1
|
adhesion G protein-coupled receptor G1 |
| chr3_-_49815223 | 0.10 |
ENSDART00000181208
|
gcgra
|
glucagon receptor a |
| chr8_-_7475917 | 0.10 |
ENSDART00000082157
|
gata1b
|
GATA binding protein 1b |
| chr6_+_27339962 | 0.10 |
ENSDART00000193726
|
klhl30
|
kelch-like family member 30 |
| chr5_-_3684402 | 0.10 |
ENSDART00000184354
|
FO704648.1
|
|
| chr14_+_21042178 | 0.10 |
ENSDART00000168551
|
pimr192
|
Pim proto-oncogene, serine/threonine kinase, related 192 |
| chr25_+_16646113 | 0.09 |
ENSDART00000110426
|
cecr2
|
cat eye syndrome chromosome region, candidate 2 |
| chr23_+_25822742 | 0.09 |
ENSDART00000184436
|
r3hdml
|
R3H domain containing-like |
| chr13_+_33368140 | 0.09 |
ENSDART00000033848
|
brf1a
|
BRF1, RNA polymerase III transcription initiation factor a |
| chr18_+_3022188 | 0.08 |
ENSDART00000170004
|
CU570791.1
|
|
| chr19_+_7124337 | 0.08 |
ENSDART00000031380
|
tap2a
|
transporter associated with antigen processing, subunit type a |
| chr25_-_3347418 | 0.08 |
ENSDART00000082385
|
golt1bb
|
golgi transport 1Bb |
| chr15_-_41677689 | 0.08 |
ENSDART00000187063
|
spsb4b
|
splA/ryanodine receptor domain and SOCS box containing 4b |
| chr8_+_52377516 | 0.08 |
ENSDART00000115398
|
arid5a
|
AT rich interactive domain 5A (MRF1-like) |
| chr25_+_29161609 | 0.08 |
ENSDART00000180752
|
pkmb
|
pyruvate kinase M1/2b |
| chr4_-_12914163 | 0.07 |
ENSDART00000140002
ENSDART00000145917 ENSDART00000141355 ENSDART00000067135 |
msrb3
|
methionine sulfoxide reductase B3 |
| chr3_+_25220334 | 0.07 |
ENSDART00000055440
|
c1qtnf6a
|
C1q and TNF related 6a |
| chr12_-_3903886 | 0.07 |
ENSDART00000184214
ENSDART00000041082 |
gdpd3b
|
glycerophosphodiester phosphodiesterase domain containing 3b |
| chr17_-_20202725 | 0.07 |
ENSDART00000133650
|
ecd
|
ecdysoneless homolog (Drosophila) |
| chr16_-_22747125 | 0.07 |
ENSDART00000179865
|
tlr19
|
toll-like receptor 19 |
Network of associatons between targets according to the STRING database.
First level regulatory network of hivep1
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.7 | GO:0015677 | copper ion import(GO:0015677) |
| 0.2 | 0.5 | GO:0046415 | urate catabolic process(GO:0019628) urate metabolic process(GO:0046415) |
| 0.1 | 0.4 | GO:0002468 | dendritic cell antigen processing and presentation(GO:0002468) |
| 0.1 | 0.1 | GO:0072679 | thymocyte migration(GO:0072679) |
| 0.1 | 0.3 | GO:0002320 | lymphoid progenitor cell differentiation(GO:0002320) |
| 0.1 | 0.2 | GO:1901533 | negative regulation of hematopoietic progenitor cell differentiation(GO:1901533) |
| 0.1 | 0.2 | GO:0045649 | regulation of macrophage differentiation(GO:0045649) |
| 0.1 | 0.2 | GO:0034138 | chemokine production(GO:0032602) toll-like receptor 3 signaling pathway(GO:0034138) |
| 0.1 | 0.2 | GO:1903232 | melanosome assembly(GO:1903232) |
| 0.0 | 0.1 | GO:0016554 | cytidine to uridine editing(GO:0016554) positive regulation of DNA binding(GO:0043388) |
| 0.0 | 0.3 | GO:0050907 | detection of chemical stimulus involved in sensory perception(GO:0050907) |
| 0.0 | 0.2 | GO:0048903 | anterior lateral line neuromast hair cell differentiation(GO:0048903) |
| 0.0 | 0.2 | GO:0090161 | Golgi ribbon formation(GO:0090161) |
| 0.0 | 1.1 | GO:0006383 | transcription from RNA polymerase III promoter(GO:0006383) |
| 0.0 | 0.2 | GO:0005513 | detection of calcium ion(GO:0005513) |
| 0.0 | 0.2 | GO:0007141 | male meiosis I(GO:0007141) |
| 0.0 | 0.2 | GO:0044209 | AMP salvage(GO:0044209) |
| 0.0 | 0.2 | GO:0090497 | mesenchymal cell migration(GO:0090497) |
| 0.0 | 0.6 | GO:0046686 | response to cadmium ion(GO:0046686) |
| 0.0 | 0.3 | GO:0046548 | retinal rod cell development(GO:0046548) |
| 0.0 | 0.7 | GO:0035329 | hippo signaling(GO:0035329) |
| 0.0 | 0.5 | GO:0015701 | bicarbonate transport(GO:0015701) |
| 0.0 | 0.1 | GO:0070445 | regulation of oligodendrocyte progenitor proliferation(GO:0070445) |
| 0.0 | 0.6 | GO:0032526 | response to retinoic acid(GO:0032526) |
| 0.0 | 0.3 | GO:0000463 | maturation of LSU-rRNA from tricistronic rRNA transcript (SSU-rRNA, 5.8S rRNA, LSU-rRNA)(GO:0000463) |
| 0.0 | 0.1 | GO:0000098 | sulfur amino acid catabolic process(GO:0000098) |
| 0.0 | 0.1 | GO:0030091 | protein repair(GO:0030091) |
| 0.0 | 1.5 | GO:0010951 | negative regulation of endopeptidase activity(GO:0010951) |
| 0.0 | 0.3 | GO:0036230 | granulocyte activation(GO:0036230) neutrophil activation(GO:0042119) |
| 0.0 | 0.3 | GO:0030168 | platelet activation(GO:0030168) |
| 0.0 | 0.1 | GO:0001836 | release of cytochrome c from mitochondria(GO:0001836) |
| 0.0 | 0.1 | GO:0007080 | mitotic metaphase plate congression(GO:0007080) |
| 0.0 | 0.2 | GO:0043114 | regulation of vascular permeability(GO:0043114) |
| 0.0 | 0.2 | GO:0002430 | complement receptor mediated signaling pathway(GO:0002430) |
| 0.0 | 0.2 | GO:0001516 | prostaglandin biosynthetic process(GO:0001516) prostanoid biosynthetic process(GO:0046457) |
| 0.0 | 0.5 | GO:0019226 | transmission of nerve impulse(GO:0019226) |
| 0.0 | 0.2 | GO:0033209 | tumor necrosis factor-mediated signaling pathway(GO:0033209) |
| 0.0 | 0.3 | GO:0016601 | Rac protein signal transduction(GO:0016601) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.2 | GO:0031085 | BLOC-3 complex(GO:0031085) |
| 0.0 | 1.0 | GO:0005666 | DNA-directed RNA polymerase III complex(GO:0005666) |
| 0.0 | 0.3 | GO:0030688 | preribosome, small subunit precursor(GO:0030688) |
| 0.0 | 0.4 | GO:0005751 | mitochondrial respiratory chain complex IV(GO:0005751) |
| 0.0 | 0.1 | GO:1990923 | PET complex(GO:1990923) |
| 0.0 | 0.2 | GO:0005892 | acetylcholine-gated channel complex(GO:0005892) |
| 0.0 | 0.5 | GO:0000421 | autophagosome membrane(GO:0000421) |
| 0.0 | 0.2 | GO:0000178 | exosome (RNase complex)(GO:0000178) |
| 0.0 | 0.1 | GO:0000126 | transcription factor TFIIIB complex(GO:0000126) |
| 0.0 | 0.2 | GO:0005885 | Arp2/3 protein complex(GO:0005885) |
| 0.0 | 0.3 | GO:0045180 | basal cortex(GO:0045180) |
| 0.0 | 0.3 | GO:0005839 | proteasome core complex(GO:0005839) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.7 | GO:0008823 | cupric reductase activity(GO:0008823) ferric-chelate reductase (NADPH) activity(GO:0052851) |
| 0.2 | 0.5 | GO:0004736 | pyruvate carboxylase activity(GO:0004736) |
| 0.1 | 0.3 | GO:0016436 | rRNA (uridine) methyltransferase activity(GO:0016436) |
| 0.1 | 0.2 | GO:0001729 | ceramide kinase activity(GO:0001729) |
| 0.1 | 0.3 | GO:0005153 | interleukin-8 receptor binding(GO:0005153) |
| 0.0 | 0.2 | GO:0098639 | C-X3-C chemokine binding(GO:0019960) protein binding involved in cell-matrix adhesion(GO:0098634) collagen binding involved in cell-matrix adhesion(GO:0098639) |
| 0.0 | 0.2 | GO:0004001 | adenosine kinase activity(GO:0004001) |
| 0.0 | 0.1 | GO:0042809 | vitamin D receptor binding(GO:0042809) |
| 0.0 | 0.5 | GO:0019531 | oxalate transmembrane transporter activity(GO:0019531) |
| 0.0 | 0.4 | GO:0070915 | lysophosphatidic acid receptor activity(GO:0070915) |
| 0.0 | 0.1 | GO:0004126 | cytidine deaminase activity(GO:0004126) |
| 0.0 | 0.2 | GO:0008511 | sodium:potassium:chloride symporter activity(GO:0008511) |
| 0.0 | 0.2 | GO:0004035 | alkaline phosphatase activity(GO:0004035) |
| 0.0 | 0.1 | GO:0016519 | gastric inhibitory peptide receptor activity(GO:0016519) |
| 0.0 | 0.2 | GO:0005332 | gamma-aminobutyric acid:sodium symporter activity(GO:0005332) |
| 0.0 | 0.5 | GO:0032266 | phosphatidylinositol-3-phosphate binding(GO:0032266) |
| 0.0 | 0.2 | GO:0005223 | intracellular cyclic nucleotide activated cation channel activity(GO:0005221) intracellular cAMP activated cation channel activity(GO:0005222) intracellular cGMP activated cation channel activity(GO:0005223) cyclic nucleotide-gated ion channel activity(GO:0043855) |
| 0.0 | 0.1 | GO:0004096 | catalase activity(GO:0004096) |
| 0.0 | 0.2 | GO:0008191 | metalloendopeptidase inhibitor activity(GO:0008191) |
| 0.0 | 0.5 | GO:0005164 | tumor necrosis factor receptor binding(GO:0005164) |
| 0.0 | 0.3 | GO:0070003 | threonine-type endopeptidase activity(GO:0004298) threonine-type peptidase activity(GO:0070003) |
| 0.0 | 1.5 | GO:0004867 | serine-type endopeptidase inhibitor activity(GO:0004867) |
| 0.0 | 0.4 | GO:0004129 | cytochrome-c oxidase activity(GO:0004129) heme-copper terminal oxidase activity(GO:0015002) oxidoreductase activity, acting on a heme group of donors(GO:0016675) oxidoreductase activity, acting on a heme group of donors, oxygen as acceptor(GO:0016676) |
| 0.0 | 0.3 | GO:0035591 | signaling adaptor activity(GO:0035591) |
| 0.0 | 0.1 | GO:0004743 | pyruvate kinase activity(GO:0004743) |
| 0.0 | 0.1 | GO:0000995 | transcription factor activity, core RNA polymerase III binding(GO:0000995) |
| 0.0 | 0.2 | GO:0070530 | K63-linked polyubiquitin binding(GO:0070530) |
| 0.0 | 0.1 | GO:0043515 | kinetochore binding(GO:0043515) |
| 0.0 | 0.3 | GO:0048306 | calcium-dependent protein binding(GO:0048306) |
| 0.0 | 0.1 | GO:0033743 | peptide-methionine (R)-S-oxide reductase activity(GO:0033743) |
| 0.0 | 0.2 | GO:0015269 | calcium-activated potassium channel activity(GO:0015269) |
| 0.0 | 0.5 | GO:0048020 | CCR chemokine receptor binding(GO:0048020) |
| 0.0 | 0.3 | GO:0031683 | G-protein beta/gamma-subunit complex binding(GO:0031683) |
| 0.0 | 0.5 | GO:0016831 | carboxy-lyase activity(GO:0016831) |
| 0.0 | 0.2 | GO:0022848 | acetylcholine-gated cation channel activity(GO:0022848) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.8 | PID NFKAPPAB ATYPICAL PATHWAY | Atypical NF-kappaB pathway |
| 0.0 | 0.3 | PID CONE PATHWAY | Visual signal transduction: Cones |
| 0.0 | 0.2 | PID RHODOPSIN PATHWAY | Visual signal transduction: Rods |
| 0.0 | 0.5 | ST TUMOR NECROSIS FACTOR PATHWAY | Tumor Necrosis Factor Pathway. |
| 0.0 | 0.1 | SA G2 AND M PHASES | Cdc25 activates the cdc2/cyclin B complex to induce the G2/M transition. |
| 0.0 | 0.2 | PID TOLL ENDOGENOUS PATHWAY | Endogenous TLR signaling |
| 0.0 | 0.2 | PID HIV NEF PATHWAY | HIV-1 Nef: Negative effector of Fas and TNF-alpha |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.5 | REACTOME RIP MEDIATED NFKB ACTIVATION VIA DAI | Genes involved in RIP-mediated NFkB activation via DAI |
| 0.0 | 0.3 | REACTOME INTRINSIC PATHWAY | Genes involved in Intrinsic Pathway |
| 0.0 | 0.3 | REACTOME TRAF6 MEDIATED IRF7 ACTIVATION | Genes involved in TRAF6 mediated IRF7 activation |
| 0.0 | 0.3 | REACTOME G PROTEIN ACTIVATION | Genes involved in G-protein activation |
| 0.0 | 0.2 | REACTOME CS DS DEGRADATION | Genes involved in CS/DS degradation |
| 0.0 | 0.1 | REACTOME G2 M DNA DAMAGE CHECKPOINT | Genes involved in G2/M DNA damage checkpoint |