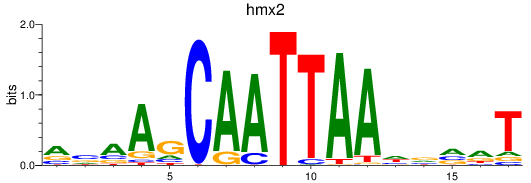
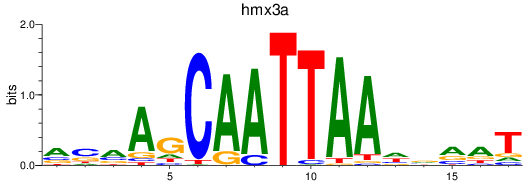
Project
PRJNA207719: Tissue specific transcriptome profiling
Navigation
Downloads
Results for hmx2_hmx3a
Z-value: 1.13
Transcription factors associated with hmx2_hmx3a
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
hmx2
|
ENSDARG00000070954 | H6 family homeobox 2 |
|
hmx2
|
ENSDARG00000115364 | H6 family homeobox 2 |
|
hmx3a
|
ENSDARG00000070955 | H6 family homeobox 3a |
|
hmx3a
|
ENSDARG00000115051 | H6 family homeobox 3a |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| hmx3a | dr11_v1_chr17_-_21793113_21793113 | -0.72 | 1.7e-01 | Click! |
| hmx2 | dr11_v1_chr17_-_21784152_21784152 | -0.08 | 9.0e-01 | Click! |
Activity profile of hmx2_hmx3a motif
Sorted Z-values of hmx2_hmx3a motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr24_+_38301080 | 1.63 |
ENSDART00000105672
|
mybpc2b
|
myosin binding protein C, fast type b |
| chr5_-_25723079 | 1.13 |
ENSDART00000014013
|
gda
|
guanine deaminase |
| chr17_+_50701748 | 0.54 |
ENSDART00000191938
ENSDART00000183220 ENSDART00000049464 |
fermt2
|
fermitin family member 2 |
| chr8_-_53166975 | 0.53 |
ENSDART00000114683
|
rbsn
|
rabenosyn, RAB effector |
| chr11_+_36243774 | 0.41 |
ENSDART00000023323
|
zgc:172270
|
zgc:172270 |
| chr8_-_39739627 | 0.38 |
ENSDART00000135422
ENSDART00000067844 |
si:ch211-170d8.5
|
si:ch211-170d8.5 |
| chr8_-_51293265 | 0.37 |
ENSDART00000127875
ENSDART00000181145 |
bmp1a
|
bone morphogenetic protein 1a |
| chr8_-_39739056 | 0.35 |
ENSDART00000147992
|
si:ch211-170d8.5
|
si:ch211-170d8.5 |
| chr25_-_26018240 | 0.35 |
ENSDART00000150800
|
acsbg1
|
acyl-CoA synthetase bubblegum family member 1 |
| chr16_-_13516745 | 0.34 |
ENSDART00000145410
|
si:dkeyp-69b9.3
|
si:dkeyp-69b9.3 |
| chr1_-_53750522 | 0.32 |
ENSDART00000190755
|
akt3b
|
v-akt murine thymoma viral oncogene homolog 3b |
| chr5_+_37032038 | 0.32 |
ENSDART00000045036
|
nfkbib
|
nuclear factor of kappa light polypeptide gene enhancer in B-cells inhibitor, beta |
| chr9_+_30720048 | 0.31 |
ENSDART00000146115
|
klf12b
|
Kruppel-like factor 12b |
| chr25_-_10503043 | 0.31 |
ENSDART00000155404
|
cox8b
|
cytochrome c oxidase subunit 8b |
| chr20_-_40755614 | 0.31 |
ENSDART00000061247
|
cx32.3
|
connexin 32.3 |
| chr6_+_29410986 | 0.30 |
ENSDART00000065293
|
usp13
|
ubiquitin specific peptidase 13 |
| chr24_-_14711597 | 0.30 |
ENSDART00000131830
|
jph1a
|
junctophilin 1a |
| chr5_-_43959972 | 0.30 |
ENSDART00000180517
|
si:ch211-204c21.1
|
si:ch211-204c21.1 |
| chr3_+_60721342 | 0.29 |
ENSDART00000157772
|
foxj1a
|
forkhead box J1a |
| chr24_-_9997948 | 0.29 |
ENSDART00000136274
|
si:ch211-146l10.7
|
si:ch211-146l10.7 |
| chr5_+_65157576 | 0.28 |
ENSDART00000159599
|
cfap157
|
cilia and flagella associated protein 157 |
| chr21_+_40484088 | 0.28 |
ENSDART00000161919
|
coro6
|
coronin 6 |
| chr18_+_36582815 | 0.27 |
ENSDART00000059311
|
siae
|
sialic acid acetylesterase |
| chr25_+_3326885 | 0.27 |
ENSDART00000104866
|
ldhbb
|
lactate dehydrogenase Bb |
| chr3_-_26017592 | 0.27 |
ENSDART00000030890
|
hmox1a
|
heme oxygenase 1a |
| chr25_+_31267268 | 0.26 |
ENSDART00000181239
|
tnni2a.3
|
troponin I type 2a (skeletal, fast), tandem duplicate 3 |
| chr10_+_9372702 | 0.26 |
ENSDART00000021100
|
lhx6
|
LIM homeobox 6 |
| chr10_-_27046639 | 0.26 |
ENSDART00000041841
|
cnih2
|
cornichon family AMPA receptor auxiliary protein 2 |
| chr14_-_33044955 | 0.25 |
ENSDART00000170626
|
kdrl
|
kinase insert domain receptor like |
| chr12_-_3756405 | 0.25 |
ENSDART00000150839
|
fam57bb
|
family with sequence similarity 57, member Bb |
| chr15_-_19677511 | 0.24 |
ENSDART00000043743
|
si:dkey-4p15.3
|
si:dkey-4p15.3 |
| chr8_-_25120231 | 0.24 |
ENSDART00000147308
|
amigo1
|
adhesion molecule with Ig-like domain 1 |
| chr18_+_8346920 | 0.24 |
ENSDART00000083421
|
cpt1b
|
carnitine palmitoyltransferase 1B (muscle) |
| chr9_-_22821901 | 0.24 |
ENSDART00000101711
|
neb
|
nebulin |
| chr8_+_49778756 | 0.24 |
ENSDART00000083790
|
ntrk2a
|
neurotrophic tyrosine kinase, receptor, type 2a |
| chr2_+_22702488 | 0.23 |
ENSDART00000076647
|
kif1ab
|
kinesin family member 1Ab |
| chr23_-_26652273 | 0.23 |
ENSDART00000112124
ENSDART00000111029 |
hspg2
|
heparan sulfate proteoglycan 2 |
| chr6_-_23931442 | 0.23 |
ENSDART00000160547
|
sec16b
|
SEC16 homolog B, endoplasmic reticulum export factor |
| chr22_-_24757785 | 0.23 |
ENSDART00000078225
|
vtg5
|
vitellogenin 5 |
| chr2_+_47623202 | 0.22 |
ENSDART00000154465
|
si:ch211-165b10.3
|
si:ch211-165b10.3 |
| chr2_+_56657804 | 0.22 |
ENSDART00000113964
|
POLR2E (1 of many)
|
RNA polymerase II subunit E |
| chr18_-_15532016 | 0.22 |
ENSDART00000165279
|
ppfibp1b
|
PTPRF interacting protein, binding protein 1b (liprin beta 1) |
| chr18_+_13792490 | 0.21 |
ENSDART00000136754
|
cdh13
|
cadherin 13, H-cadherin (heart) |
| chr8_-_19051906 | 0.21 |
ENSDART00000089024
|
sema6bb
|
sema domain, transmembrane domain (TM), and cytoplasmic domain, (semaphorin) 6Bb |
| chr10_-_36591511 | 0.21 |
ENSDART00000063347
|
slc1a3b
|
solute carrier family 1 (glial high affinity glutamate transporter), member 3b |
| chr11_-_4235811 | 0.20 |
ENSDART00000121716
|
si:ch211-236d3.4
|
si:ch211-236d3.4 |
| chr17_+_45737992 | 0.20 |
ENSDART00000135073
ENSDART00000143525 ENSDART00000158165 ENSDART00000184167 ENSDART00000109525 |
aspg
|
asparaginase homolog (S. cerevisiae) |
| chr16_-_5873866 | 0.20 |
ENSDART00000136936
|
trak1a
|
trafficking protein, kinesin binding 1a |
| chr5_+_24287927 | 0.20 |
ENSDART00000143563
|
zdhhc23a
|
zinc finger, DHHC-type containing 23a |
| chr2_+_9821757 | 0.19 |
ENSDART00000018408
ENSDART00000141227 ENSDART00000144681 ENSDART00000148227 |
anxa13l
|
annexin A13, like |
| chr3_-_35800221 | 0.19 |
ENSDART00000031390
|
caskin1
|
CASK interacting protein 1 |
| chr23_-_27345425 | 0.19 |
ENSDART00000022042
ENSDART00000191870 |
scn8aa
|
sodium channel, voltage gated, type VIII, alpha subunit a |
| chr24_-_24959607 | 0.19 |
ENSDART00000146930
ENSDART00000184482 |
pdk3a
|
pyruvate dehydrogenase kinase, isozyme 3a |
| chr13_+_24280380 | 0.19 |
ENSDART00000184115
|
acta1b
|
actin, alpha 1b, skeletal muscle |
| chr17_-_23727978 | 0.19 |
ENSDART00000079600
|
minpp1a
|
multiple inositol-polyphosphate phosphatase 1a |
| chr19_-_34873566 | 0.19 |
ENSDART00000016057
|
ctnnal1
|
catenin (cadherin-associated protein), alpha-like 1 |
| chr15_-_18361475 | 0.19 |
ENSDART00000155866
|
zbtb16b
|
zinc finger and BTB domain containing 16b |
| chr25_-_7683316 | 0.18 |
ENSDART00000128820
|
si:ch211-286c4.6
|
si:ch211-286c4.6 |
| chr5_+_44806374 | 0.18 |
ENSDART00000184237
|
ctsla
|
cathepsin La |
| chr10_+_21780250 | 0.18 |
ENSDART00000183782
|
pcdh1g9
|
protocadherin 1 gamma 9 |
| chr1_-_51465730 | 0.18 |
ENSDART00000074284
|
spred2a
|
sprouty-related, EVH1 domain containing 2a |
| chr5_-_65662996 | 0.18 |
ENSDART00000147707
|
camsap1b
|
calmodulin regulated spectrin-associated protein 1b |
| chr6_+_11250316 | 0.18 |
ENSDART00000137122
|
atg9a
|
ATG9 autophagy related 9 homolog A (S. cerevisiae) |
| chr4_-_6806632 | 0.18 |
ENSDART00000139242
|
ifrd1
|
interferon-related developmental regulator 1 |
| chr14_-_33454595 | 0.18 |
ENSDART00000109615
ENSDART00000173267 ENSDART00000185737 ENSDART00000190989 |
tmem255a
|
transmembrane protein 255A |
| chr7_-_19420334 | 0.17 |
ENSDART00000173838
|
si:ch73-71d17.2
|
si:ch73-71d17.2 |
| chr8_+_50534948 | 0.17 |
ENSDART00000174435
|
PEBP4
|
phosphatidylethanolamine binding protein 4 |
| chr16_+_46995398 | 0.17 |
ENSDART00000177679
|
thsd7ab
|
thrombospondin, type I, domain containing 7Ab |
| chr22_-_7129631 | 0.17 |
ENSDART00000171359
|
asic1b
|
acid-sensing (proton-gated) ion channel 1b |
| chr19_-_5865766 | 0.17 |
ENSDART00000191007
|
LO018585.1
|
|
| chr3_-_40945710 | 0.17 |
ENSDART00000138719
ENSDART00000102416 |
cyp3c4
|
cytochrome P450, family 3, subfamily C, polypeptide 4 |
| chr7_-_26270014 | 0.16 |
ENSDART00000079347
|
serpine1
|
serpin peptidase inhibitor, clade E (nexin, plasminogen activator inhibitor type 1), member 1 |
| chr13_+_15656042 | 0.16 |
ENSDART00000134240
|
mark3a
|
MAP/microtubule affinity-regulating kinase 3a |
| chr20_-_40367493 | 0.16 |
ENSDART00000075096
|
smpdl3a
|
sphingomyelin phosphodiesterase, acid-like 3A |
| chr11_+_30057762 | 0.16 |
ENSDART00000164139
|
nhsb
|
Nance-Horan syndrome b (congenital cataracts and dental anomalies) |
| chr7_+_3229828 | 0.16 |
ENSDART00000135367
|
CR855393.2
|
|
| chr21_+_9628854 | 0.16 |
ENSDART00000161753
ENSDART00000160711 |
mapk10
|
mitogen-activated protein kinase 10 |
| chr13_-_21688176 | 0.16 |
ENSDART00000063825
|
sprn
|
shadow of prion protein |
| chr3_+_17314997 | 0.16 |
ENSDART00000139763
|
stat5a
|
signal transducer and activator of transcription 5a |
| chr15_-_30857350 | 0.16 |
ENSDART00000138988
|
akap1b
|
A kinase (PRKA) anchor protein 1b |
| chr24_+_25692802 | 0.16 |
ENSDART00000190493
|
camk2n2
|
calcium/calmodulin-dependent protein kinase II inhibitor 2 |
| chr4_+_25607101 | 0.16 |
ENSDART00000133929
|
acot14
|
acyl-CoA thioesterase 14 |
| chr4_-_10599062 | 0.16 |
ENSDART00000048003
|
tspan12
|
tetraspanin 12 |
| chr2_+_37140448 | 0.16 |
ENSDART00000045016
ENSDART00000142940 |
pex19
|
peroxisomal biogenesis factor 19 |
| chr12_+_46605600 | 0.15 |
ENSDART00000123533
|
FADS6
|
fatty acid desaturase 6 |
| chr15_+_627619 | 0.15 |
ENSDART00000157207
|
si:ch211-210b2.1
|
si:ch211-210b2.1 |
| chr6_-_14146979 | 0.15 |
ENSDART00000089564
|
cacnb4b
|
calcium channel, voltage-dependent, beta 4b subunit |
| chr10_+_14963898 | 0.15 |
ENSDART00000187363
ENSDART00000175732 |
si:dkey-88l16.3
|
si:dkey-88l16.3 |
| chr18_-_8313686 | 0.15 |
ENSDART00000182187
|
mapk8ip2
|
mitogen-activated protein kinase 8 interacting protein 2 |
| chr21_+_44857293 | 0.15 |
ENSDART00000134365
ENSDART00000168217 ENSDART00000065083 |
fstl4
|
follistatin-like 4 |
| chr19_-_31522625 | 0.15 |
ENSDART00000158438
ENSDART00000035049 |
necab1
|
N-terminal EF-hand calcium binding protein 1 |
| chr23_+_20803270 | 0.15 |
ENSDART00000097381
|
zgc:154075
|
zgc:154075 |
| chr14_+_15155684 | 0.14 |
ENSDART00000167966
|
zgc:158852
|
zgc:158852 |
| chr4_+_25607743 | 0.14 |
ENSDART00000028297
|
acot14
|
acyl-CoA thioesterase 14 |
| chr16_+_28932038 | 0.14 |
ENSDART00000149480
|
npr1b
|
natriuretic peptide receptor 1b |
| chr1_+_22002649 | 0.14 |
ENSDART00000141317
|
dnah6
|
dynein, axonemal, heavy chain 6 |
| chr16_-_45854882 | 0.14 |
ENSDART00000027013
ENSDART00000128068 |
ntrk1
|
neurotrophic tyrosine kinase, receptor, type 1 |
| chr13_+_681628 | 0.14 |
ENSDART00000016604
|
abcc2
|
ATP-binding cassette, sub-family C (CFTR/MRP), member 2 |
| chr7_-_4461104 | 0.14 |
ENSDART00000023090
ENSDART00000140770 |
slc12a10.1
|
solute carrier family 12 (sodium/potassium/chloride transporters), member 10, tandem duplicate 1 |
| chr23_+_21424747 | 0.14 |
ENSDART00000135440
ENSDART00000104214 |
tas1r1
|
taste receptor, type 1, member 1 |
| chr9_-_22822084 | 0.14 |
ENSDART00000142020
|
neb
|
nebulin |
| chr11_+_37178271 | 0.14 |
ENSDART00000161771
|
itih3b
|
inter-alpha-trypsin inhibitor heavy chain 3b |
| chr14_+_29581710 | 0.14 |
ENSDART00000188820
ENSDART00000193874 |
TENM2
|
si:dkey-34l15.2 |
| chr16_-_2818170 | 0.14 |
ENSDART00000081918
|
acot22
|
acyl-CoA thioesterase 22 |
| chr24_+_5912635 | 0.13 |
ENSDART00000153736
|
pimr63
|
Pim proto-oncogene, serine/threonine kinase, related 63 |
| chr17_+_15433518 | 0.13 |
ENSDART00000026180
|
fabp7a
|
fatty acid binding protein 7, brain, a |
| chr5_+_19314574 | 0.13 |
ENSDART00000133247
|
rusc2
|
RUN and SH3 domain containing 2 |
| chr23_-_39784368 | 0.13 |
ENSDART00000110282
|
si:ch211-286f9.2
|
si:ch211-286f9.2 |
| chr20_-_1174266 | 0.13 |
ENSDART00000023304
|
gabrr2b
|
gamma-aminobutyric acid (GABA) A receptor, rho 2b |
| chr1_+_54908895 | 0.13 |
ENSDART00000145652
|
golga7ba
|
golgin A7 family, member Ba |
| chr5_+_31036911 | 0.13 |
ENSDART00000141463
ENSDART00000162121 |
zzef1
|
zinc finger, ZZ-type with EF hand domain 1 |
| chr17_+_15433671 | 0.13 |
ENSDART00000149568
|
fabp7a
|
fatty acid binding protein 7, brain, a |
| chr9_+_24106469 | 0.13 |
ENSDART00000039295
|
lrrfip1a
|
leucine rich repeat (in FLII) interacting protein 1a |
| chr11_-_34628789 | 0.13 |
ENSDART00000192433
|
mapkapk3
|
mitogen-activated protein kinase-activated protein kinase 3 |
| chr21_-_23307653 | 0.13 |
ENSDART00000140284
ENSDART00000134103 |
zbtb16a
|
zinc finger and BTB domain containing 16a |
| chr2_+_20332044 | 0.13 |
ENSDART00000112131
|
plppr4a
|
phospholipid phosphatase related 4a |
| chr17_-_1407593 | 0.13 |
ENSDART00000157622
ENSDART00000159458 |
zbtb42
|
zinc finger and BTB domain containing 42 |
| chr25_+_13498188 | 0.13 |
ENSDART00000015710
|
snrkb
|
SNF related kinase b |
| chr13_+_2894536 | 0.13 |
ENSDART00000183678
|
CABZ01087629.1
|
|
| chr16_-_28268201 | 0.13 |
ENSDART00000121671
ENSDART00000141911 |
si:dkey-12j5.1
|
si:dkey-12j5.1 |
| chr9_-_48397702 | 0.13 |
ENSDART00000147169
|
zgc:172182
|
zgc:172182 |
| chr25_+_3327071 | 0.13 |
ENSDART00000136131
ENSDART00000133243 |
ldhbb
|
lactate dehydrogenase Bb |
| chr9_-_14273652 | 0.12 |
ENSDART00000135458
|
abcb6b
|
ATP-binding cassette, sub-family B (MDR/TAP), member 6b |
| chr9_-_390874 | 0.12 |
ENSDART00000165324
|
si:dkey-11f4.7
|
si:dkey-11f4.7 |
| chr16_+_28881235 | 0.12 |
ENSDART00000146525
|
chtopb
|
chromatin target of PRMT1b |
| chr12_+_1139690 | 0.12 |
ENSDART00000160442
|
CABZ01072885.1
|
|
| chr18_+_41495841 | 0.12 |
ENSDART00000098671
|
si:ch211-203b8.6
|
si:ch211-203b8.6 |
| chr1_-_25966068 | 0.12 |
ENSDART00000137869
ENSDART00000134192 |
synpo2b
|
synaptopodin 2b |
| chr2_-_47431205 | 0.12 |
ENSDART00000014350
ENSDART00000038828 |
pax3a
|
paired box 3a |
| chr5_+_38913621 | 0.12 |
ENSDART00000137112
|
fras1
|
Fraser extracellular matrix complex subunit 1 |
| chr4_-_16824231 | 0.12 |
ENSDART00000014007
|
gys2
|
glycogen synthase 2 |
| chr18_-_7677208 | 0.12 |
ENSDART00000092456
|
shank3a
|
SH3 and multiple ankyrin repeat domains 3a |
| chr13_+_30169681 | 0.12 |
ENSDART00000138326
|
pald1b
|
phosphatase domain containing, paladin 1b |
| chr15_-_33172246 | 0.12 |
ENSDART00000158666
|
nbeab
|
neurobeachin b |
| chr13_-_29421331 | 0.12 |
ENSDART00000150228
|
chata
|
choline O-acetyltransferase a |
| chr11_-_1509773 | 0.12 |
ENSDART00000050762
|
phactr3b
|
phosphatase and actin regulator 3b |
| chr3_-_50177658 | 0.12 |
ENSDART00000135309
|
zgc:114118
|
zgc:114118 |
| chr10_+_33406819 | 0.12 |
ENSDART00000137089
|
ryr1a
|
ryanodine receptor 1a (skeletal) |
| chr3_-_40965328 | 0.12 |
ENSDART00000102393
|
cyp3c2
|
cytochrome P450, family 3, subfamily c, polypeptide 2 |
| chr23_+_10469955 | 0.12 |
ENSDART00000140557
|
tns2a
|
tensin 2a |
| chr13_+_41917606 | 0.12 |
ENSDART00000114741
|
polr1b
|
polymerase (RNA) I polypeptide B |
| chr2_+_24304854 | 0.12 |
ENSDART00000078972
|
fitm1
|
fat storage-inducing transmembrane protein 1 |
| chr12_+_34953038 | 0.11 |
ENSDART00000187022
ENSDART00000123988 ENSDART00000027034 |
qki2
|
QKI, KH domain containing, RNA binding 2 |
| chr18_+_14564085 | 0.11 |
ENSDART00000009363
ENSDART00000141813 ENSDART00000136120 |
si:dkey-246g23.4
|
si:dkey-246g23.4 |
| chr15_+_478524 | 0.11 |
ENSDART00000018062
|
si:ch1073-280e3.1
|
si:ch1073-280e3.1 |
| chr1_-_38709551 | 0.11 |
ENSDART00000128794
|
gpm6ab
|
glycoprotein M6Ab |
| chr23_+_9353552 | 0.11 |
ENSDART00000163298
|
BX511246.1
|
|
| chr6_+_8315050 | 0.11 |
ENSDART00000189987
|
gcdha
|
glutaryl-CoA dehydrogenase a |
| chr1_+_45085194 | 0.11 |
ENSDART00000193863
|
si:ch211-151p13.8
|
si:ch211-151p13.8 |
| chr9_+_40939336 | 0.11 |
ENSDART00000100386
|
mstnb
|
myostatin b |
| chr18_+_1145571 | 0.11 |
ENSDART00000135055
|
rec114
|
REC114 meiotic recombination protein |
| chr21_-_19314618 | 0.11 |
ENSDART00000188744
|
gpat3
|
glycerol-3-phosphate acyltransferase 3 |
| chr5_-_12407194 | 0.11 |
ENSDART00000125291
|
ksr2
|
kinase suppressor of ras 2 |
| chr11_-_41966854 | 0.11 |
ENSDART00000055709
|
her2
|
hairy-related 2 |
| chr3_+_54761569 | 0.11 |
ENSDART00000135913
ENSDART00000180983 |
si:ch211-74m13.1
|
si:ch211-74m13.1 |
| chr8_+_42941555 | 0.11 |
ENSDART00000183206
|
slc23a2
|
solute carrier family 23 (ascorbic acid transporter), member 2 |
| chr8_-_14375890 | 0.11 |
ENSDART00000090306
|
xpr1a
|
xenotropic and polytropic retrovirus receptor 1a |
| chr21_-_14803366 | 0.11 |
ENSDART00000190872
|
si:dkey-11o18.5
|
si:dkey-11o18.5 |
| chr1_-_53407448 | 0.11 |
ENSDART00000160033
ENSDART00000172322 |
elmod2
|
ELMO/CED-12 domain containing 2 |
| chr3_-_40965596 | 0.11 |
ENSDART00000137188
|
cyp3c2
|
cytochrome P450, family 3, subfamily c, polypeptide 2 |
| chr23_+_26946744 | 0.11 |
ENSDART00000115141
|
cacnb3b
|
calcium channel, voltage-dependent, beta 3b |
| chr11_-_23332592 | 0.11 |
ENSDART00000125024
|
golt1a
|
golgi transport 1A |
| chr21_+_39197628 | 0.11 |
ENSDART00000113607
|
cpdb
|
carboxypeptidase D, b |
| chr7_-_24005268 | 0.11 |
ENSDART00000173608
|
si:dkey-183c6.9
|
si:dkey-183c6.9 |
| chr10_-_43392267 | 0.11 |
ENSDART00000142872
|
edil3b
|
EGF-like repeats and discoidin I-like domains 3b |
| chr5_-_7834582 | 0.11 |
ENSDART00000162626
ENSDART00000157661 |
pdlim5a
|
PDZ and LIM domain 5a |
| chr8_+_41035230 | 0.10 |
ENSDART00000141349
|
gpat2
|
glycerol-3-phosphate acyltransferase 2, mitochondrial |
| chr4_-_1720648 | 0.10 |
ENSDART00000103484
|
gas2l3
|
growth arrest-specific 2 like 3 |
| chr24_+_5912898 | 0.10 |
ENSDART00000132686
|
pimr63
|
Pim proto-oncogene, serine/threonine kinase, related 63 |
| chr22_-_20289948 | 0.10 |
ENSDART00000132951
|
si:dkey-110c1.10
|
si:dkey-110c1.10 |
| chr22_+_9027884 | 0.10 |
ENSDART00000171839
|
si:ch211-213a13.2
|
si:ch211-213a13.2 |
| chr15_+_45563656 | 0.10 |
ENSDART00000157501
|
cldn15lb
|
claudin 15-like b |
| chr24_-_6898302 | 0.10 |
ENSDART00000158646
|
dpp6a
|
dipeptidyl-peptidase 6a |
| chr4_+_4267451 | 0.10 |
ENSDART00000192069
|
ano2
|
anoctamin 2 |
| chr24_-_16904803 | 0.10 |
ENSDART00000165972
ENSDART00000149974 ENSDART00000149706 |
mtrr
|
5-methyltetrahydrofolate-homocysteine methyltransferase reductase |
| chr20_-_8443425 | 0.10 |
ENSDART00000083908
|
dab1a
|
Dab, reelin signal transducer, homolog 1a (Drosophila) |
| chr24_-_5911973 | 0.10 |
ENSDART00000077933
ENSDART00000077922 |
pimr64
|
Pim proto-oncogene, serine/threonine kinase, related 64 |
| chr16_-_33816082 | 0.10 |
ENSDART00000181769
|
rspo1
|
R-spondin 1 |
| chr4_-_11751037 | 0.10 |
ENSDART00000102301
|
podxl
|
podocalyxin-like |
| chr24_-_16905018 | 0.10 |
ENSDART00000066759
|
mtrr
|
5-methyltetrahydrofolate-homocysteine methyltransferase reductase |
| chr1_-_19845378 | 0.10 |
ENSDART00000139314
ENSDART00000132958 ENSDART00000147502 |
grhprb
|
glyoxylate reductase/hydroxypyruvate reductase b |
| chr12_+_24561832 | 0.10 |
ENSDART00000088159
|
nrxn1a
|
neurexin 1a |
| chr20_+_34717403 | 0.10 |
ENSDART00000034252
|
pnocb
|
prepronociceptin b |
| chr25_-_3469576 | 0.10 |
ENSDART00000186738
|
hbp1
|
HMG-box transcription factor 1 |
| chr3_+_22059066 | 0.10 |
ENSDART00000155739
|
kansl1b
|
KAT8 regulatory NSL complex subunit 1b |
| chr21_+_45502773 | 0.10 |
ENSDART00000160059
ENSDART00000165704 |
si:dkey-223p19.2
|
si:dkey-223p19.2 |
| chr19_+_31532043 | 0.10 |
ENSDART00000136289
|
tmem64
|
transmembrane protein 64 |
| chr18_+_45573251 | 0.10 |
ENSDART00000191309
|
kifc3
|
kinesin family member C3 |
| chr15_-_21014270 | 0.10 |
ENSDART00000154019
|
si:ch211-212c13.10
|
si:ch211-212c13.10 |
| chr8_+_38415374 | 0.10 |
ENSDART00000085395
|
nkx6.3
|
NK6 homeobox 3 |
| chr15_-_21155641 | 0.10 |
ENSDART00000061098
ENSDART00000046443 |
A2ML1 (1 of many)
|
si:dkey-105h12.2 |
| chr10_+_45089820 | 0.10 |
ENSDART00000175481
|
camk2b2
|
calcium/calmodulin-dependent protein kinase (CaM kinase) II beta 2 |
| chr15_+_14856307 | 0.10 |
ENSDART00000167213
|
diabloa
|
diablo, IAP-binding mitochondrial protein a |
| chr1_-_29758947 | 0.10 |
ENSDART00000049514
ENSDART00000183571 ENSDART00000140345 |
alg11
|
asparagine-linked glycosylation 11 (alpha-1,2-mannosyltransferase) |
| chr14_-_46213571 | 0.10 |
ENSDART00000056734
|
setd7
|
SET domain containing (lysine methyltransferase) 7 |
| chr1_+_53321878 | 0.10 |
ENSDART00000143909
|
tbc1d9
|
TBC1 domain family, member 9 (with GRAM domain) |
| chr25_-_19433244 | 0.10 |
ENSDART00000154778
|
map1ab
|
microtubule-associated protein 1Ab |
| chr7_+_13382852 | 0.09 |
ENSDART00000166318
|
dagla
|
diacylglycerol lipase, alpha |
| chr13_+_23176330 | 0.09 |
ENSDART00000168351
|
sorbs1
|
sorbin and SH3 domain containing 1 |
| chr23_+_19675780 | 0.09 |
ENSDART00000124335
|
smim4
|
small integral membrane protein 4 |
Network of associatons between targets according to the STRING database.
First level regulatory network of hmx2_hmx3a
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.4 | 1.1 | GO:0006145 | purine nucleobase catabolic process(GO:0006145) |
| 0.1 | 0.3 | GO:0007585 | respiratory gaseous exchange(GO:0007585) regulation of respiratory gaseous exchange(GO:0043576) |
| 0.1 | 0.2 | GO:0099538 | synaptic signaling via neuropeptide(GO:0099538) trans-synaptic signaling by neuropeptide(GO:0099540) trans-synaptic signaling by neuropeptide, modulating synaptic transmission(GO:0099551) cellular response to brain-derived neurotrophic factor stimulus(GO:1990416) |
| 0.1 | 0.3 | GO:0051145 | smooth muscle cell differentiation(GO:0051145) |
| 0.1 | 0.4 | GO:0071691 | cardiac muscle thin filament assembly(GO:0071691) |
| 0.1 | 0.4 | GO:0032964 | collagen biosynthetic process(GO:0032964) |
| 0.1 | 0.2 | GO:0010755 | regulation of plasminogen activation(GO:0010755) |
| 0.0 | 0.2 | GO:0007638 | mechanosensory behavior(GO:0007638) |
| 0.0 | 0.2 | GO:0070863 | positive regulation of protein exit from endoplasmic reticulum(GO:0070863) protein localization to endoplasmic reticulum exit site(GO:0070973) |
| 0.0 | 0.2 | GO:0060296 | regulation of cilium movement involved in cell motility(GO:0060295) regulation of cilium beat frequency involved in ciliary motility(GO:0060296) regulation of cilium-dependent cell motility(GO:1902019) |
| 0.0 | 0.2 | GO:0098920 | endocannabinoid signaling pathway(GO:0071926) retrograde trans-synaptic signaling by lipid(GO:0098920) retrograde trans-synaptic signaling by endocannabinoid(GO:0098921) |
| 0.0 | 0.3 | GO:0007288 | sperm axoneme assembly(GO:0007288) |
| 0.0 | 0.1 | GO:1900620 | acetylcholine biosynthetic process(GO:0008292) acetate ester biosynthetic process(GO:1900620) |
| 0.0 | 0.2 | GO:0015919 | peroxisomal membrane transport(GO:0015919) protein import into peroxisome membrane(GO:0045046) |
| 0.0 | 0.2 | GO:1901862 | negative regulation of striated muscle tissue development(GO:0045843) negative regulation of muscle tissue development(GO:1901862) |
| 0.0 | 0.1 | GO:0019852 | L-ascorbic acid transport(GO:0015882) L-ascorbic acid metabolic process(GO:0019852) |
| 0.0 | 0.2 | GO:0098957 | anterograde axonal transport of mitochondrion(GO:0098957) |
| 0.0 | 0.3 | GO:0070836 | membrane raft assembly(GO:0001765) plasma membrane raft assembly(GO:0044854) plasma membrane raft organization(GO:0044857) caveola assembly(GO:0070836) |
| 0.0 | 0.1 | GO:0033631 | cell-cell adhesion mediated by integrin(GO:0033631) regulation of cell-cell adhesion mediated by integrin(GO:0033632) |
| 0.0 | 0.1 | GO:1903673 | mitotic cytokinetic process(GO:1902410) mitotic cleavage furrow formation(GO:1903673) |
| 0.0 | 0.2 | GO:0010801 | regulation of peptidyl-threonine phosphorylation(GO:0010799) negative regulation of peptidyl-threonine phosphorylation(GO:0010801) |
| 0.0 | 0.1 | GO:0060958 | cell proliferation involved in heart valve morphogenesis(GO:0003249) regulation of cell proliferation involved in heart valve morphogenesis(GO:0003250) endocardial cell development(GO:0060958) cell proliferation involved in heart valve development(GO:2000793) |
| 0.0 | 0.1 | GO:0001778 | plasma membrane repair(GO:0001778) |
| 0.0 | 0.1 | GO:0072045 | convergent extension involved in nephron morphogenesis(GO:0072045) |
| 0.0 | 0.1 | GO:0003403 | optic vesicle formation(GO:0003403) |
| 0.0 | 0.3 | GO:0006685 | sphingomyelin catabolic process(GO:0006685) |
| 0.0 | 0.2 | GO:1902262 | apoptotic process involved in patterning of blood vessels(GO:1902262) |
| 0.0 | 0.1 | GO:0042730 | fibrinolysis(GO:0042730) |
| 0.0 | 0.2 | GO:0060397 | JAK-STAT cascade involved in growth hormone signaling pathway(GO:0060397) |
| 0.0 | 0.3 | GO:0034497 | protein localization to pre-autophagosomal structure(GO:0034497) |
| 0.0 | 0.1 | GO:0010265 | SCF complex assembly(GO:0010265) |
| 0.0 | 0.3 | GO:0032481 | positive regulation of type I interferon production(GO:0032481) |
| 0.0 | 0.2 | GO:0071260 | cellular response to mechanical stimulus(GO:0071260) |
| 0.0 | 0.1 | GO:0034729 | histone H3-K79 methylation(GO:0034729) regulation of transcription regulatory region DNA binding(GO:2000677) |
| 0.0 | 0.2 | GO:0031113 | regulation of microtubule polymerization(GO:0031113) |
| 0.0 | 0.1 | GO:0051725 | protein de-ADP-ribosylation(GO:0051725) |
| 0.0 | 0.2 | GO:0071467 | cellular response to pH(GO:0071467) |
| 0.0 | 0.1 | GO:0042984 | amyloid precursor protein biosynthetic process(GO:0042983) regulation of amyloid precursor protein biosynthetic process(GO:0042984) |
| 0.0 | 0.1 | GO:0071918 | urea transmembrane transport(GO:0071918) |
| 0.0 | 0.1 | GO:0000066 | mitochondrial ornithine transport(GO:0000066) mitochondrial L-ornithine transmembrane transport(GO:1990575) |
| 0.0 | 0.1 | GO:0038003 | opioid receptor signaling pathway(GO:0038003) |
| 0.0 | 0.1 | GO:0036076 | ligamentous ossification(GO:0036076) |
| 0.0 | 0.1 | GO:0008635 | activation of cysteine-type endopeptidase activity involved in apoptotic process by cytochrome c(GO:0008635) |
| 0.0 | 0.1 | GO:0097477 | spinal cord motor neuron migration(GO:0097476) lateral motor column neuron migration(GO:0097477) |
| 0.0 | 0.1 | GO:0002446 | neutrophil mediated immunity(GO:0002446) |
| 0.0 | 0.1 | GO:0007405 | neuroblast proliferation(GO:0007405) |
| 0.0 | 0.1 | GO:0021534 | cell proliferation in hindbrain(GO:0021534) |
| 0.0 | 0.1 | GO:0051503 | intracellular nucleoside transport(GO:0015859) purine nucleoside transmembrane transport(GO:0015860) purine nucleotide transport(GO:0015865) ATP transport(GO:0015867) purine ribonucleotide transport(GO:0015868) adenine nucleotide transport(GO:0051503) mitochondrial ATP transmembrane transport(GO:1990544) |
| 0.0 | 0.1 | GO:2000463 | postsynaptic density assembly(GO:0097107) modulation of excitatory postsynaptic potential(GO:0098815) positive regulation of excitatory postsynaptic potential(GO:2000463) |
| 0.0 | 0.1 | GO:0006686 | sphingomyelin biosynthetic process(GO:0006686) |
| 0.0 | 0.1 | GO:1903964 | monounsaturated fatty acid metabolic process(GO:1903964) monounsaturated fatty acid biosynthetic process(GO:1903966) |
| 0.0 | 0.1 | GO:0098795 | mRNA cleavage involved in gene silencing by miRNA(GO:0035279) mRNA cleavage involved in gene silencing(GO:0098795) |
| 0.0 | 0.2 | GO:0071678 | olfactory bulb axon guidance(GO:0071678) |
| 0.0 | 0.0 | GO:0010893 | positive regulation of steroid biosynthetic process(GO:0010893) |
| 0.0 | 0.5 | GO:2000311 | regulation of alpha-amino-3-hydroxy-5-methyl-4-isoxazole propionate selective glutamate receptor activity(GO:2000311) |
| 0.0 | 0.2 | GO:0050667 | homocysteine metabolic process(GO:0050667) |
| 0.0 | 0.1 | GO:1990511 | piRNA biosynthetic process(GO:1990511) |
| 0.0 | 0.2 | GO:0009437 | carnitine metabolic process(GO:0009437) |
| 0.0 | 0.0 | GO:0021985 | neurohypophysis development(GO:0021985) |
| 0.0 | 0.1 | GO:0060217 | hemangioblast cell differentiation(GO:0060217) |
| 0.0 | 0.2 | GO:1902547 | regulation of vascular endothelial growth factor signaling pathway(GO:1900746) regulation of cellular response to vascular endothelial growth factor stimulus(GO:1902547) |
| 0.0 | 0.1 | GO:0016122 | tetraterpenoid metabolic process(GO:0016108) tetraterpenoid biosynthetic process(GO:0016109) carotenoid metabolic process(GO:0016116) carotenoid biosynthetic process(GO:0016117) xanthophyll metabolic process(GO:0016122) xanthophyll biosynthetic process(GO:0016123) zeaxanthin metabolic process(GO:1901825) zeaxanthin biosynthetic process(GO:1901827) |
| 0.0 | 0.1 | GO:0035434 | copper ion transmembrane transport(GO:0035434) |
| 0.0 | 0.1 | GO:1901678 | heme transport(GO:0015886) iron coordination entity transport(GO:1901678) |
| 0.0 | 0.1 | GO:0033539 | fatty acid beta-oxidation using acyl-CoA dehydrogenase(GO:0033539) |
| 0.0 | 0.1 | GO:0046341 | CDP-diacylglycerol biosynthetic process(GO:0016024) CDP-diacylglycerol metabolic process(GO:0046341) |
| 0.0 | 0.1 | GO:0035889 | otolith tethering(GO:0035889) |
| 0.0 | 0.1 | GO:0061469 | regulation of type B pancreatic cell proliferation(GO:0061469) |
| 0.0 | 0.3 | GO:0030878 | thyroid gland development(GO:0030878) |
| 0.0 | 0.2 | GO:0007413 | axonal fasciculation(GO:0007413) |
| 0.0 | 0.4 | GO:0055008 | cardiac muscle tissue morphogenesis(GO:0055008) |
| 0.0 | 0.3 | GO:0042759 | long-chain fatty acid biosynthetic process(GO:0042759) |
| 0.0 | 0.0 | GO:0060571 | invagination involved in gastrulation with mouth forming second(GO:0055109) morphogenesis of an epithelial fold(GO:0060571) |
| 0.0 | 0.1 | GO:0018026 | peptidyl-lysine monomethylation(GO:0018026) |
| 0.0 | 0.2 | GO:0006490 | oligosaccharide-lipid intermediate biosynthetic process(GO:0006490) |
| 0.0 | 0.1 | GO:0045604 | regulation of epidermal cell differentiation(GO:0045604) |
| 0.0 | 0.1 | GO:0051013 | microtubule severing(GO:0051013) |
| 0.0 | 0.0 | GO:0070314 | G1 to G0 transition(GO:0070314) |
| 0.0 | 0.2 | GO:0045162 | clustering of voltage-gated sodium channels(GO:0045162) |
| 0.0 | 0.0 | GO:0034154 | toll-like receptor 7 signaling pathway(GO:0034154) |
| 0.0 | 0.0 | GO:0032782 | canalicular bile acid transport(GO:0015722) bile acid secretion(GO:0032782) |
| 0.0 | 0.1 | GO:0050848 | regulation of calcium-mediated signaling(GO:0050848) |
| 0.0 | 0.1 | GO:1902101 | positive regulation of mitotic metaphase/anaphase transition(GO:0045842) positive regulation of mitotic sister chromatid separation(GO:1901970) positive regulation of metaphase/anaphase transition of cell cycle(GO:1902101) |
| 0.0 | 0.3 | GO:0070536 | protein K63-linked deubiquitination(GO:0070536) |
| 0.0 | 0.1 | GO:0006570 | tyrosine metabolic process(GO:0006570) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.5 | GO:0031258 | lamellipodium membrane(GO:0031258) |
| 0.1 | 0.3 | GO:0030314 | junctional membrane complex(GO:0030314) |
| 0.0 | 0.2 | GO:0036449 | microtubule minus-end(GO:0036449) |
| 0.0 | 0.3 | GO:0045277 | respiratory chain complex IV(GO:0045277) |
| 0.0 | 0.1 | GO:0005790 | smooth endoplasmic reticulum(GO:0005790) |
| 0.0 | 0.1 | GO:0005845 | mRNA cap binding complex(GO:0005845) |
| 0.0 | 0.1 | GO:0000938 | GARP complex(GO:0000938) |
| 0.0 | 0.1 | GO:0005899 | insulin receptor complex(GO:0005899) |
| 0.0 | 0.2 | GO:0005869 | dynactin complex(GO:0005869) |
| 0.0 | 0.2 | GO:0002178 | palmitoyltransferase complex(GO:0002178) |
| 0.0 | 0.1 | GO:0031528 | microvillus membrane(GO:0031528) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.2 | GO:0048403 | brain-derived neurotrophic factor binding(GO:0048403) brain-derived neurotrophic factor-activated receptor activity(GO:0060175) |
| 0.1 | 0.2 | GO:0004377 | GDP-Man:Man3GlcNAc2-PP-Dol alpha-1,2-mannosyltransferase activity(GO:0004377) |
| 0.1 | 0.4 | GO:0004459 | L-lactate dehydrogenase activity(GO:0004459) |
| 0.1 | 0.2 | GO:0030586 | [methionine synthase] reductase activity(GO:0030586) |
| 0.0 | 0.1 | GO:0005030 | neurotrophin receptor activity(GO:0005030) |
| 0.0 | 0.2 | GO:0034416 | bisphosphoglycerate phosphatase activity(GO:0034416) |
| 0.0 | 0.3 | GO:0008126 | acetylesterase activity(GO:0008126) |
| 0.0 | 0.3 | GO:0004392 | heme oxygenase (decyclizing) activity(GO:0004392) |
| 0.0 | 0.2 | GO:0016416 | carnitine O-palmitoyltransferase activity(GO:0004095) O-palmitoyltransferase activity(GO:0016416) |
| 0.0 | 0.2 | GO:0044736 | acid-sensing ion channel activity(GO:0044736) |
| 0.0 | 0.1 | GO:0016618 | hydroxypyruvate reductase activity(GO:0016618) glyoxylate reductase (NADP) activity(GO:0030267) |
| 0.0 | 0.2 | GO:0045735 | nutrient reservoir activity(GO:0045735) |
| 0.0 | 0.1 | GO:0004373 | glycogen (starch) synthase activity(GO:0004373) |
| 0.0 | 0.2 | GO:0004067 | asparaginase activity(GO:0004067) |
| 0.0 | 0.1 | GO:0004361 | glutaryl-CoA dehydrogenase activity(GO:0004361) |
| 0.0 | 0.2 | GO:0004366 | glycerol-3-phosphate O-acyltransferase activity(GO:0004366) |
| 0.0 | 0.1 | GO:0038046 | enkephalin receptor activity(GO:0038046) |
| 0.0 | 0.2 | GO:0008427 | calcium-dependent protein kinase inhibitor activity(GO:0008427) calcium-dependent protein kinase regulator activity(GO:0010858) |
| 0.0 | 0.1 | GO:0048763 | ryanodine-sensitive calcium-release channel activity(GO:0005219) calcium-induced calcium release activity(GO:0048763) |
| 0.0 | 0.2 | GO:0004740 | pyruvate dehydrogenase (acetyl-transferring) kinase activity(GO:0004740) |
| 0.0 | 0.1 | GO:0031151 | histone methyltransferase activity (H3-K79 specific)(GO:0031151) |
| 0.0 | 0.1 | GO:0033188 | sphingomyelin synthase activity(GO:0033188) ceramide cholinephosphotransferase activity(GO:0047493) |
| 0.0 | 0.1 | GO:0016509 | long-chain-3-hydroxyacyl-CoA dehydrogenase activity(GO:0016509) |
| 0.0 | 1.1 | GO:0016814 | hydrolase activity, acting on carbon-nitrogen (but not peptide) bonds, in cyclic amidines(GO:0016814) |
| 0.0 | 0.1 | GO:0003875 | ADP-ribosylarginine hydrolase activity(GO:0003875) |
| 0.0 | 0.1 | GO:0031782 | type 3 melanocortin receptor binding(GO:0031781) type 4 melanocortin receptor binding(GO:0031782) |
| 0.0 | 0.1 | GO:0070883 | pre-miRNA binding(GO:0070883) |
| 0.0 | 0.3 | GO:0016405 | CoA-ligase activity(GO:0016405) |
| 0.0 | 0.3 | GO:0004767 | sphingomyelin phosphodiesterase activity(GO:0004767) |
| 0.0 | 0.4 | GO:0047617 | acyl-CoA hydrolase activity(GO:0047617) |
| 0.0 | 0.1 | GO:0016941 | natriuretic peptide receptor activity(GO:0016941) |
| 0.0 | 0.1 | GO:0015232 | heme transporter activity(GO:0015232) |
| 0.0 | 0.1 | GO:0015018 | galactosylgalactosylxylosylprotein 3-beta-glucuronosyltransferase activity(GO:0015018) |
| 0.0 | 0.3 | GO:0005504 | fatty acid binding(GO:0005504) |
| 0.0 | 0.1 | GO:0005471 | ATP:ADP antiporter activity(GO:0005471) |
| 0.0 | 0.1 | GO:0030160 | GKAP/Homer scaffold activity(GO:0030160) |
| 0.0 | 0.1 | GO:0032896 | stearoyl-CoA 9-desaturase activity(GO:0004768) acyl-CoA desaturase activity(GO:0016215) palmitoyl-CoA 9-desaturase activity(GO:0032896) |
| 0.0 | 0.2 | GO:0016909 | JUN kinase activity(GO:0004705) SAP kinase activity(GO:0016909) |
| 0.0 | 0.1 | GO:0008511 | sodium:potassium:chloride symporter activity(GO:0008511) |
| 0.0 | 0.1 | GO:0030228 | lipoprotein particle receptor activity(GO:0030228) |
| 0.0 | 0.0 | GO:0033130 | acetylcholine receptor binding(GO:0033130) |
| 0.0 | 0.1 | GO:0016262 | protein N-acetylglucosaminyltransferase activity(GO:0016262) protein O-GlcNAc transferase activity(GO:0097363) |
| 0.0 | 0.2 | GO:0000268 | peroxisome targeting sequence binding(GO:0000268) |
| 0.0 | 0.1 | GO:0050251 | retinol isomerase activity(GO:0050251) all-trans-retinyl-ester hydrolase, 11-cis retinol forming activity(GO:0052885) |
| 0.0 | 0.1 | GO:0005375 | copper ion transmembrane transporter activity(GO:0005375) |
| 0.0 | 0.8 | GO:0005178 | integrin binding(GO:0005178) |
| 0.0 | 0.1 | GO:0043560 | insulin-activated receptor activity(GO:0005009) insulin receptor substrate binding(GO:0043560) |
| 0.0 | 0.1 | GO:0043139 | 5'-3' DNA helicase activity(GO:0043139) |
| 0.0 | 0.3 | GO:0050811 | GABA receptor binding(GO:0050811) |
| 0.0 | 0.0 | GO:0000822 | inositol hexakisphosphate binding(GO:0000822) |
| 0.0 | 0.1 | GO:0072518 | Rho-dependent protein serine/threonine kinase activity(GO:0072518) |
| 0.0 | 0.1 | GO:0042030 | ATPase inhibitor activity(GO:0042030) |
| 0.0 | 0.1 | GO:0008281 | sulfonylurea receptor activity(GO:0008281) |
| 0.0 | 0.1 | GO:0017150 | tRNA dihydrouridine synthase activity(GO:0017150) |
| 0.0 | 0.1 | GO:0070697 | activin receptor binding(GO:0070697) |
| 0.0 | 0.0 | GO:0015432 | canalicular bile acid transmembrane transporter activity(GO:0015126) bile acid-exporting ATPase activity(GO:0015432) |
| 0.0 | 0.1 | GO:0003980 | UDP-glucose:glycoprotein glucosyltransferase activity(GO:0003980) |
| 0.0 | 0.3 | GO:0042577 | lipid phosphatase activity(GO:0042577) |
| 0.0 | 0.1 | GO:0035035 | histone acetyltransferase binding(GO:0035035) |
| 0.0 | 0.0 | GO:1901612 | cardiolipin binding(GO:1901612) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.2 | PID IL5 PATHWAY | IL5-mediated signaling events |
| 0.0 | 0.6 | SIG CD40PATHWAYMAP | Genes related to CD40 signaling |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 1.1 | REACTOME PURINE CATABOLISM | Genes involved in Purine catabolism |
| 0.0 | 0.5 | REACTOME CELL EXTRACELLULAR MATRIX INTERACTIONS | Genes involved in Cell-extracellular matrix interactions |
| 0.0 | 0.1 | REACTOME ACETYLCHOLINE NEUROTRANSMITTER RELEASE CYCLE | Genes involved in Acetylcholine Neurotransmitter Release Cycle |
| 0.0 | 0.2 | REACTOME CHYLOMICRON MEDIATED LIPID TRANSPORT | Genes involved in Chylomicron-mediated lipid transport |
| 0.0 | 0.1 | REACTOME P38MAPK EVENTS | Genes involved in p38MAPK events |
| 0.0 | 0.2 | REACTOME CLASS C 3 METABOTROPIC GLUTAMATE PHEROMONE RECEPTORS | Genes involved in Class C/3 (Metabotropic glutamate/pheromone receptors) |
| 0.0 | 0.1 | REACTOME ARMS MEDIATED ACTIVATION | Genes involved in ARMS-mediated activation |
| 0.0 | 0.3 | REACTOME RIP MEDIATED NFKB ACTIVATION VIA DAI | Genes involved in RIP-mediated NFkB activation via DAI |
| 0.0 | 0.5 | REACTOME STRIATED MUSCLE CONTRACTION | Genes involved in Striated Muscle Contraction |
| 0.0 | 0.2 | REACTOME ACTIVATION OF THE AP1 FAMILY OF TRANSCRIPTION FACTORS | Genes involved in Activation of the AP-1 family of transcription factors |
| 0.0 | 0.2 | REACTOME ABCA TRANSPORTERS IN LIPID HOMEOSTASIS | Genes involved in ABCA transporters in lipid homeostasis |
| 0.0 | 0.2 | REACTOME PROLACTIN RECEPTOR SIGNALING | Genes involved in Prolactin receptor signaling |
| 0.0 | 0.2 | REACTOME ABC FAMILY PROTEINS MEDIATED TRANSPORT | Genes involved in ABC-family proteins mediated transport |
| 0.0 | 0.2 | REACTOME ACTIVATED AMPK STIMULATES FATTY ACID OXIDATION IN MUSCLE | Genes involved in Activated AMPK stimulates fatty-acid oxidation in muscle |