Project
PRJNA207719: Tissue specific transcriptome profiling
Navigation
Downloads
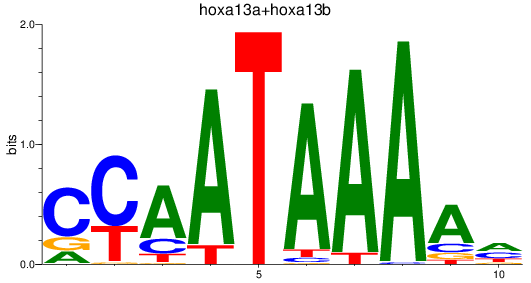
Results for hoxa13a+hoxa13b
Z-value: 1.20
Transcription factors associated with hoxa13a+hoxa13b
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
hoxa13b
|
ENSDARG00000036254 | homeobox A13b |
|
hoxa13a
|
ENSDARG00000100312 | homeobox A13a |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| hoxa13b | dr11_v1_chr16_+_20895904_20895904 | -0.14 | 8.2e-01 | Click! |
| hoxa13a | dr11_v1_chr19_+_19729506_19729506 | -0.12 | 8.5e-01 | Click! |
Activity profile of hoxa13a+hoxa13b motif
Sorted Z-values of hoxa13a+hoxa13b motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr10_+_26612321 | 0.91 |
ENSDART00000134322
|
fhl1b
|
four and a half LIM domains 1b |
| chr11_-_30611814 | 0.64 |
ENSDART00000089803
|
slc8a1a
|
solute carrier family 8 (sodium/calcium exchanger), member 1a |
| chr2_-_24289641 | 0.60 |
ENSDART00000128784
ENSDART00000123565 ENSDART00000141922 ENSDART00000184550 ENSDART00000191469 |
myh7l
|
myosin heavy chain 7-like |
| chr24_-_39826865 | 0.59 |
ENSDART00000089232
|
slc12a7b
|
solute carrier family 12 (potassium/chloride transporter), member 7b |
| chr11_-_18253111 | 0.49 |
ENSDART00000125984
|
mustn1b
|
musculoskeletal, embryonic nuclear protein 1b |
| chr5_+_21211135 | 0.48 |
ENSDART00000088492
|
bmp10
|
bone morphogenetic protein 10 |
| chr1_+_55140970 | 0.46 |
ENSDART00000039807
|
mb
|
myoglobin |
| chr23_+_18722715 | 0.44 |
ENSDART00000137438
|
myh7bb
|
myosin, heavy chain 7B, cardiac muscle, beta b |
| chr10_+_17026870 | 0.43 |
ENSDART00000184529
ENSDART00000157480 |
CR855996.2
|
|
| chr23_+_18722915 | 0.42 |
ENSDART00000025057
|
myh7bb
|
myosin, heavy chain 7B, cardiac muscle, beta b |
| chr7_-_16562200 | 0.42 |
ENSDART00000169093
ENSDART00000173491 |
csrp3
|
cysteine and glycine-rich protein 3 (cardiac LIM protein) |
| chr23_+_7379728 | 0.41 |
ENSDART00000012194
|
gata5
|
GATA binding protein 5 |
| chr4_+_7391400 | 0.40 |
ENSDART00000169111
ENSDART00000186395 |
tnni4a
|
troponin I4a |
| chr22_+_15624371 | 0.37 |
ENSDART00000124868
|
lpl
|
lipoprotein lipase |
| chr16_-_45917322 | 0.37 |
ENSDART00000060822
|
afp4
|
antifreeze protein type IV |
| chr4_-_2380173 | 0.37 |
ENSDART00000177727
|
nap1l1
|
nucleosome assembly protein 1-like 1 |
| chr12_-_20373058 | 0.36 |
ENSDART00000066382
|
aqp8a.1
|
aquaporin 8a, tandem duplicate 1 |
| chr16_-_21785261 | 0.35 |
ENSDART00000078858
|
si:ch73-86n18.1
|
si:ch73-86n18.1 |
| chr5_-_42272517 | 0.35 |
ENSDART00000137692
ENSDART00000164363 |
si:ch211-207c6.2
|
si:ch211-207c6.2 |
| chr13_+_23132666 | 0.35 |
ENSDART00000164639
|
sorbs1
|
sorbin and SH3 domain containing 1 |
| chr13_+_844150 | 0.34 |
ENSDART00000058260
|
gsta.1
|
glutathione S-transferase, alpha tandem duplicate 1 |
| chr13_-_37619159 | 0.34 |
ENSDART00000186348
|
zgc:152791
|
zgc:152791 |
| chr4_+_7391110 | 0.34 |
ENSDART00000160708
ENSDART00000187823 |
tnni4a
|
troponin I4a |
| chr21_-_25756119 | 0.33 |
ENSDART00000002341
|
cldnc
|
claudin c |
| chr11_+_24314148 | 0.33 |
ENSDART00000171491
|
rem1
|
RAS (RAD and GEM)-like GTP-binding 1 |
| chr11_+_24313931 | 0.32 |
ENSDART00000017599
ENSDART00000166045 |
rem1
|
RAS (RAD and GEM)-like GTP-binding 1 |
| chr6_+_43450221 | 0.32 |
ENSDART00000075521
|
zgc:113054
|
zgc:113054 |
| chr23_+_42810055 | 0.32 |
ENSDART00000186647
|
myl9a
|
myosin, light chain 9a, regulatory |
| chr15_+_14856307 | 0.32 |
ENSDART00000167213
|
diabloa
|
diablo, IAP-binding mitochondrial protein a |
| chr6_-_15101477 | 0.30 |
ENSDART00000187713
ENSDART00000124132 |
fhl2b
|
four and a half LIM domains 2b |
| chr7_-_69025306 | 0.30 |
ENSDART00000180796
|
CABZ01057488.2
|
|
| chr18_+_7151783 | 0.30 |
ENSDART00000134872
|
vwf
|
von Willebrand factor |
| chr14_-_21123551 | 0.29 |
ENSDART00000171679
ENSDART00000165882 |
si:dkey-74k8.4
|
si:dkey-74k8.4 |
| chr2_+_52232630 | 0.28 |
ENSDART00000006216
|
plpp2a
|
phospholipid phosphatase 2a |
| chr23_+_17417539 | 0.28 |
ENSDART00000182605
|
BX649300.2
|
|
| chr14_+_36738069 | 0.28 |
ENSDART00000105590
|
tdo2a
|
tryptophan 2,3-dioxygenase a |
| chr6_+_13201358 | 0.27 |
ENSDART00000190290
|
CT009620.1
|
|
| chr13_+_37022601 | 0.27 |
ENSDART00000131800
ENSDART00000041300 |
esr2b
|
estrogen receptor 2b |
| chr10_-_21545091 | 0.27 |
ENSDART00000029122
ENSDART00000132207 |
zgc:165539
|
zgc:165539 |
| chr17_+_53294228 | 0.26 |
ENSDART00000158172
|
si:ch1073-416d2.3
|
si:ch1073-416d2.3 |
| chr10_+_22775253 | 0.26 |
ENSDART00000190141
|
tmem88a
|
transmembrane protein 88 a |
| chr17_-_50022827 | 0.26 |
ENSDART00000161008
|
filip1a
|
filamin A interacting protein 1a |
| chr12_+_27704015 | 0.25 |
ENSDART00000153256
|
cacna1g
|
calcium channel, voltage-dependent, T type, alpha 1G subunit |
| chr15_-_21132480 | 0.25 |
ENSDART00000078734
ENSDART00000157481 |
a2ml
|
alpha-2-macroglobulin-like |
| chr9_-_32300611 | 0.25 |
ENSDART00000127938
|
hspd1
|
heat shock 60 protein 1 |
| chr24_-_36723116 | 0.24 |
ENSDART00000088204
|
si:ch73-334d15.1
|
si:ch73-334d15.1 |
| chr23_+_1078072 | 0.24 |
ENSDART00000159263
ENSDART00000053527 |
slc34a2b
|
solute carrier family 34 (type II sodium/phosphate cotransporter), member 2b |
| chr8_-_19280856 | 0.24 |
ENSDART00000100473
|
zgc:77486
|
zgc:77486 |
| chr20_+_25586099 | 0.24 |
ENSDART00000063122
ENSDART00000134047 |
cyp2p10
|
cytochrome P450, family 2, subfamily P, polypeptide 10 |
| chr19_+_20778011 | 0.24 |
ENSDART00000024208
|
nutf2l
|
nuclear transport factor 2, like |
| chr24_+_21640635 | 0.24 |
ENSDART00000105925
|
gtf3ab
|
general transcription factor IIIA, b |
| chr21_-_2707768 | 0.24 |
ENSDART00000165384
|
CABZ01101739.1
|
|
| chr9_-_32300783 | 0.23 |
ENSDART00000078596
|
hspd1
|
heat shock 60 protein 1 |
| chr24_+_1042594 | 0.23 |
ENSDART00000109117
|
si:dkey-192l18.9
|
si:dkey-192l18.9 |
| chr8_+_30709685 | 0.23 |
ENSDART00000133989
|
upb1
|
ureidopropionase, beta |
| chr8_+_40477264 | 0.23 |
ENSDART00000085559
|
gck
|
glucokinase (hexokinase 4) |
| chr10_-_1276046 | 0.22 |
ENSDART00000169779
|
pdlim5b
|
PDZ and LIM domain 5b |
| chr1_-_33647138 | 0.22 |
ENSDART00000142111
ENSDART00000015547 |
cldng
|
claudin g |
| chr25_+_34641536 | 0.22 |
ENSDART00000167033
|
CABZ01079011.1
|
|
| chr17_+_15788100 | 0.22 |
ENSDART00000027667
|
rragd
|
ras-related GTP binding D |
| chr7_+_48761646 | 0.22 |
ENSDART00000017467
|
acana
|
aggrecan a |
| chr7_+_67429185 | 0.22 |
ENSDART00000162553
ENSDART00000178646 |
kars
|
lysyl-tRNA synthetase |
| chr16_+_23921610 | 0.21 |
ENSDART00000143855
|
apoa4b.3
|
apolipoprotein A-IV b, tandem duplicate 3 |
| chr10_-_22803740 | 0.21 |
ENSDART00000079469
ENSDART00000187968 ENSDART00000122543 |
pcolcea
|
procollagen C-endopeptidase enhancer a |
| chr16_-_33001153 | 0.20 |
ENSDART00000147941
|
me1
|
malic enzyme 1, NADP(+)-dependent, cytosolic |
| chr2_+_16781015 | 0.20 |
ENSDART00000155147
ENSDART00000003845 |
tfa
|
transferrin-a |
| chr7_+_48761875 | 0.20 |
ENSDART00000003690
|
acana
|
aggrecan a |
| chr5_+_1493767 | 0.20 |
ENSDART00000022132
|
haus4
|
HAUS augmin-like complex, subunit 4 |
| chr16_+_16826504 | 0.20 |
ENSDART00000108691
|
kcnj14
|
potassium inwardly-rectifying channel, subfamily J, member 14 |
| chr3_-_60316118 | 0.20 |
ENSDART00000171458
|
si:ch211-214b16.2
|
si:ch211-214b16.2 |
| chr22_-_38819603 | 0.20 |
ENSDART00000104437
|
si:ch211-262h13.5
|
si:ch211-262h13.5 |
| chr20_-_25533739 | 0.20 |
ENSDART00000063064
|
cyp2ad6
|
cytochrome P450, family 2, subfamily AD, polypeptide 6 |
| chr21_-_20840714 | 0.19 |
ENSDART00000144861
ENSDART00000139430 |
c6
|
complement component 6 |
| chr9_+_32301017 | 0.19 |
ENSDART00000127916
ENSDART00000183298 ENSDART00000143103 |
hspe1
|
heat shock 10 protein 1 |
| chr11_-_16394971 | 0.19 |
ENSDART00000180981
ENSDART00000179925 |
lrig1
|
leucine-rich repeats and immunoglobulin-like domains 1 |
| chr23_+_17865953 | 0.19 |
ENSDART00000014723
ENSDART00000140302 ENSDART00000144800 |
naca
|
nascent polypeptide-associated complex alpha subunit |
| chr11_-_45185792 | 0.19 |
ENSDART00000171328
|
si:dkey-93h22.7
|
si:dkey-93h22.7 |
| chr20_+_40150612 | 0.19 |
ENSDART00000143680
ENSDART00000109681 ENSDART00000101041 ENSDART00000121818 |
trdn
|
triadin |
| chr2_+_16780643 | 0.19 |
ENSDART00000125647
ENSDART00000108611 ENSDART00000181245 ENSDART00000163194 |
tfa
|
transferrin-a |
| chr16_-_17300030 | 0.19 |
ENSDART00000149267
|
kel
|
Kell blood group, metallo-endopeptidase |
| chr21_-_27195256 | 0.19 |
ENSDART00000133152
ENSDART00000065401 |
zgc:110782
|
zgc:110782 |
| chr21_+_13301798 | 0.19 |
ENSDART00000140206
|
adora2ab
|
adenosine A2a receptor b |
| chr10_-_35410518 | 0.19 |
ENSDART00000048430
|
gabrr3a
|
gamma-aminobutyric acid (GABA) A receptor, rho 3a |
| chr8_-_18899427 | 0.19 |
ENSDART00000079840
|
rorca
|
RAR-related orphan receptor C a |
| chr21_+_5531138 | 0.19 |
ENSDART00000163825
|
ly6m6
|
lymphocyte antigen 6 family member M6 |
| chr3_+_1005059 | 0.18 |
ENSDART00000132460
|
zgc:172051
|
zgc:172051 |
| chr13_+_7442023 | 0.18 |
ENSDART00000080975
|
tnfaip2b
|
tumor necrosis factor, alpha-induced protein 2b |
| chr7_+_37372479 | 0.18 |
ENSDART00000173652
|
sall1a
|
spalt-like transcription factor 1a |
| chr18_+_35861930 | 0.18 |
ENSDART00000185223
|
ppp1r13l
|
protein phosphatase 1, regulatory subunit 13 like |
| chr5_-_42883761 | 0.18 |
ENSDART00000167374
|
BX323596.2
|
|
| chr7_+_48555400 | 0.18 |
ENSDART00000174474
|
kcnq1
|
potassium voltage-gated channel, KQT-like subfamily, member 1 |
| chr10_+_41199660 | 0.18 |
ENSDART00000125314
|
adrb3b
|
adrenoceptor beta 3b |
| chr1_-_9249943 | 0.17 |
ENSDART00000055011
|
zgc:136472
|
zgc:136472 |
| chr7_-_39537292 | 0.17 |
ENSDART00000173965
|
otog
|
otogelin |
| chr13_-_35892243 | 0.17 |
ENSDART00000002750
ENSDART00000122810 ENSDART00000162399 |
tacc3
|
transforming, acidic coiled-coil containing protein 3 |
| chr5_-_42071505 | 0.17 |
ENSDART00000137224
ENSDART00000193721 |
cxcl11.7
cenpv
|
chemokine (C-X-C motif) ligand 11, duplicate 7 centromere protein V |
| chr6_-_53426773 | 0.17 |
ENSDART00000162791
|
mst1
|
macrophage stimulating 1 |
| chr15_+_17030941 | 0.17 |
ENSDART00000062069
|
plin2
|
perilipin 2 |
| chr20_+_48116476 | 0.17 |
ENSDART00000043938
|
tram2
|
translocation associated membrane protein 2 |
| chr7_+_33132074 | 0.17 |
ENSDART00000073554
|
zgc:153219
|
zgc:153219 |
| chr11_-_25418856 | 0.17 |
ENSDART00000013714
|
gata1a
|
GATA binding protein 1a |
| chr6_-_33916756 | 0.17 |
ENSDART00000137447
ENSDART00000138488 |
nasp
|
nuclear autoantigenic sperm protein (histone-binding) |
| chr5_-_33281046 | 0.17 |
ENSDART00000051344
ENSDART00000138116 |
surf6
|
surfeit 6 |
| chr20_+_51199666 | 0.17 |
ENSDART00000169321
|
hsp90ab1
|
heat shock protein 90, alpha (cytosolic), class B member 1 |
| chr2_-_54639964 | 0.16 |
ENSDART00000100103
|
acss2l
|
acyl-CoA synthetase short chain family member 2 like |
| chr15_-_34567370 | 0.16 |
ENSDART00000099793
|
sostdc1a
|
sclerostin domain containing 1a |
| chr18_-_10713230 | 0.16 |
ENSDART00000183646
|
rbm28
|
RNA binding motif protein 28 |
| chr7_-_30367650 | 0.16 |
ENSDART00000075519
|
aldh1a2
|
aldehyde dehydrogenase 1 family, member A2 |
| chr19_-_42557416 | 0.16 |
ENSDART00000163217
ENSDART00000128278 ENSDART00000162304 ENSDART00000166556 |
si:dkey-267n13.1
|
si:dkey-267n13.1 |
| chr20_-_30369598 | 0.16 |
ENSDART00000144549
|
allc
|
allantoicase |
| chr13_+_24584401 | 0.16 |
ENSDART00000057599
|
fuom
|
fucose mutarotase |
| chr9_+_426392 | 0.16 |
ENSDART00000172515
|
bzw1b
|
basic leucine zipper and W2 domains 1b |
| chr16_+_50969248 | 0.16 |
ENSDART00000172068
|
si:dkeyp-97a10.2
|
si:dkeyp-97a10.2 |
| chr17_-_50020672 | 0.16 |
ENSDART00000188942
|
filip1a
|
filamin A interacting protein 1a |
| chr4_-_77331797 | 0.16 |
ENSDART00000162325
|
slco1f3
|
solute carrier organic anion transporter family, member 1F3 |
| chr11_+_13223625 | 0.16 |
ENSDART00000161275
|
abcb11b
|
ATP-binding cassette, sub-family B (MDR/TAP), member 11b |
| chr4_+_77981553 | 0.16 |
ENSDART00000174108
ENSDART00000122459 |
terfa
|
telomeric repeat binding factor a |
| chr17_-_2690083 | 0.16 |
ENSDART00000135374
|
ptpn21
|
protein tyrosine phosphatase, non-receptor type 21 |
| chr3_+_46764278 | 0.15 |
ENSDART00000136051
ENSDART00000164930 |
prkcsh
|
protein kinase C substrate 80K-H |
| chr20_+_32501748 | 0.15 |
ENSDART00000152944
ENSDART00000021035 |
sec63
|
SEC63 homolog, protein translocation regulator |
| chr19_+_17385561 | 0.15 |
ENSDART00000141397
ENSDART00000143913 ENSDART00000133626 |
rpl15
|
ribosomal protein L15 |
| chr11_-_24016761 | 0.15 |
ENSDART00000153601
ENSDART00000067817 ENSDART00000170531 |
chia.3
|
chitinase, acidic.3 |
| chr10_+_35526528 | 0.15 |
ENSDART00000184110
|
phldb2a
|
pleckstrin homology-like domain, family B, member 2a |
| chr24_+_7782313 | 0.15 |
ENSDART00000111090
|
ptprh
|
protein tyrosine phosphatase, receptor type, h |
| chr16_-_563235 | 0.15 |
ENSDART00000016303
|
irx2a
|
iroquois homeobox 2a |
| chr25_+_9003230 | 0.15 |
ENSDART00000180330
ENSDART00000142917 |
rag1
|
recombination activating gene 1 |
| chr1_-_58562129 | 0.15 |
ENSDART00000159070
|
slc27a1b
|
solute carrier family 27 (fatty acid transporter), member 1b |
| chr7_+_34592526 | 0.15 |
ENSDART00000173959
|
fhod1
|
formin homology 2 domain containing 1 |
| chr22_+_10781894 | 0.15 |
ENSDART00000081183
|
enc3
|
ectodermal-neural cortex 3 |
| chr13_-_36844945 | 0.15 |
ENSDART00000129562
ENSDART00000150899 |
nin
|
ninein (GSK3B interacting protein) |
| chr21_-_26114886 | 0.15 |
ENSDART00000139320
|
nipal4
|
NIPA-like domain containing 4 |
| chr2_+_45548890 | 0.15 |
ENSDART00000113994
|
fndc7a
|
fibronectin type III domain containing 7a |
| chr15_+_45586471 | 0.15 |
ENSDART00000165699
|
cldn15lb
|
claudin 15-like b |
| chr2_+_25315591 | 0.15 |
ENSDART00000161386
|
fndc3ba
|
fibronectin type III domain containing 3Ba |
| chr17_+_7513673 | 0.14 |
ENSDART00000156674
|
klhl10b.1
|
kelch-like family member 10b, tandem duplicate 1 |
| chr4_-_16836006 | 0.14 |
ENSDART00000010777
|
ldhba
|
lactate dehydrogenase Ba |
| chr16_-_560574 | 0.14 |
ENSDART00000148452
|
irx2a
|
iroquois homeobox 2a |
| chr21_-_17290941 | 0.14 |
ENSDART00000147993
|
gfi1b
|
growth factor independent 1B transcription repressor |
| chr10_+_14963898 | 0.14 |
ENSDART00000187363
ENSDART00000175732 |
si:dkey-88l16.3
|
si:dkey-88l16.3 |
| chr18_+_46151505 | 0.14 |
ENSDART00000015034
ENSDART00000141287 |
blvrb
|
biliverdin reductase B |
| chr20_+_52546186 | 0.14 |
ENSDART00000110777
ENSDART00000153377 ENSDART00000153013 ENSDART00000042704 |
eef1db
|
eukaryotic translation elongation factor 1 delta b (guanine nucleotide exchange protein) |
| chr25_+_5035343 | 0.14 |
ENSDART00000011751
|
parvb
|
parvin, beta |
| chr2_+_21048661 | 0.14 |
ENSDART00000156876
|
rreb1b
|
ras responsive element binding protein 1b |
| chr24_-_23671383 | 0.14 |
ENSDART00000144193
|
hnf4g
|
hepatocyte nuclear factor 4, gamma |
| chr14_+_36886950 | 0.14 |
ENSDART00000183719
|
si:ch211-132p1.3
|
si:ch211-132p1.3 |
| chr9_+_41080029 | 0.14 |
ENSDART00000141179
ENSDART00000019289 |
zgc:136439
|
zgc:136439 |
| chr1_-_35924495 | 0.14 |
ENSDART00000184424
|
smad1
|
SMAD family member 1 |
| chr5_-_68244564 | 0.14 |
ENSDART00000169350
|
CABZ01083944.1
|
|
| chr2_+_10147029 | 0.14 |
ENSDART00000139064
ENSDART00000053426 ENSDART00000153678 |
pfn2l
|
profilin 2 like |
| chr7_-_23777445 | 0.14 |
ENSDART00000173527
|
si:ch211-200p22.4
|
si:ch211-200p22.4 |
| chr25_+_13620555 | 0.14 |
ENSDART00000163642
|
si:ch211-172l8.4
|
si:ch211-172l8.4 |
| chr12_+_27231607 | 0.14 |
ENSDART00000066270
|
tmem106a
|
transmembrane protein 106A |
| chr20_-_25522911 | 0.14 |
ENSDART00000063058
|
cyp2n13
|
cytochrome P450, family 2, subfamily N, polypeptide 13 |
| chr25_-_24046870 | 0.14 |
ENSDART00000047569
|
igf2b
|
insulin-like growth factor 2b |
| chr19_-_24555623 | 0.14 |
ENSDART00000176022
|
polr3gla
|
polymerase (RNA) III (DNA directed) polypeptide G like a |
| chr8_+_40476811 | 0.14 |
ENSDART00000129772
|
gck
|
glucokinase (hexokinase 4) |
| chr21_+_20396858 | 0.14 |
ENSDART00000003299
ENSDART00000146615 |
zgc:103482
|
zgc:103482 |
| chr25_-_15040369 | 0.13 |
ENSDART00000159342
ENSDART00000166490 |
pax6a
|
paired box 6a |
| chr2_-_30182353 | 0.13 |
ENSDART00000019149
|
rpl7
|
ribosomal protein L7 |
| chr2_+_49457626 | 0.13 |
ENSDART00000129967
|
sh3gl1a
|
SH3-domain GRB2-like 1a |
| chr11_+_25539698 | 0.13 |
ENSDART00000035602
|
cxxc1b
|
CXXC finger protein 1b |
| chr9_-_38036984 | 0.13 |
ENSDART00000134574
|
hacd2
|
3-hydroxyacyl-CoA dehydratase 2 |
| chr5_-_23200880 | 0.13 |
ENSDART00000051531
|
iqcd
|
IQ motif containing D |
| chr6_-_52348562 | 0.13 |
ENSDART00000142565
ENSDART00000145369 ENSDART00000016890 |
eif6
|
eukaryotic translation initiation factor 6 |
| chr5_-_34993242 | 0.13 |
ENSDART00000134516
ENSDART00000051295 |
btf3
|
basic transcription factor 3 |
| chr2_-_50225411 | 0.13 |
ENSDART00000147117
ENSDART00000000042 |
mcm6l
|
MCM6 minichromosome maintenance deficient 6, like |
| chr4_+_31293362 | 0.13 |
ENSDART00000169781
|
si:dkey-57l19.1
|
si:dkey-57l19.1 |
| chr16_+_11992498 | 0.13 |
ENSDART00000143442
|
p3h3
|
prolyl 3-hydroxylase 3 |
| chr18_+_619619 | 0.13 |
ENSDART00000159846
|
prtga
|
protogenin homolog a (Gallus gallus) |
| chr25_+_336503 | 0.13 |
ENSDART00000160395
|
CU929262.1
|
|
| chr1_+_44395976 | 0.13 |
ENSDART00000159686
ENSDART00000189905 ENSDART00000025145 |
unc93b1
|
unc-93 homolog B1, TLR signaling regulator |
| chr24_+_21621654 | 0.13 |
ENSDART00000002595
|
rpl21
|
ribosomal protein L21 |
| chr22_+_6674992 | 0.13 |
ENSDART00000144054
|
si:ch211-209l18.2
|
si:ch211-209l18.2 |
| chr16_-_7793457 | 0.13 |
ENSDART00000113483
|
trim71
|
tripartite motif containing 71, E3 ubiquitin protein ligase |
| chr6_+_52804267 | 0.13 |
ENSDART00000065681
|
matn4
|
matrilin 4 |
| chr14_-_15171435 | 0.13 |
ENSDART00000159148
ENSDART00000166622 |
si:dkey-77g12.1
|
si:dkey-77g12.1 |
| chr2_-_10877765 | 0.13 |
ENSDART00000100607
|
cdc7
|
cell division cycle 7 homolog (S. cerevisiae) |
| chr6_-_12588044 | 0.13 |
ENSDART00000047896
|
slc15a1b
|
solute carrier family 15 (oligopeptide transporter), member 1b |
| chr23_+_39963599 | 0.12 |
ENSDART00000166539
|
fyco1a
|
FYVE and coiled-coil domain containing 1a |
| chr1_+_51827046 | 0.12 |
ENSDART00000052992
|
dand5
|
DAN domain family, member 5 |
| chr1_+_12177195 | 0.12 |
ENSDART00000146842
ENSDART00000142081 |
stra6l
|
STRA6-like |
| chr14_+_26247319 | 0.12 |
ENSDART00000192793
|
CCDC69
|
coiled-coil domain containing 69 |
| chr11_-_44484952 | 0.12 |
ENSDART00000166674
ENSDART00000188016 |
mfn1b
|
mitofusin 1b |
| chr11_-_25539323 | 0.12 |
ENSDART00000155785
|
si:dkey-245f22.3
|
si:dkey-245f22.3 |
| chr9_+_22364997 | 0.12 |
ENSDART00000188054
ENSDART00000046116 |
crygs3
|
crystallin, gamma S3 |
| chr16_+_29509133 | 0.12 |
ENSDART00000112116
|
ctss2.1
|
cathepsin S, ortholog2, tandem duplicate 1 |
| chr16_+_42464613 | 0.12 |
ENSDART00000162454
|
si:ch211-215k15.4
|
si:ch211-215k15.4 |
| chr12_+_19199735 | 0.12 |
ENSDART00000066393
|
pdap1a
|
pdgfa associated protein 1a |
| chr7_+_38507006 | 0.12 |
ENSDART00000173830
|
slc7a9
|
solute carrier family 7 (amino acid transporter light chain, bo,+ system), member 9 |
| chr23_+_37482727 | 0.12 |
ENSDART00000162737
|
agmat
|
agmatine ureohydrolase (agmatinase) |
| chr18_+_40462445 | 0.12 |
ENSDART00000087645
|
ugt5c2
|
UDP glucuronosyltransferase 5 family, polypeptide C2 |
| chr2_+_49457449 | 0.12 |
ENSDART00000185470
|
sh3gl1a
|
SH3-domain GRB2-like 1a |
| chr1_+_47486104 | 0.12 |
ENSDART00000114746
|
lrrc58a
|
leucine rich repeat containing 58a |
| chr9_+_27348809 | 0.12 |
ENSDART00000147540
|
tlr20.1
|
toll-like receptor 20, tandem duplicate 1 |
| chr7_+_58751504 | 0.12 |
ENSDART00000024185
|
zgc:56231
|
zgc:56231 |
| chr4_-_73190246 | 0.12 |
ENSDART00000170842
|
LO018260.1
|
|
| chr18_+_6558146 | 0.12 |
ENSDART00000169401
|
b4galnt3b
|
beta-1,4-N-acetyl-galactosaminyl transferase 3b |
| chr6_-_46768040 | 0.12 |
ENSDART00000154071
|
igfn1.2
|
immunoglobulin-like and fibronectin type III domain containing 1, tandem duplicate 2 |
| chr1_-_26664840 | 0.12 |
ENSDART00000102500
|
anp32b
|
acidic (leucine-rich) nuclear phosphoprotein 32 family, member B |
| chr13_+_30692669 | 0.12 |
ENSDART00000187818
|
CR762483.1
|
|
Network of associatons between targets according to the STRING database.
First level regulatory network of hoxa13a+hoxa13b
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.5 | GO:0032755 | positive regulation of interleukin-6 production(GO:0032755) |
| 0.1 | 0.6 | GO:1901842 | regulation of high voltage-gated calcium channel activity(GO:1901841) negative regulation of high voltage-gated calcium channel activity(GO:1901842) |
| 0.1 | 0.4 | GO:0048618 | post-embryonic foregut morphogenesis(GO:0048618) |
| 0.1 | 0.5 | GO:0035988 | chondrocyte proliferation(GO:0035988) |
| 0.1 | 0.4 | GO:0035994 | response to muscle stretch(GO:0035994) |
| 0.1 | 0.3 | GO:0061317 | negative regulation of striated muscle cell differentiation(GO:0051154) negative regulation of cardiac muscle tissue development(GO:0055026) cardiac muscle cell fate commitment(GO:0060923) canonical Wnt signaling pathway involved in cardiac muscle cell fate commitment(GO:0061317) regulation of canonical Wnt signaling pathway involved in cardiac muscle cell fate commitment(GO:1901295) negative regulation of canonical Wnt signaling pathway involved in cardiac muscle cell fate commitment(GO:1901296) negative regulation of cardiocyte differentiation(GO:1905208) negative regulation of cardiac muscle cell differentiation(GO:2000726) |
| 0.1 | 0.4 | GO:0003151 | outflow tract morphogenesis(GO:0003151) |
| 0.1 | 0.3 | GO:0008635 | activation of cysteine-type endopeptidase activity involved in apoptotic process by cytochrome c(GO:0008635) |
| 0.1 | 0.5 | GO:0010890 | positive regulation of sequestering of triglyceride(GO:0010890) |
| 0.1 | 1.4 | GO:1990798 | pancreas regeneration(GO:1990798) |
| 0.1 | 0.2 | GO:0042256 | mature ribosome assembly(GO:0042256) |
| 0.1 | 0.3 | GO:0071072 | heat generation(GO:0031649) regulation of heat generation(GO:0031650) positive regulation of heat generation(GO:0031652) regulation of phospholipid biosynthetic process(GO:0071071) negative regulation of phospholipid biosynthetic process(GO:0071072) |
| 0.1 | 0.2 | GO:0008344 | adult locomotory behavior(GO:0008344) |
| 0.1 | 0.2 | GO:0071635 | transforming growth factor beta activation(GO:0036363) regulation of transforming growth factor beta production(GO:0071634) negative regulation of transforming growth factor beta production(GO:0071635) |
| 0.1 | 0.2 | GO:0000256 | allantoin catabolic process(GO:0000256) |
| 0.0 | 0.1 | GO:0018171 | peptidyl-cysteine oxidation(GO:0018171) |
| 0.0 | 0.1 | GO:0032814 | regulation of natural killer cell activation(GO:0032814) positive regulation of natural killer cell activation(GO:0032816) |
| 0.0 | 0.4 | GO:0019441 | tryptophan catabolic process to kynurenine(GO:0019441) |
| 0.0 | 0.2 | GO:0019483 | beta-alanine metabolic process(GO:0019482) beta-alanine biosynthetic process(GO:0019483) |
| 0.0 | 0.2 | GO:0016338 | calcium-independent cell-cell adhesion via plasma membrane cell-adhesion molecules(GO:0016338) |
| 0.0 | 0.4 | GO:0019731 | antibacterial humoral response(GO:0019731) |
| 0.0 | 0.1 | GO:0034154 | toll-like receptor 3 signaling pathway(GO:0034138) toll-like receptor 7 signaling pathway(GO:0034154) |
| 0.0 | 0.2 | GO:0030224 | monocyte differentiation(GO:0030224) |
| 0.0 | 0.2 | GO:0034723 | DNA replication-dependent nucleosome assembly(GO:0006335) DNA replication-dependent nucleosome organization(GO:0034723) |
| 0.0 | 0.1 | GO:0065001 | specification of axis polarity(GO:0065001) |
| 0.0 | 0.2 | GO:0044341 | sodium-dependent phosphate transport(GO:0044341) |
| 0.0 | 0.2 | GO:0061113 | pancreas morphogenesis(GO:0061113) |
| 0.0 | 0.2 | GO:0032782 | canalicular bile acid transport(GO:0015722) bile acid secretion(GO:0032782) |
| 0.0 | 0.2 | GO:0032208 | regulation of telomere maintenance via recombination(GO:0032207) negative regulation of telomere maintenance via recombination(GO:0032208) |
| 0.0 | 0.2 | GO:0002025 | vasodilation by norepinephrine-epinephrine involved in regulation of systemic arterial blood pressure(GO:0002025) |
| 0.0 | 0.1 | GO:0030857 | negative regulation of epithelial cell differentiation(GO:0030857) regulation of lens fiber cell differentiation(GO:1902746) |
| 0.0 | 0.7 | GO:0098703 | calcium ion import across plasma membrane(GO:0098703) calcium ion import into cell(GO:1990035) |
| 0.0 | 0.1 | GO:0030237 | female sex determination(GO:0030237) |
| 0.0 | 0.1 | GO:0003249 | cell proliferation involved in heart valve morphogenesis(GO:0003249) regulation of cell proliferation involved in heart valve morphogenesis(GO:0003250) endocardial cell development(GO:0060958) cell proliferation involved in heart valve development(GO:2000793) |
| 0.0 | 0.2 | GO:0006616 | SRP-dependent cotranslational protein targeting to membrane, translocation(GO:0006616) |
| 0.0 | 0.1 | GO:0033512 | lysine biosynthetic process(GO:0009085) lysine biosynthetic process via aminoadipic acid(GO:0019878) L-lysine catabolic process to acetyl-CoA via saccharopine(GO:0033512) |
| 0.0 | 0.2 | GO:0019427 | acetate metabolic process(GO:0006083) acetyl-CoA biosynthetic process from acetate(GO:0019427) |
| 0.0 | 0.2 | GO:0006004 | fucose metabolic process(GO:0006004) |
| 0.0 | 0.1 | GO:0032776 | DNA methylation on cytosine(GO:0032776) |
| 0.0 | 0.1 | GO:0015682 | ferric iron transport(GO:0015682) transferrin transport(GO:0033572) trivalent inorganic cation transport(GO:0072512) |
| 0.0 | 0.1 | GO:0010586 | miRNA metabolic process(GO:0010586) |
| 0.0 | 0.1 | GO:1903069 | regulation of ER-associated ubiquitin-dependent protein catabolic process(GO:1903069) |
| 0.0 | 0.1 | GO:1901098 | positive regulation of autophagosome maturation(GO:1901098) |
| 0.0 | 0.1 | GO:0071939 | vitamin A transport(GO:0071938) vitamin A import(GO:0071939) |
| 0.0 | 0.1 | GO:0006750 | glutathione biosynthetic process(GO:0006750) |
| 0.0 | 0.1 | GO:0042364 | water-soluble vitamin biosynthetic process(GO:0042364) |
| 0.0 | 0.0 | GO:0003097 | renal water homeostasis(GO:0003091) renal water transport(GO:0003097) |
| 0.0 | 0.5 | GO:0014823 | response to activity(GO:0014823) |
| 0.0 | 0.1 | GO:0034454 | microtubule anchoring at centrosome(GO:0034454) |
| 0.0 | 0.2 | GO:0006032 | chitin catabolic process(GO:0006032) |
| 0.0 | 0.3 | GO:0030520 | intracellular estrogen receptor signaling pathway(GO:0030520) |
| 0.0 | 0.2 | GO:0031204 | posttranslational protein targeting to membrane, translocation(GO:0031204) |
| 0.0 | 0.4 | GO:0019433 | triglyceride catabolic process(GO:0019433) |
| 0.0 | 0.6 | GO:0006884 | cell volume homeostasis(GO:0006884) |
| 0.0 | 0.1 | GO:0014707 | branchiomeric skeletal muscle development(GO:0014707) |
| 0.0 | 0.0 | GO:0030326 | embryonic limb morphogenesis(GO:0030326) |
| 0.0 | 0.1 | GO:0038093 | Fc receptor signaling pathway(GO:0038093) |
| 0.0 | 0.1 | GO:0097039 | protein linear polyubiquitination(GO:0097039) |
| 0.0 | 0.2 | GO:0001973 | adenosine receptor signaling pathway(GO:0001973) purinergic receptor signaling pathway(GO:0035587) G-protein coupled purinergic receptor signaling pathway(GO:0035588) |
| 0.0 | 0.1 | GO:0072386 | plus-end-directed vesicle transport along microtubule(GO:0072383) plus-end-directed organelle transport along microtubule(GO:0072386) |
| 0.0 | 0.1 | GO:0051561 | positive regulation of mitochondrial calcium ion concentration(GO:0051561) |
| 0.0 | 0.2 | GO:0051601 | exocyst localization(GO:0051601) |
| 0.0 | 0.1 | GO:0007635 | chemosensory behavior(GO:0007635) detection of chemical stimulus involved in sensory perception of pain(GO:0050968) |
| 0.0 | 0.1 | GO:0045648 | positive regulation of erythrocyte differentiation(GO:0045648) |
| 0.0 | 0.1 | GO:0009957 | epidermal cell fate specification(GO:0009957) |
| 0.0 | 0.2 | GO:0006108 | malate metabolic process(GO:0006108) |
| 0.0 | 0.1 | GO:0035513 | oxidative RNA demethylation(GO:0035513) |
| 0.0 | 0.4 | GO:0032008 | positive regulation of TOR signaling(GO:0032008) |
| 0.0 | 0.5 | GO:0060030 | dorsal convergence(GO:0060030) |
| 0.0 | 0.3 | GO:0000727 | double-strand break repair via break-induced replication(GO:0000727) |
| 0.0 | 0.1 | GO:0045329 | carnitine biosynthetic process(GO:0045329) |
| 0.0 | 0.1 | GO:0035372 | protein localization to microtubule(GO:0035372) protein localization to microtubule plus-end(GO:1904825) |
| 0.0 | 0.0 | GO:0045041 | protein import into mitochondrial intermembrane space(GO:0045041) |
| 0.0 | 0.1 | GO:0006857 | oligopeptide transport(GO:0006857) |
| 0.0 | 0.2 | GO:0009263 | deoxyribonucleotide biosynthetic process(GO:0009263) |
| 0.0 | 0.2 | GO:0033151 | V(D)J recombination(GO:0033151) |
| 0.0 | 0.1 | GO:0043649 | dicarboxylic acid catabolic process(GO:0043649) |
| 0.0 | 0.1 | GO:0045948 | positive regulation of translational initiation(GO:0045948) |
| 0.0 | 0.1 | GO:0090134 | mesendoderm migration(GO:0090133) cell migration involved in mesendoderm migration(GO:0090134) |
| 0.0 | 0.1 | GO:0071260 | cellular response to mechanical stimulus(GO:0071260) |
| 0.0 | 0.1 | GO:0032218 | riboflavin transport(GO:0032218) |
| 0.0 | 0.0 | GO:0021563 | glossopharyngeal nerve development(GO:0021563) adenohypophysis morphogenesis(GO:0048855) |
| 0.0 | 0.0 | GO:0007638 | mechanosensory behavior(GO:0007638) |
| 0.0 | 0.3 | GO:0043252 | sodium-independent organic anion transport(GO:0043252) |
| 0.0 | 0.1 | GO:0034434 | steroid esterification(GO:0034433) sterol esterification(GO:0034434) cholesterol esterification(GO:0034435) |
| 0.0 | 0.1 | GO:0048210 | Golgi vesicle fusion to target membrane(GO:0048210) |
| 0.0 | 0.1 | GO:0006685 | sphingomyelin catabolic process(GO:0006685) |
| 0.0 | 0.1 | GO:0006552 | leucine catabolic process(GO:0006552) |
| 0.0 | 0.0 | GO:1903961 | regulation of plasma membrane long-chain fatty acid transport(GO:0010746) positive regulation of plasma membrane long-chain fatty acid transport(GO:0010747) plasma membrane long-chain fatty acid transport(GO:0015911) fatty acid transmembrane transport(GO:1902001) positive regulation of anion transmembrane transport(GO:1903961) regulation of fatty acid transport(GO:2000191) positive regulation of fatty acid transport(GO:2000193) |
| 0.0 | 0.1 | GO:0002082 | regulation of oxidative phosphorylation(GO:0002082) |
| 0.0 | 0.0 | GO:0031120 | snRNA pseudouridine synthesis(GO:0031120) |
| 0.0 | 0.0 | GO:0071421 | manganese ion transmembrane transport(GO:0071421) |
| 0.0 | 0.1 | GO:0009446 | putrescine biosynthetic process(GO:0009446) |
| 0.0 | 0.0 | GO:2001244 | positive regulation of intrinsic apoptotic signaling pathway(GO:2001244) |
| 0.0 | 0.1 | GO:0038083 | peptidyl-tyrosine autophosphorylation(GO:0038083) |
| 0.0 | 0.2 | GO:0000463 | maturation of LSU-rRNA from tricistronic rRNA transcript (SSU-rRNA, 5.8S rRNA, LSU-rRNA)(GO:0000463) |
| 0.0 | 0.0 | GO:0010481 | epidermal cell division(GO:0010481) regulation of epidermal cell division(GO:0010482) |
| 0.0 | 0.2 | GO:0086010 | membrane depolarization during action potential(GO:0086010) |
| 0.0 | 0.1 | GO:0001887 | selenium compound metabolic process(GO:0001887) |
| 0.0 | 0.2 | GO:0015693 | magnesium ion transport(GO:0015693) |
| 0.0 | 0.6 | GO:0042737 | drug metabolic process(GO:0017144) drug catabolic process(GO:0042737) exogenous drug catabolic process(GO:0042738) |
| 0.0 | 0.0 | GO:0034080 | chromatin remodeling at centromere(GO:0031055) CENP-A containing nucleosome assembly(GO:0034080) CENP-A containing chromatin organization(GO:0061641) |
| 0.0 | 0.1 | GO:0045638 | negative regulation of myeloid cell differentiation(GO:0045638) |
| 0.0 | 0.1 | GO:0046247 | carotene metabolic process(GO:0016119) carotene catabolic process(GO:0016121) terpene metabolic process(GO:0042214) terpene catabolic process(GO:0046247) |
| 0.0 | 0.1 | GO:0006833 | water transport(GO:0006833) fluid transport(GO:0042044) |
| 0.0 | 0.1 | GO:0045039 | protein import into mitochondrial inner membrane(GO:0045039) |
| 0.0 | 0.5 | GO:0010862 | positive regulation of pathway-restricted SMAD protein phosphorylation(GO:0010862) |
| 0.0 | 0.1 | GO:0018904 | glycerol ether metabolic process(GO:0006662) ether metabolic process(GO:0018904) |
| 0.0 | 0.1 | GO:0043928 | exonucleolytic nuclear-transcribed mRNA catabolic process involved in deadenylation-dependent decay(GO:0043928) |
| 0.0 | 0.4 | GO:0001878 | response to yeast(GO:0001878) |
| 0.0 | 0.0 | GO:0006419 | alanyl-tRNA aminoacylation(GO:0006419) |
| 0.0 | 0.2 | GO:0045743 | positive regulation of fibroblast growth factor receptor signaling pathway(GO:0045743) |
| 0.0 | 0.1 | GO:0060282 | positive regulation of oocyte development(GO:0060282) |
| 0.0 | 0.1 | GO:0042790 | transcription of nuclear large rRNA transcript from RNA polymerase I promoter(GO:0042790) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.3 | GO:0005854 | nascent polypeptide-associated complex(GO:0005854) |
| 0.0 | 0.2 | GO:1990131 | EGO complex(GO:0034448) Gtr1-Gtr2 GTPase complex(GO:1990131) |
| 0.0 | 0.2 | GO:0017177 | glucosidase II complex(GO:0017177) |
| 0.0 | 0.2 | GO:0097519 | DNA recombinase complex(GO:0097519) |
| 0.0 | 0.3 | GO:0070652 | HAUS complex(GO:0070652) |
| 0.0 | 0.4 | GO:0042627 | chylomicron(GO:0042627) |
| 0.0 | 0.2 | GO:0005853 | eukaryotic translation elongation factor 1 complex(GO:0005853) |
| 0.0 | 0.1 | GO:0031166 | integral component of vacuolar membrane(GO:0031166) intrinsic component of vacuolar membrane(GO:0031310) |
| 0.0 | 0.2 | GO:0044613 | nuclear pore central transport channel(GO:0044613) |
| 0.0 | 0.1 | GO:0031428 | box C/D snoRNP complex(GO:0031428) |
| 0.0 | 0.1 | GO:0005797 | Golgi medial cisterna(GO:0005797) |
| 0.0 | 0.1 | GO:0031213 | RSF complex(GO:0031213) |
| 0.0 | 0.2 | GO:0071797 | LUBAC complex(GO:0071797) |
| 0.0 | 0.1 | GO:0042721 | mitochondrial inner membrane protein insertion complex(GO:0042721) |
| 0.0 | 0.1 | GO:0071541 | eukaryotic translation initiation factor 3 complex, eIF3m(GO:0071541) |
| 0.0 | 0.1 | GO:0005662 | DNA replication factor A complex(GO:0005662) |
| 0.0 | 0.3 | GO:0005666 | DNA-directed RNA polymerase III complex(GO:0005666) |
| 0.0 | 1.6 | GO:0016459 | myosin complex(GO:0016459) |
| 0.0 | 0.1 | GO:0030891 | VCB complex(GO:0030891) |
| 0.0 | 0.2 | GO:0005903 | brush border(GO:0005903) |
| 0.0 | 0.1 | GO:0001405 | presequence translocase-associated import motor(GO:0001405) |
| 0.0 | 0.1 | GO:0098888 | presynaptic endocytic zone(GO:0098833) presynaptic endocytic zone membrane(GO:0098835) extrinsic component of presynaptic membrane(GO:0098888) extrinsic component of presynaptic endocytic zone(GO:0098894) |
| 0.0 | 0.1 | GO:0097431 | mitotic spindle pole(GO:0097431) |
| 0.0 | 0.2 | GO:0005579 | membrane attack complex(GO:0005579) |
| 0.0 | 0.2 | GO:0034751 | aryl hydrocarbon receptor complex(GO:0034751) |
| 0.0 | 0.2 | GO:0030015 | CCR4-NOT core complex(GO:0030015) |
| 0.0 | 0.2 | GO:0000276 | mitochondrial proton-transporting ATP synthase complex, coupling factor F(o)(GO:0000276) |
| 0.0 | 0.1 | GO:0030915 | Smc5-Smc6 complex(GO:0030915) |
| 0.0 | 0.2 | GO:0033017 | sarcoplasmic reticulum membrane(GO:0033017) |
| 0.0 | 1.1 | GO:0030018 | Z disc(GO:0030018) |
| 0.0 | 0.1 | GO:0005736 | DNA-directed RNA polymerase I complex(GO:0005736) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.4 | GO:0004833 | tryptophan 2,3-dioxygenase activity(GO:0004833) |
| 0.1 | 0.4 | GO:0034185 | apolipoprotein binding(GO:0034185) |
| 0.1 | 0.4 | GO:0004340 | glucokinase activity(GO:0004340) hexokinase activity(GO:0004396) glucose binding(GO:0005536) fructokinase activity(GO:0008865) mannokinase activity(GO:0019158) |
| 0.1 | 0.2 | GO:0042806 | fucose binding(GO:0042806) |
| 0.0 | 0.6 | GO:0005432 | calcium:sodium antiporter activity(GO:0005432) |
| 0.0 | 0.4 | GO:0015250 | water channel activity(GO:0015250) |
| 0.0 | 0.2 | GO:0004470 | malic enzyme activity(GO:0004470) malate dehydrogenase (decarboxylating) (NAD+) activity(GO:0004471) malate dehydrogenase (decarboxylating) (NADP+) activity(GO:0004473) |
| 0.0 | 0.2 | GO:0051380 | norepinephrine binding(GO:0051380) |
| 0.0 | 0.2 | GO:0046556 | alpha-L-arabinofuranosidase activity(GO:0046556) |
| 0.0 | 0.3 | GO:0031957 | very long-chain fatty acid-CoA ligase activity(GO:0031957) |
| 0.0 | 0.2 | GO:0005436 | sodium:phosphate symporter activity(GO:0005436) |
| 0.0 | 0.2 | GO:0015432 | canalicular bile acid transmembrane transporter activity(GO:0015126) bile acid-exporting ATPase activity(GO:0015432) |
| 0.0 | 0.3 | GO:1903924 | estradiol binding(GO:1903924) |
| 0.0 | 0.2 | GO:1990238 | double-stranded DNA endodeoxyribonuclease activity(GO:1990238) |
| 0.0 | 0.1 | GO:0008336 | gamma-butyrobetaine dioxygenase activity(GO:0008336) |
| 0.0 | 0.1 | GO:0102344 | 3-hydroxyacyl-CoA dehydratase activity(GO:0018812) 3-hydroxy-behenoyl-CoA dehydratase activity(GO:0102344) 3-hydroxy-lignoceroyl-CoA dehydratase activity(GO:0102345) |
| 0.0 | 0.1 | GO:0031544 | procollagen-proline 3-dioxygenase activity(GO:0019797) peptidyl-proline 3-dioxygenase activity(GO:0031544) |
| 0.0 | 0.2 | GO:0004104 | cholinesterase activity(GO:0004104) |
| 0.0 | 0.2 | GO:0001609 | G-protein coupled adenosine receptor activity(GO:0001609) |
| 0.0 | 0.1 | GO:0004485 | methylcrotonoyl-CoA carboxylase activity(GO:0004485) |
| 0.0 | 0.1 | GO:0034736 | sterol O-acyltransferase activity(GO:0004772) cholesterol O-acyltransferase activity(GO:0034736) |
| 0.0 | 0.2 | GO:0043531 | ADP binding(GO:0043531) |
| 0.0 | 0.1 | GO:0016427 | tRNA (cytosine) methyltransferase activity(GO:0016427) |
| 0.0 | 0.2 | GO:0003987 | acetate-CoA ligase activity(GO:0003987) |
| 0.0 | 0.1 | GO:0031704 | apelin receptor binding(GO:0031704) |
| 0.0 | 0.7 | GO:0042805 | actinin binding(GO:0042805) |
| 0.0 | 0.1 | GO:1990518 | ATP-dependent 3'-5' DNA helicase activity(GO:0043140) single-stranded DNA-dependent ATP-dependent 3'-5' DNA helicase activity(GO:1990518) |
| 0.0 | 0.1 | GO:0004998 | transferrin receptor activity(GO:0004998) |
| 0.0 | 0.2 | GO:0008332 | low voltage-gated calcium channel activity(GO:0008332) |
| 0.0 | 0.1 | GO:0016972 | thiol oxidase activity(GO:0016972) |
| 0.0 | 0.6 | GO:0015379 | cation:chloride symporter activity(GO:0015377) potassium:chloride symporter activity(GO:0015379) |
| 0.0 | 0.1 | GO:0033842 | N-acetyl-beta-glucosaminyl-glycoprotein 4-beta-N-acetylgalactosaminyltransferase activity(GO:0033842) |
| 0.0 | 0.1 | GO:1902945 | metalloendopeptidase activity involved in amyloid precursor protein catabolic process(GO:1902945) |
| 0.0 | 0.5 | GO:0005344 | oxygen transporter activity(GO:0005344) oxygen binding(GO:0019825) |
| 0.0 | 0.9 | GO:0051087 | chaperone binding(GO:0051087) |
| 0.0 | 0.2 | GO:0004568 | chitinase activity(GO:0004568) |
| 0.0 | 0.1 | GO:0043023 | ribosomal large subunit binding(GO:0043023) |
| 0.0 | 0.2 | GO:0016728 | ribonucleoside-diphosphate reductase activity, thioredoxin disulfide as acceptor(GO:0004748) oxidoreductase activity, acting on CH or CH2 groups, disulfide as acceptor(GO:0016728) ribonucleoside-diphosphate reductase activity(GO:0061731) |
| 0.0 | 0.2 | GO:0016813 | hydrolase activity, acting on carbon-nitrogen (but not peptide) bonds, in linear amidines(GO:0016813) |
| 0.0 | 0.2 | GO:0005159 | insulin-like growth factor receptor binding(GO:0005159) |
| 0.0 | 0.2 | GO:0003691 | double-stranded telomeric DNA binding(GO:0003691) |
| 0.0 | 0.1 | GO:0004459 | L-lactate dehydrogenase activity(GO:0004459) |
| 0.0 | 0.2 | GO:0036122 | BMP binding(GO:0036122) |
| 0.0 | 0.1 | GO:0035516 | oxidative DNA demethylase activity(GO:0035516) |
| 0.0 | 0.1 | GO:1990756 | protein binding, bridging involved in substrate recognition for ubiquitination(GO:1990756) |
| 0.0 | 0.0 | GO:0016725 | oxidoreductase activity, acting on CH or CH2 groups(GO:0016725) |
| 0.0 | 0.1 | GO:0071916 | dipeptide transporter activity(GO:0042936) dipeptide transmembrane transporter activity(GO:0071916) |
| 0.0 | 0.1 | GO:0001729 | ceramide kinase activity(GO:0001729) D-erythro-sphingosine kinase activity(GO:0017050) |
| 0.0 | 0.1 | GO:0001096 | TFIIF-class transcription factor binding(GO:0001096) |
| 0.0 | 0.3 | GO:0048038 | quinone binding(GO:0048038) |
| 0.0 | 0.1 | GO:0070324 | thyroid hormone binding(GO:0070324) |
| 0.0 | 0.1 | GO:0098639 | C-X3-C chemokine binding(GO:0019960) protein binding involved in cell-matrix adhesion(GO:0098634) collagen binding involved in cell-matrix adhesion(GO:0098639) |
| 0.0 | 0.5 | GO:0005540 | hyaluronic acid binding(GO:0005540) |
| 0.0 | 0.1 | GO:0032217 | riboflavin transporter activity(GO:0032217) |
| 0.0 | 0.3 | GO:0015347 | sodium-independent organic anion transmembrane transporter activity(GO:0015347) |
| 0.0 | 0.3 | GO:0005246 | calcium channel regulator activity(GO:0005246) |
| 0.0 | 0.1 | GO:0070698 | type I activin receptor binding(GO:0070698) |
| 0.0 | 0.1 | GO:0035198 | miRNA binding(GO:0035198) |
| 0.0 | 0.2 | GO:0004535 | poly(A)-specific ribonuclease activity(GO:0004535) |
| 0.0 | 0.0 | GO:0034513 | box H/ACA snoRNA binding(GO:0034513) |
| 0.0 | 0.1 | GO:0050431 | transforming growth factor beta binding(GO:0050431) |
| 0.0 | 0.2 | GO:0004029 | aldehyde dehydrogenase (NAD) activity(GO:0004029) |
| 0.0 | 0.1 | GO:1990050 | phosphatidic acid transporter activity(GO:1990050) |
| 0.0 | 0.1 | GO:0010436 | beta-carotene 15,15'-monooxygenase activity(GO:0003834) carotenoid dioxygenase activity(GO:0010436) |
| 0.0 | 0.1 | GO:0005545 | 1-phosphatidylinositol binding(GO:0005545) |
| 0.0 | 0.3 | GO:0030165 | PDZ domain binding(GO:0030165) |
| 0.0 | 0.1 | GO:0008798 | beta-aspartyl-peptidase activity(GO:0008798) |
| 0.0 | 0.1 | GO:0015168 | glycerol transmembrane transporter activity(GO:0015168) |
| 0.0 | 0.2 | GO:0008320 | protein transmembrane transporter activity(GO:0008320) |
| 0.0 | 0.0 | GO:0004813 | alanine-tRNA ligase activity(GO:0004813) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.6 | PID TOLL ENDOGENOUS PATHWAY | Endogenous TLR signaling |
| 0.0 | 0.1 | ST JAK STAT PATHWAY | Jak-STAT Pathway |
| 0.0 | 0.1 | ST PHOSPHOINOSITIDE 3 KINASE PATHWAY | PI3K Pathway |
| 0.0 | 0.0 | PID FAS PATHWAY | FAS (CD95) signaling pathway |
| 0.0 | 0.1 | PID ALK2 PATHWAY | ALK2 signaling events |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.2 | REACTOME THE NLRP3 INFLAMMASOME | Genes involved in The NLRP3 inflammasome |
| 0.0 | 0.4 | REACTOME CHYLOMICRON MEDIATED LIPID TRANSPORT | Genes involved in Chylomicron-mediated lipid transport |
| 0.0 | 0.3 | REACTOME PLATELET ADHESION TO EXPOSED COLLAGEN | Genes involved in Platelet Adhesion to exposed collagen |
| 0.0 | 0.7 | REACTOME REGULATION OF GENE EXPRESSION IN BETA CELLS | Genes involved in Regulation of gene expression in beta cells |
| 0.0 | 0.2 | REACTOME PYRIMIDINE CATABOLISM | Genes involved in Pyrimidine catabolism |
| 0.0 | 0.4 | REACTOME SMOOTH MUSCLE CONTRACTION | Genes involved in Smooth Muscle Contraction |
| 0.0 | 0.2 | REACTOME ADVANCED GLYCOSYLATION ENDPRODUCT RECEPTOR SIGNALING | Genes involved in Advanced glycosylation endproduct receptor signaling |
| 0.0 | 0.3 | REACTOME BRANCHED CHAIN AMINO ACID CATABOLISM | Genes involved in Branched-chain amino acid catabolism |
| 0.0 | 0.2 | REACTOME TRAFFICKING AND PROCESSING OF ENDOSOMAL TLR | Genes involved in Trafficking and processing of endosomal TLR |
| 0.0 | 0.1 | REACTOME G BETA GAMMA SIGNALLING THROUGH PI3KGAMMA | Genes involved in G beta:gamma signalling through PI3Kgamma |
| 0.0 | 0.0 | REACTOME RAF MAP KINASE CASCADE | Genes involved in RAF/MAP kinase cascade |
| 0.0 | 0.1 | REACTOME IKK COMPLEX RECRUITMENT MEDIATED BY RIP1 | Genes involved in IKK complex recruitment mediated by RIP1 |
| 0.0 | 0.2 | REACTOME MITOCHONDRIAL TRNA AMINOACYLATION | Genes involved in Mitochondrial tRNA aminoacylation |
| 0.0 | 0.1 | REACTOME REGULATION OF INSULIN LIKE GROWTH FACTOR IGF ACTIVITY BY INSULIN LIKE GROWTH FACTOR BINDING PROTEINS IGFBPS | Genes involved in Regulation of Insulin-like Growth Factor (IGF) Activity by Insulin-like Growth Factor Binding Proteins (IGFBPs) |
| 0.0 | 0.5 | REACTOME MITOCHONDRIAL PROTEIN IMPORT | Genes involved in Mitochondrial Protein Import |
| 0.0 | 0.1 | REACTOME SIGNALING BY BMP | Genes involved in Signaling by BMP |
| 0.0 | 0.1 | REACTOME METABOLISM OF PORPHYRINS | Genes involved in Metabolism of porphyrins |
| 0.0 | 0.1 | REACTOME CELL EXTRACELLULAR MATRIX INTERACTIONS | Genes involved in Cell-extracellular matrix interactions |