Project
PRJNA207719: Tissue specific transcriptome profiling
Navigation
Downloads
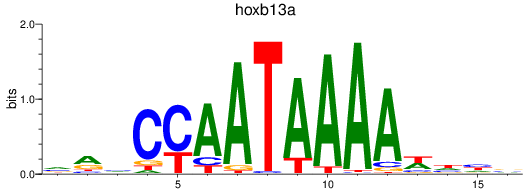
Results for hoxb13a
Z-value: 0.59
Transcription factors associated with hoxb13a
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
hoxb13a
|
ENSDARG00000056015 | homeobox B13a |
Activity profile of hoxb13a motif
Sorted Z-values of hoxb13a motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr20_+_40150612 | 0.74 |
ENSDART00000143680
ENSDART00000109681 ENSDART00000101041 ENSDART00000121818 |
trdn
|
triadin |
| chr11_-_18253111 | 0.66 |
ENSDART00000125984
|
mustn1b
|
musculoskeletal, embryonic nuclear protein 1b |
| chr16_+_14002944 | 0.40 |
ENSDART00000059926
|
si:dkey-85k15.7
|
si:dkey-85k15.7 |
| chr11_-_30611814 | 0.39 |
ENSDART00000089803
|
slc8a1a
|
solute carrier family 8 (sodium/calcium exchanger), member 1a |
| chr11_+_24314148 | 0.38 |
ENSDART00000171491
|
rem1
|
RAS (RAD and GEM)-like GTP-binding 1 |
| chr17_+_15788100 | 0.30 |
ENSDART00000027667
|
rragd
|
ras-related GTP binding D |
| chr1_-_44940830 | 0.24 |
ENSDART00000097500
ENSDART00000134464 ENSDART00000137216 |
tmem176
|
transmembrane protein 176 |
| chr7_-_30367650 | 0.23 |
ENSDART00000075519
|
aldh1a2
|
aldehyde dehydrogenase 1 family, member A2 |
| chr1_+_44941031 | 0.23 |
ENSDART00000141145
|
si:dkey-9i23.16
|
si:dkey-9i23.16 |
| chr12_+_27704015 | 0.23 |
ENSDART00000153256
|
cacna1g
|
calcium channel, voltage-dependent, T type, alpha 1G subunit |
| chr16_-_563235 | 0.21 |
ENSDART00000016303
|
irx2a
|
iroquois homeobox 2a |
| chr20_-_27733683 | 0.19 |
ENSDART00000103317
ENSDART00000138139 |
zgc:153157
|
zgc:153157 |
| chr12_+_41697664 | 0.19 |
ENSDART00000162302
|
bnip3
|
BCL2 interacting protein 3 |
| chr1_-_1627487 | 0.19 |
ENSDART00000166094
|
clic6
|
chloride intracellular channel 6 |
| chr17_-_15382704 | 0.15 |
ENSDART00000005313
|
zgc:85722
|
zgc:85722 |
| chr5_+_27434601 | 0.14 |
ENSDART00000064701
|
loxl2b
|
lysyl oxidase-like 2b |
| chr14_-_24761132 | 0.14 |
ENSDART00000146299
|
slit3
|
slit homolog 3 (Drosophila) |
| chr18_+_44649804 | 0.13 |
ENSDART00000059063
|
ehd2b
|
EH-domain containing 2b |
| chr20_-_52338782 | 0.12 |
ENSDART00000109735
ENSDART00000132941 |
si:ch1073-287p18.1
|
si:ch1073-287p18.1 |
| chr14_+_23184517 | 0.11 |
ENSDART00000181410
|
enox2
|
ecto-NOX disulfide-thiol exchanger 2 |
| chr23_-_19682971 | 0.11 |
ENSDART00000048891
|
zgc:193598
|
zgc:193598 |
| chr9_+_2333550 | 0.11 |
ENSDART00000016417
|
atp5mc3a
|
ATP synthase membrane subunit c locus 3a |
| chr9_-_14683574 | 0.11 |
ENSDART00000144022
|
pard3bb
|
par-3 family cell polarity regulator beta b |
| chr24_+_1042594 | 0.10 |
ENSDART00000109117
|
si:dkey-192l18.9
|
si:dkey-192l18.9 |
| chr6_-_6258451 | 0.10 |
ENSDART00000081966
ENSDART00000125918 |
rtn4a
|
reticulon 4a |
| chr7_-_20865005 | 0.09 |
ENSDART00000190752
|
fis1
|
fission, mitochondrial 1 |
| chr1_+_52481332 | 0.09 |
ENSDART00000074231
|
cldnd1b
|
claudin domain containing 1b |
| chr6_-_12644563 | 0.09 |
ENSDART00000153797
|
dock9b
|
dedicator of cytokinesis 9b |
| chr20_+_29743904 | 0.09 |
ENSDART00000146366
ENSDART00000153154 |
kidins220b
|
kinase D-interacting substrate 220b |
| chr17_-_23412705 | 0.09 |
ENSDART00000126995
|
si:ch211-149k12.3
|
si:ch211-149k12.3 |
| chr3_+_46271911 | 0.08 |
ENSDART00000186557
ENSDART00000113531 |
mkl2b
|
MKL/myocardin-like 2b |
| chr5_-_46980651 | 0.08 |
ENSDART00000181022
ENSDART00000168038 |
edil3a
|
EGF-like repeats and discoidin I-like domains 3a |
| chr16_-_41717063 | 0.08 |
ENSDART00000084610
|
cep85
|
centrosomal protein 85 |
| chr21_-_37733287 | 0.07 |
ENSDART00000157826
|
mpp1
|
membrane protein, palmitoylated 1 |
| chr21_-_27273147 | 0.07 |
ENSDART00000143239
|
mark2a
|
MAP/microtubule affinity-regulating kinase 2a |
| chr2_-_31661609 | 0.07 |
ENSDART00000144383
|
si:ch211-106h4.9
|
si:ch211-106h4.9 |
| chr7_-_56831621 | 0.07 |
ENSDART00000182912
|
senp8
|
SUMO/sentrin peptidase family member, NEDD8 specific |
| chr15_-_28596507 | 0.07 |
ENSDART00000156800
|
si:ch211-225b7.5
|
si:ch211-225b7.5 |
| chr19_-_17526735 | 0.07 |
ENSDART00000189391
|
thrb
|
thyroid hormone receptor beta |
| chr21_-_27272657 | 0.07 |
ENSDART00000040754
ENSDART00000175009 |
mark2a
|
MAP/microtubule affinity-regulating kinase 2a |
| chr7_-_31781339 | 0.06 |
ENSDART00000142666
|
nap1l4b
|
nucleosome assembly protein 1-like 4b |
| chr11_-_16394971 | 0.06 |
ENSDART00000180981
ENSDART00000179925 |
lrig1
|
leucine-rich repeats and immunoglobulin-like domains 1 |
| chr15_+_11381532 | 0.06 |
ENSDART00000124172
|
si:ch73-321d9.2
|
si:ch73-321d9.2 |
| chr23_-_24226533 | 0.06 |
ENSDART00000109134
|
plekhm2
|
pleckstrin homology domain containing, family M (with RUN domain) member 2 |
| chr22_+_11857356 | 0.05 |
ENSDART00000179540
|
mras
|
muscle RAS oncogene homolog |
| chr13_-_49802194 | 0.05 |
ENSDART00000148722
|
b3galnt2
|
beta-1,3-N-acetylgalactosaminyltransferase 2 |
| chr3_+_32443395 | 0.05 |
ENSDART00000188447
|
prr12b
|
proline rich 12b |
| chr7_+_33130639 | 0.05 |
ENSDART00000142450
ENSDART00000173967 ENSDART00000173832 |
zgc:153219
si:ch211-194p6.7
|
zgc:153219 si:ch211-194p6.7 |
| chr16_+_39485608 | 0.05 |
ENSDART00000188085
|
CR855318.1
|
|
| chr17_-_11439815 | 0.04 |
ENSDART00000130105
|
psma3
|
proteasome subunit alpha 3 |
| chr2_-_4070850 | 0.04 |
ENSDART00000159990
|
yme1l1b
|
YME1-like 1b |
| chr5_+_30520249 | 0.04 |
ENSDART00000013431
|
hmbsa
|
hydroxymethylbilane synthase a |
| chr16_-_27628994 | 0.04 |
ENSDART00000157407
|
nacad
|
NAC alpha domain containing |
| chr25_-_35599887 | 0.04 |
ENSDART00000153827
|
clpxb
|
caseinolytic mitochondrial matrix peptidase chaperone subunit b |
| chr25_+_25453221 | 0.04 |
ENSDART00000176822
|
rassf7a
|
Ras association (RalGDS/AF-6) domain family (N-terminal) member 7a |
| chr6_+_35362225 | 0.04 |
ENSDART00000133783
ENSDART00000102483 |
rgs4
|
regulator of G protein signaling 4 |
| chr7_-_40082053 | 0.04 |
ENSDART00000083719
ENSDART00000173687 |
shrprbck1r
|
sharpin and rbck1 related |
| chr1_-_19079957 | 0.04 |
ENSDART00000141795
|
phox2ba
|
paired-like homeobox 2ba |
| chr8_+_50953776 | 0.04 |
ENSDART00000013870
|
zgc:56596
|
zgc:56596 |
| chr22_+_15624371 | 0.03 |
ENSDART00000124868
|
lpl
|
lipoprotein lipase |
| chr12_-_3705862 | 0.03 |
ENSDART00000193864
ENSDART00000185857 |
CABZ01069040.1
|
|
| chr23_-_7799184 | 0.03 |
ENSDART00000190946
ENSDART00000165427 |
myt1b
|
myelin transcription factor 1b |
| chr24_-_20808283 | 0.03 |
ENSDART00000143759
|
vipr1b
|
vasoactive intestinal peptide receptor 1b |
| chr17_-_45150763 | 0.03 |
ENSDART00000155043
ENSDART00000156786 ENSDART00000191147 |
tmed8
|
transmembrane p24 trafficking protein 8 |
| chr17_+_31820401 | 0.03 |
ENSDART00000192607
ENSDART00000157490 |
eef1akmt2
|
EEF1A lysine methyltransferase 2 |
| chr24_-_36238054 | 0.03 |
ENSDART00000155725
|
tmem241
|
transmembrane protein 241 |
| chr6_+_45300746 | 0.02 |
ENSDART00000159176
|
BX942846.1
|
|
| chr19_-_42557416 | 0.02 |
ENSDART00000163217
ENSDART00000128278 ENSDART00000162304 ENSDART00000166556 |
si:dkey-267n13.1
|
si:dkey-267n13.1 |
| chr4_-_4387012 | 0.02 |
ENSDART00000191836
|
CU468826.3
|
Danio rerio U2 small nuclear ribonucleoprotein auxiliary factor 35 kDa subunit-related protein 1-like (LOC100331497), mRNA. |
| chr19_+_42071033 | 0.02 |
ENSDART00000169067
ENSDART00000183436 |
nfyc
|
nuclear transcription factor Y, gamma |
| chr7_-_8324927 | 0.02 |
ENSDART00000102535
|
f13a1b
|
coagulation factor XIII, A1 polypeptide b |
| chr25_+_19106574 | 0.02 |
ENSDART00000067332
|
rlbp1b
|
retinaldehyde binding protein 1b |
| chr19_-_2582858 | 0.02 |
ENSDART00000113829
|
cdca7b
|
cell division cycle associated 7b |
| chr5_-_61970674 | 0.01 |
ENSDART00000143161
|
znf541
|
zinc finger protein 541 |
| chr22_+_2228919 | 0.01 |
ENSDART00000133475
|
znf1161
|
zinc finger protein 1161 |
| chr15_-_17406056 | 0.01 |
ENSDART00000146735
|
tubd1
|
tubulin, delta 1 |
| chr5_-_26181863 | 0.00 |
ENSDART00000098500
|
ccdc125
|
coiled-coil domain containing 125 |
| chr3_-_31069776 | 0.00 |
ENSDART00000167462
|
elob
|
elongin B |
| chr1_+_19515228 | 0.00 |
ENSDART00000103091
|
clrn2
|
clarin 2 |
| chr3_+_60044780 | 0.00 |
ENSDART00000080437
|
zgc:113030
|
zgc:113030 |
| chr16_+_21738194 | 0.00 |
ENSDART00000163688
|
FP085428.1
|
Danio rerio si:ch211-154o6.4 (si:ch211-154o6.4), mRNA. |
| chr25_+_28823952 | 0.00 |
ENSDART00000067072
|
nfybb
|
nuclear transcription factor Y, beta b |
| chr10_-_21953643 | 0.00 |
ENSDART00000188921
ENSDART00000193569 |
FO744833.2
|
|
Network of associatons between targets according to the STRING database.
First level regulatory network of hoxb13a
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.7 | GO:0035988 | chondrocyte proliferation(GO:0035988) |
| 0.1 | 0.4 | GO:1901842 | regulation of high voltage-gated calcium channel activity(GO:1901841) negative regulation of high voltage-gated calcium channel activity(GO:1901842) |
| 0.1 | 0.2 | GO:0061113 | pancreas morphogenesis(GO:0061113) |
| 0.0 | 0.1 | GO:1902455 | negative regulation of stem cell population maintenance(GO:1902455) |
| 0.0 | 0.1 | GO:0051660 | establishment of centrosome localization(GO:0051660) |
| 0.0 | 0.1 | GO:0007624 | ultradian rhythm(GO:0007624) |
| 0.0 | 0.2 | GO:0097345 | mitochondrial outer membrane permeabilization(GO:0097345) |
| 0.0 | 0.1 | GO:0060231 | mesenchymal to epithelial transition(GO:0060231) |
| 0.0 | 0.4 | GO:1990035 | calcium ion import across plasma membrane(GO:0098703) calcium ion import into cell(GO:1990035) |
| 0.0 | 0.1 | GO:0060220 | retinal cone cell fate determination(GO:0042671) eye photoreceptor cell fate commitment(GO:0042706) photoreceptor cell fate determination(GO:0043703) retinal cone cell fate commitment(GO:0046551) photoreceptor cell fate commitment(GO:0046552) camera-type eye photoreceptor cell fate commitment(GO:0060220) |
| 0.0 | 0.1 | GO:0016559 | peroxisome fission(GO:0016559) mitochondrial fragmentation involved in apoptotic process(GO:0043653) |
| 0.0 | 0.2 | GO:0086010 | membrane depolarization during action potential(GO:0086010) |
| 0.0 | 0.2 | GO:0039014 | pronephric nephron tubule epithelial cell differentiation(GO:0035778) cell differentiation involved in pronephros development(GO:0039014) |
| 0.0 | 0.3 | GO:0032008 | positive regulation of TOR signaling(GO:0032008) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.3 | GO:0034448 | EGO complex(GO:0034448) Gtr1-Gtr2 GTPase complex(GO:1990131) |
| 0.0 | 0.7 | GO:0033017 | sarcoplasmic reticulum membrane(GO:0033017) |
| 0.0 | 0.1 | GO:0031231 | integral component of peroxisomal membrane(GO:0005779) intrinsic component of peroxisomal membrane(GO:0031231) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.2 | GO:0008332 | low voltage-gated calcium channel activity(GO:0008332) |
| 0.0 | 0.4 | GO:0005432 | calcium:sodium antiporter activity(GO:0005432) |
| 0.0 | 0.6 | GO:0005246 | calcium channel regulator activity(GO:0005246) |
| 0.0 | 0.1 | GO:0070324 | thyroid hormone binding(GO:0070324) |
| 0.0 | 0.2 | GO:0004029 | aldehyde dehydrogenase (NAD) activity(GO:0004029) |
| 0.0 | 0.0 | GO:0034185 | apolipoprotein binding(GO:0034185) |