Project
PRJNA207719: Tissue specific transcriptome profiling
Navigation
Downloads
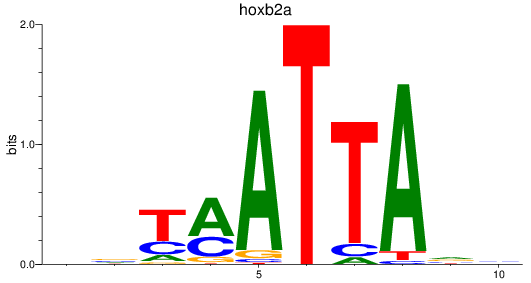
Results for hoxb2a
Z-value: 0.72
Transcription factors associated with hoxb2a
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
hoxb2a
|
ENSDARG00000000175 | homeobox B2a |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| hoxb2a | dr11_v1_chr3_+_23752150_23752150 | 0.68 | 2.1e-01 | Click! |
Activity profile of hoxb2a motif
Sorted Z-values of hoxb2a motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr9_-_21918963 | 0.61 |
ENSDART00000090782
|
lmo7a
|
LIM domain 7a |
| chr23_+_20110086 | 0.55 |
ENSDART00000054664
|
tnnc1b
|
troponin C type 1b (slow) |
| chr20_-_9462433 | 0.51 |
ENSDART00000152674
ENSDART00000040557 |
zgc:101840
|
zgc:101840 |
| chr6_-_54826061 | 0.42 |
ENSDART00000149982
|
tnni1b
|
troponin I type 1b (skeletal, slow) |
| chr22_-_15593824 | 0.41 |
ENSDART00000123125
|
tpm4a
|
tropomyosin 4a |
| chr12_+_23912074 | 0.37 |
ENSDART00000152864
|
svila
|
supervillin a |
| chr4_-_16833518 | 0.36 |
ENSDART00000179867
|
ldhba
|
lactate dehydrogenase Ba |
| chr14_+_17376940 | 0.35 |
ENSDART00000054590
ENSDART00000010148 |
spon2b
|
spondin 2b, extracellular matrix protein |
| chr3_+_30922947 | 0.35 |
ENSDART00000184060
|
cldni
|
claudin i |
| chr11_-_25418856 | 0.35 |
ENSDART00000013714
|
gata1a
|
GATA binding protein 1a |
| chr19_+_43297546 | 0.33 |
ENSDART00000168002
|
laptm5
|
lysosomal protein transmembrane 5 |
| chr14_-_15171435 | 0.32 |
ENSDART00000159148
ENSDART00000166622 |
si:dkey-77g12.1
|
si:dkey-77g12.1 |
| chr9_+_41156818 | 0.31 |
ENSDART00000105764
ENSDART00000147052 |
stat4
|
signal transducer and activator of transcription 4 |
| chr24_-_6078222 | 0.31 |
ENSDART00000146830
|
apbb1ip
|
amyloid beta (A4) precursor protein-binding, family B, member 1 interacting protein |
| chr11_-_20096018 | 0.31 |
ENSDART00000030420
|
ogfrl2
|
opioid growth factor receptor-like 2 |
| chr18_+_18000887 | 0.31 |
ENSDART00000147797
|
si:ch211-212o1.2
|
si:ch211-212o1.2 |
| chr21_-_44565236 | 0.30 |
ENSDART00000159323
|
fundc2
|
fun14 domain containing 2 |
| chr7_-_25895189 | 0.29 |
ENSDART00000173599
ENSDART00000079235 ENSDART00000173786 ENSDART00000173602 ENSDART00000079245 ENSDART00000187568 ENSDART00000173505 |
cd99l2
|
CD99 molecule-like 2 |
| chr5_-_63302944 | 0.29 |
ENSDART00000047110
|
gsnb
|
gelsolin b |
| chr3_+_13624815 | 0.29 |
ENSDART00000161451
|
pglyrp6
|
peptidoglycan recognition protein 6 |
| chr17_+_53425037 | 0.28 |
ENSDART00000154983
|
fabp10b
|
fatty acid binding protein 10b, liver basic |
| chr25_+_28825657 | 0.27 |
ENSDART00000153625
|
nfybb
|
nuclear transcription factor Y, beta b |
| chr2_+_33326522 | 0.26 |
ENSDART00000056655
|
klf17
|
Kruppel-like factor 17 |
| chr16_-_51271962 | 0.26 |
ENSDART00000164021
ENSDART00000046420 |
serpinb1l1
|
serpin peptidase inhibitor, clade B (ovalbumin), member 1, like 1 |
| chr22_-_19552796 | 0.26 |
ENSDART00000148088
ENSDART00000105485 |
si:dkey-78l4.14
|
si:dkey-78l4.14 |
| chr4_+_9177997 | 0.26 |
ENSDART00000057254
ENSDART00000154614 |
nfyba
|
nuclear transcription factor Y, beta a |
| chr5_-_30615901 | 0.25 |
ENSDART00000147769
|
si:ch211-117m20.5
|
si:ch211-117m20.5 |
| chr5_-_41494831 | 0.24 |
ENSDART00000051081
|
eef2l2
|
eukaryotic translation elongation factor 2, like 2 |
| chr1_-_7951002 | 0.24 |
ENSDART00000138187
|
si:dkey-79f11.8
|
si:dkey-79f11.8 |
| chr16_-_28878080 | 0.24 |
ENSDART00000149501
|
si:dkey-239n17.4
|
si:dkey-239n17.4 |
| chr23_-_24493768 | 0.23 |
ENSDART00000135559
ENSDART00000046933 |
sult1st5
|
sulfotransferase family 1, cytosolic sulfotransferase 5 |
| chr16_-_17347727 | 0.22 |
ENSDART00000144392
|
zyx
|
zyxin |
| chr23_-_23401305 | 0.22 |
ENSDART00000078936
|
her9
|
hairy-related 9 |
| chr21_-_11575626 | 0.22 |
ENSDART00000133171
|
cast
|
calpastatin |
| chr9_+_52398531 | 0.21 |
ENSDART00000126215
|
dap1b
|
death associated protein 1b |
| chr12_+_20352400 | 0.21 |
ENSDART00000066383
|
hbae5
|
hemoglobin, alpha embryonic 5 |
| chr10_-_21955231 | 0.21 |
ENSDART00000183695
|
FO744833.2
|
|
| chr3_+_32832538 | 0.21 |
ENSDART00000139410
|
cd2bp2
|
CD2 (cytoplasmic tail) binding protein 2 |
| chr25_+_35553542 | 0.21 |
ENSDART00000113723
|
spi1a
|
Spi-1 proto-oncogene a |
| chr3_-_32596394 | 0.21 |
ENSDART00000103239
|
tspan4b
|
tetraspanin 4b |
| chr21_-_11654422 | 0.20 |
ENSDART00000081614
ENSDART00000132699 |
cast
|
calpastatin |
| chr18_+_18000695 | 0.20 |
ENSDART00000146898
|
si:ch211-212o1.2
|
si:ch211-212o1.2 |
| chr16_+_7242610 | 0.20 |
ENSDART00000081477
|
sri
|
sorcin |
| chr19_-_3931917 | 0.19 |
ENSDART00000162532
|
map7d1b
|
MAP7 domain containing 1b |
| chr21_+_20383837 | 0.19 |
ENSDART00000026430
|
hspb11
|
heat shock protein, alpha-crystallin-related, b11 |
| chr17_-_25649079 | 0.19 |
ENSDART00000130955
|
ppp1cb
|
protein phosphatase 1, catalytic subunit, beta isozyme |
| chr22_+_38037530 | 0.19 |
ENSDART00000012212
|
commd2
|
COMM domain containing 2 |
| chr13_+_27232848 | 0.19 |
ENSDART00000138043
|
rin2
|
Ras and Rab interactor 2 |
| chr23_-_35064785 | 0.19 |
ENSDART00000172240
|
BX294434.1
|
|
| chr4_+_14727018 | 0.18 |
ENSDART00000124189
|
cmasa
|
cytidine monophosphate N-acetylneuraminic acid synthetase a |
| chr23_-_39751234 | 0.18 |
ENSDART00000160853
|
minos1
|
mitochondrial inner membrane organizing system 1 |
| chr17_-_20118145 | 0.18 |
ENSDART00000149737
ENSDART00000165606 |
ryr2b
|
ryanodine receptor 2b (cardiac) |
| chr6_-_3982783 | 0.18 |
ENSDART00000171944
|
slc25a12
|
solute carrier family 25 (aspartate/glutamate carrier), member 12 |
| chr1_+_513986 | 0.18 |
ENSDART00000109083
ENSDART00000081945 |
txnl4b
|
thioredoxin-like 4B |
| chr20_+_37393134 | 0.18 |
ENSDART00000128321
|
adgrg6
|
adhesion G protein-coupled receptor G6 |
| chr22_+_39096911 | 0.18 |
ENSDART00000157127
ENSDART00000153841 |
lmcd1
|
LIM and cysteine-rich domains 1 |
| chr20_+_2039518 | 0.18 |
ENSDART00000043157
|
CABZ01088134.1
|
|
| chr23_-_24394719 | 0.17 |
ENSDART00000044918
|
epha2b
|
eph receptor A2 b |
| chr15_+_6109861 | 0.17 |
ENSDART00000185154
|
PCP4 (1 of many)
|
Purkinje cell protein 4 |
| chr5_-_68093169 | 0.17 |
ENSDART00000051849
|
slc25a11
|
solute carrier family 25 (mitochondrial carrier; oxoglutarate carrier), member 11 |
| chr16_+_21330634 | 0.17 |
ENSDART00000191285
ENSDART00000183267 |
osbpl3b
|
oxysterol binding protein-like 3b |
| chr24_+_17007407 | 0.16 |
ENSDART00000110652
|
zfx
|
zinc finger protein, X-linked |
| chr19_+_2631565 | 0.16 |
ENSDART00000171487
|
fam126a
|
family with sequence similarity 126, member A |
| chr3_+_27798094 | 0.16 |
ENSDART00000075100
ENSDART00000151437 |
carhsp1
|
calcium regulated heat stable protein 1 |
| chr16_+_28994709 | 0.16 |
ENSDART00000088023
|
gon4l
|
gon-4-like (C. elegans) |
| chr10_-_4375190 | 0.16 |
ENSDART00000016102
|
CABZ01073795.1
|
|
| chr23_+_19813677 | 0.16 |
ENSDART00000139192
ENSDART00000142308 |
emd
|
emerin (Emery-Dreifuss muscular dystrophy) |
| chr1_-_45662774 | 0.16 |
ENSDART00000042158
|
serhl
|
serine hydrolase-like |
| chr7_+_17816006 | 0.16 |
ENSDART00000080834
|
eml3
|
echinoderm microtubule associated protein like 3 |
| chr25_+_16116740 | 0.16 |
ENSDART00000139778
|
far1
|
fatty acyl CoA reductase 1 |
| chr24_+_16547035 | 0.16 |
ENSDART00000164319
|
sema5a
|
sema domain, seven thrombospondin repeats (type 1 and type 1-like), transmembrane domain (TM) and short cytoplasmic domain, (semaphorin) 5A |
| chr24_-_39518599 | 0.16 |
ENSDART00000145606
ENSDART00000031486 |
lyrm1
|
LYR motif containing 1 |
| chr20_+_25879826 | 0.16 |
ENSDART00000018519
|
zgc:153896
|
zgc:153896 |
| chr23_-_26522760 | 0.16 |
ENSDART00000142417
ENSDART00000135606 ENSDART00000122668 |
si:dkey-205h13.1
|
si:dkey-205h13.1 |
| chr5_-_26493253 | 0.15 |
ENSDART00000088222
|
arvcfb
|
ARVCF, delta catenin family member b |
| chr5_+_29794058 | 0.15 |
ENSDART00000045410
|
thy1
|
Thy-1 cell surface antigen |
| chr21_-_17296789 | 0.15 |
ENSDART00000192180
|
gfi1b
|
growth factor independent 1B transcription repressor |
| chr23_-_35790235 | 0.15 |
ENSDART00000142369
ENSDART00000141141 ENSDART00000011004 |
mfsd5
|
major facilitator superfamily domain containing 5 |
| chr7_+_17816470 | 0.15 |
ENSDART00000173807
|
eml3
|
echinoderm microtubule associated protein like 3 |
| chr3_+_45365098 | 0.15 |
ENSDART00000052746
ENSDART00000156555 |
ube2ia
|
ubiquitin-conjugating enzyme E2Ia |
| chr23_+_11285662 | 0.15 |
ENSDART00000111028
|
chl1a
|
cell adhesion molecule L1-like a |
| chr8_-_46894362 | 0.15 |
ENSDART00000111124
|
acot7
|
acyl-CoA thioesterase 7 |
| chr2_-_23768818 | 0.14 |
ENSDART00000148685
ENSDART00000191167 |
xirp1
|
xin actin binding repeat containing 1 |
| chr14_+_30795559 | 0.14 |
ENSDART00000006132
|
cfl1
|
cofilin 1 |
| chr5_-_26764880 | 0.14 |
ENSDART00000140392
ENSDART00000134728 |
rnf181
|
ring finger protein 181 |
| chr12_-_48477031 | 0.14 |
ENSDART00000105176
|
ndufb8
|
NADH dehydrogenase (ubiquinone) 1 beta subcomplex, 8 |
| chr22_-_20166660 | 0.14 |
ENSDART00000085913
ENSDART00000188241 |
btbd2a
|
BTB (POZ) domain containing 2a |
| chr17_-_31695217 | 0.14 |
ENSDART00000104332
ENSDART00000143090 |
lin52
|
lin-52 DREAM MuvB core complex component |
| chr10_-_41980797 | 0.13 |
ENSDART00000076575
|
rhof
|
ras homolog family member F |
| chr13_+_22295905 | 0.13 |
ENSDART00000180133
ENSDART00000181125 |
usp54a
|
ubiquitin specific peptidase 54a |
| chrM_+_9735 | 0.13 |
ENSDART00000093613
|
mt-co3
|
cytochrome c oxidase III, mitochondrial |
| chr20_+_46513651 | 0.13 |
ENSDART00000152977
|
zc3h14
|
zinc finger CCCH-type containing 14 |
| chr8_-_23612462 | 0.13 |
ENSDART00000025024
|
slc38a5b
|
solute carrier family 38, member 5b |
| chr20_+_38458084 | 0.13 |
ENSDART00000020153
ENSDART00000135912 |
coq8a
|
coenzyme Q8A |
| chr1_-_23308225 | 0.13 |
ENSDART00000137567
ENSDART00000008201 |
smim14
|
small integral membrane protein 14 |
| chr18_+_17428258 | 0.13 |
ENSDART00000010452
|
zgc:91860
|
zgc:91860 |
| chr8_-_21142550 | 0.13 |
ENSDART00000143192
ENSDART00000186820 ENSDART00000135938 |
cpt2
|
carnitine palmitoyltransferase 2 |
| chr25_-_21031007 | 0.13 |
ENSDART00000138985
|
gnaia
|
guanine nucleotide binding protein (G protein), alpha inhibiting activity polypeptide a |
| chr12_-_33817114 | 0.13 |
ENSDART00000161265
|
twnk
|
twinkle mtDNA helicase |
| chr15_-_1844048 | 0.13 |
ENSDART00000102410
|
taf15
|
TAF15 RNA polymerase II, TATA box binding protein (TBP)-associated factor |
| chr20_-_32270866 | 0.13 |
ENSDART00000153140
|
armc2
|
armadillo repeat containing 2 |
| chr23_-_36446307 | 0.13 |
ENSDART00000136623
|
zgc:174906
|
zgc:174906 |
| chr12_-_30500803 | 0.13 |
ENSDART00000153108
|
nhlrc2
|
NHL repeat containing 2 |
| chr13_+_24750078 | 0.13 |
ENSDART00000021053
|
col17a1b
|
collagen, type XVII, alpha 1b |
| chr6_-_14292307 | 0.12 |
ENSDART00000177852
ENSDART00000061745 |
inpp4ab
|
inositol polyphosphate-4-phosphatase type I Ab |
| chr17_+_37227936 | 0.12 |
ENSDART00000076009
|
hadhab
|
hydroxyacyl-CoA dehydrogenase trifunctional multienzyme complex subunit alpha b |
| chr11_-_28050559 | 0.12 |
ENSDART00000136859
|
ece1
|
endothelin converting enzyme 1 |
| chr6_-_35046735 | 0.12 |
ENSDART00000143649
|
uap1
|
UDP-N-acetylglucosamine pyrophosphorylase 1 |
| chr21_+_17768174 | 0.12 |
ENSDART00000141380
|
rxraa
|
retinoid X receptor, alpha a |
| chr15_-_9272328 | 0.12 |
ENSDART00000172114
|
calm2a
|
calmodulin 2a (phosphorylase kinase, delta) |
| chr21_+_28445052 | 0.12 |
ENSDART00000077871
|
pygma
|
phosphorylase, glycogen, muscle A |
| chr24_+_5237753 | 0.12 |
ENSDART00000106488
ENSDART00000005901 |
plod2
|
procollagen-lysine, 2-oxoglutarate 5-dioxygenase 2 |
| chr7_-_20758825 | 0.12 |
ENSDART00000156717
ENSDART00000182629 ENSDART00000179801 |
chd3
|
chromodomain helicase DNA binding protein 3 |
| chr19_-_19379084 | 0.12 |
ENSDART00000165206
|
smarcc1b
|
SWI/SNF related, matrix associated, actin dependent regulator of chromatin, subfamily c, member 1b |
| chr7_+_37742299 | 0.12 |
ENSDART00000143300
|
brd7
|
bromodomain containing 7 |
| chr14_+_23717165 | 0.12 |
ENSDART00000006373
|
ndfip1
|
Nedd4 family interacting protein 1 |
| chr9_-_38021889 | 0.11 |
ENSDART00000183482
ENSDART00000124333 |
adcy5
|
adenylate cyclase 5 |
| chr5_-_30074332 | 0.11 |
ENSDART00000147963
|
bco2a
|
beta-carotene oxygenase 2a |
| chr19_+_12406583 | 0.11 |
ENSDART00000013865
ENSDART00000151535 |
seh1l
|
SEH1-like (S. cerevisiae) |
| chr20_-_45060241 | 0.11 |
ENSDART00000185227
|
klhl29
|
kelch-like family member 29 |
| chr3_+_45364849 | 0.11 |
ENSDART00000153974
|
ube2ia
|
ubiquitin-conjugating enzyme E2Ia |
| chr13_+_9548849 | 0.11 |
ENSDART00000122695
|
pomk
|
protein-O-mannose kinase |
| chr24_+_23716918 | 0.11 |
ENSDART00000080332
|
cops5
|
COP9 signalosome subunit 5 |
| chr13_-_42400647 | 0.11 |
ENSDART00000043069
|
march5
|
membrane-associated ring finger (C3HC4) 5 |
| chr10_+_16584382 | 0.11 |
ENSDART00000112039
|
CR790388.1
|
|
| chr3_-_26244256 | 0.11 |
ENSDART00000103741
|
ppp4ca
|
protein phosphatase 4, catalytic subunit a |
| chr5_-_26765188 | 0.11 |
ENSDART00000029450
|
rnf181
|
ring finger protein 181 |
| chr6_-_43283122 | 0.11 |
ENSDART00000186022
|
frmd4ba
|
FERM domain containing 4Ba |
| chr8_-_25336589 | 0.11 |
ENSDART00000009682
|
atp5pb
|
ATP synthase peripheral stalk-membrane subunit b |
| chr10_+_22775253 | 0.11 |
ENSDART00000190141
|
tmem88a
|
transmembrane protein 88 a |
| chr3_-_31079186 | 0.11 |
ENSDART00000145636
ENSDART00000140569 |
ELOB (1 of many)
elob
|
elongin B elongin B |
| chr25_+_11456696 | 0.11 |
ENSDART00000171408
|
AGBL1
|
si:ch73-141f14.1 |
| chr2_-_32384683 | 0.11 |
ENSDART00000182942
ENSDART00000141757 |
ubtfl
|
upstream binding transcription factor, like |
| chr4_+_76637548 | 0.11 |
ENSDART00000133799
|
ms4a17a.9
|
membrane-spanning 4-domains, subfamily A, member 17A.9 |
| chr14_+_24935131 | 0.10 |
ENSDART00000170871
|
ppargc1b
|
peroxisome proliferator-activated receptor gamma, coactivator 1 beta |
| chr3_-_34337969 | 0.10 |
ENSDART00000151634
|
tnrc6c1
|
trinucleotide repeat containing 6C1 |
| chr10_-_6976645 | 0.10 |
ENSDART00000123312
|
sh2d4a
|
SH2 domain containing 4A |
| chr7_+_1467863 | 0.10 |
ENSDART00000173433
|
emc4
|
ER membrane protein complex subunit 4 |
| chr7_-_71389375 | 0.10 |
ENSDART00000128928
|
CABZ01074298.1
|
|
| chr20_-_25445237 | 0.10 |
ENSDART00000145034
ENSDART00000018049 |
mrpl19
|
mitochondrial ribosomal protein L19 |
| chr4_-_42408339 | 0.10 |
ENSDART00000172612
|
si:ch211-59d8.3
|
si:ch211-59d8.3 |
| chr9_+_38372216 | 0.10 |
ENSDART00000141895
|
plcd4b
|
phospholipase C, delta 4b |
| chr22_-_2937503 | 0.10 |
ENSDART00000092991
ENSDART00000131110 |
pigx
|
phosphatidylinositol glycan anchor biosynthesis, class X |
| chr24_-_34680956 | 0.10 |
ENSDART00000171009
|
ctnna1
|
catenin (cadherin-associated protein), alpha 1 |
| chr5_+_27897504 | 0.10 |
ENSDART00000130936
|
adam28
|
ADAM metallopeptidase domain 28 |
| chr16_-_47426482 | 0.10 |
ENSDART00000148631
ENSDART00000149723 |
sept7b
|
septin 7b |
| chr22_-_10121880 | 0.09 |
ENSDART00000002348
|
rdh5
|
retinol dehydrogenase 5 (11-cis/9-cis) |
| chr14_-_7137808 | 0.09 |
ENSDART00000054803
|
trpt1
|
tRNA phosphotransferase 1 |
| chr1_-_9123465 | 0.09 |
ENSDART00000081337
|
ndufab1a
|
NADH dehydrogenase (ubiquinone) 1, alpha/beta subcomplex, 1a |
| chr25_+_30298377 | 0.09 |
ENSDART00000153622
|
C11orf96
|
chromosome 11 open reading frame 96 |
| chr1_+_52929185 | 0.09 |
ENSDART00000147683
|
inpp4b
|
inositol polyphosphate-4-phosphatase type II B |
| chr7_+_34592526 | 0.09 |
ENSDART00000173959
|
fhod1
|
formin homology 2 domain containing 1 |
| chr16_-_43344859 | 0.09 |
ENSDART00000058680
|
psma2
|
proteasome subunit alpha 2 |
| chr1_-_10647307 | 0.09 |
ENSDART00000103548
|
si:dkey-31e10.1
|
si:dkey-31e10.1 |
| chr19_+_31904836 | 0.09 |
ENSDART00000162297
ENSDART00000088340 ENSDART00000151280 ENSDART00000151218 |
tpd52
|
tumor protein D52 |
| chr2_-_59145027 | 0.09 |
ENSDART00000128320
|
FO834803.1
|
|
| chr17_+_24722646 | 0.09 |
ENSDART00000138356
|
mtfr1l
|
mitochondrial fission regulator 1-like |
| chr21_+_34088377 | 0.09 |
ENSDART00000170070
|
mtmr1b
|
myotubularin related protein 1b |
| chr16_+_13818743 | 0.08 |
ENSDART00000090191
|
flcn
|
folliculin |
| chr12_+_10631266 | 0.08 |
ENSDART00000161455
|
csf3a
|
colony stimulating factor 3 (granulocyte) a |
| chr1_-_13989643 | 0.08 |
ENSDART00000191046
|
elf2b
|
E74-like factor 2b (ets domain transcription factor) |
| chr22_+_30543437 | 0.08 |
ENSDART00000137983
|
si:dkey-103k4.1
|
si:dkey-103k4.1 |
| chr17_-_31639845 | 0.08 |
ENSDART00000154196
|
znf839
|
zinc finger protein 839 |
| chr8_-_25034411 | 0.08 |
ENSDART00000135973
|
nfyal
|
nuclear transcription factor Y, alpha, like |
| chr21_+_26726936 | 0.08 |
ENSDART00000065392
|
calm2a
|
calmodulin 2a (phosphorylase kinase, delta) |
| chr3_+_4346854 | 0.08 |
ENSDART00000004273
|
si:dkey-73p2.3
|
si:dkey-73p2.3 |
| chr25_+_32755485 | 0.08 |
ENSDART00000162188
|
etfa
|
electron-transfer-flavoprotein, alpha polypeptide |
| chr19_-_30510259 | 0.08 |
ENSDART00000135128
ENSDART00000186169 ENSDART00000182974 ENSDART00000187797 |
bag6l
|
BCL2 associated athanogene 6, like |
| chr5_+_40837539 | 0.08 |
ENSDART00000188279
|
si:dkey-3h3.3
|
si:dkey-3h3.3 |
| chr10_-_7988396 | 0.08 |
ENSDART00000141445
ENSDART00000024282 |
ewsr1a
|
EWS RNA-binding protein 1a |
| chr13_+_21768447 | 0.08 |
ENSDART00000100941
|
chchd1
|
coiled-coil-helix-coiled-coil-helix domain containing 1 |
| chr16_+_54209504 | 0.08 |
ENSDART00000020033
|
xrcc1
|
X-ray repair complementing defective repair in Chinese hamster cells 1 |
| chr3_+_27786601 | 0.08 |
ENSDART00000086994
|
nat15
|
N-acetyltransferase 15 (GCN5-related, putative) |
| chr3_-_34089310 | 0.08 |
ENSDART00000151774
|
ighv5-5
|
immunoglobulin heavy variable 5-5 |
| chr25_-_37262220 | 0.08 |
ENSDART00000153789
ENSDART00000155182 |
rfwd3
|
ring finger and WD repeat domain 3 |
| chr21_+_382400 | 0.08 |
ENSDART00000190785
ENSDART00000124615 |
rad23b
|
RAD23 homolog B, nucleotide excision repair protein |
| chr12_-_3077395 | 0.07 |
ENSDART00000002867
ENSDART00000126315 |
rfng
|
RFNG O-fucosylpeptide 3-beta-N-acetylglucosaminyltransferase |
| chr3_+_26244353 | 0.07 |
ENSDART00000103733
|
atad5a
|
ATPase family, AAA domain containing 5a |
| chr6_+_3280939 | 0.07 |
ENSDART00000151359
|
kdm4aa
|
lysine (K)-specific demethylase 4A, genome duplicate a |
| chr7_+_2236317 | 0.07 |
ENSDART00000075859
|
zgc:172065
|
zgc:172065 |
| chr3_-_19517462 | 0.07 |
ENSDART00000162027
|
CU571315.2
|
|
| chr23_-_1017605 | 0.07 |
ENSDART00000138290
|
cdh26.1
|
cadherin 26, tandem duplicate 1 |
| chr2_-_59247811 | 0.07 |
ENSDART00000141384
|
ftr32
|
finTRIM family, member 32 |
| chr2_-_38256589 | 0.07 |
ENSDART00000173064
|
si:ch211-14a17.10
|
si:ch211-14a17.10 |
| chr11_+_45153104 | 0.07 |
ENSDART00000159204
ENSDART00000177585 |
tk1
|
thymidine kinase 1, soluble |
| chr3_-_34084387 | 0.07 |
ENSDART00000155365
|
ighv4-3
|
immunoglobulin heavy variable 4-3 |
| chr22_-_881725 | 0.07 |
ENSDART00000035514
|
cept1b
|
choline/ethanolamine phosphotransferase 1b |
| chr17_-_6514962 | 0.07 |
ENSDART00000163514
|
dnajc5gb
|
DnaJ (Hsp40) homolog, subfamily C, member 5 gamma b |
| chr19_-_32804535 | 0.07 |
ENSDART00000175613
ENSDART00000052098 |
nt5c1aa
|
5'-nucleotidase, cytosolic IAa |
| chr14_-_38889311 | 0.07 |
ENSDART00000186978
|
zgc:101583
|
zgc:101583 |
| chr8_+_48484455 | 0.07 |
ENSDART00000122737
|
PRDM16
|
si:ch211-263k4.2 |
| chr8_-_50888806 | 0.06 |
ENSDART00000053750
|
acsl2
|
acyl-CoA synthetase long chain family member 2 |
| chr23_+_4741543 | 0.06 |
ENSDART00000144761
|
raf1a
|
Raf-1 proto-oncogene, serine/threonine kinase a |
| chr20_-_1454131 | 0.06 |
ENSDART00000138059
|
ptprk
|
protein tyrosine phosphatase, receptor type, K |
| chr9_+_3175201 | 0.06 |
ENSDART00000179022
ENSDART00000007756 ENSDART00000182108 |
dync1i2a
|
dynein, cytoplasmic 1, intermediate chain 2a |
| chr24_-_40860603 | 0.06 |
ENSDART00000188032
|
CU633479.7
|
|
| chr17_+_16046314 | 0.06 |
ENSDART00000154554
ENSDART00000154338 ENSDART00000155336 |
si:ch73-204p21.2
|
si:ch73-204p21.2 |
| chr23_-_36418708 | 0.06 |
ENSDART00000132273
|
znf740b
|
zinc finger protein 740b |
| chr8_+_11425048 | 0.06 |
ENSDART00000018739
|
tjp2b
|
tight junction protein 2b (zona occludens 2) |
| chr8_-_30338872 | 0.06 |
ENSDART00000137583
|
dock8
|
dedicator of cytokinesis 8 |
Network of associatons between targets according to the STRING database.
First level regulatory network of hoxb2a
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.3 | GO:0016045 | detection of biotic stimulus(GO:0009595) detection of bacterium(GO:0016045) detection of other organism(GO:0098543) detection of external biotic stimulus(GO:0098581) |
| 0.1 | 0.3 | GO:0030224 | monocyte differentiation(GO:0030224) |
| 0.1 | 0.3 | GO:0048785 | hatching gland development(GO:0048785) |
| 0.1 | 0.3 | GO:0036306 | embryonic heart tube elongation(GO:0036306) |
| 0.1 | 0.4 | GO:0097340 | inhibition of cysteine-type endopeptidase activity(GO:0097340) zymogen inhibition(GO:0097341) |
| 0.1 | 0.2 | GO:0032290 | peripheral nervous system myelin formation(GO:0032290) semicircular canal fusion(GO:0060879) |
| 0.0 | 0.1 | GO:0030043 | actin filament fragmentation(GO:0030043) |
| 0.0 | 0.1 | GO:1900364 | negative regulation of mRNA 3'-end processing(GO:0031441) negative regulation of mRNA polyadenylation(GO:1900364) |
| 0.0 | 0.1 | GO:0052576 | carbohydrate localization(GO:0052575) carbohydrate storage(GO:0052576) |
| 0.0 | 0.1 | GO:2000726 | negative regulation of striated muscle cell differentiation(GO:0051154) negative regulation of cardiac muscle tissue development(GO:0055026) cardiac muscle cell fate commitment(GO:0060923) canonical Wnt signaling pathway involved in cardiac muscle cell fate commitment(GO:0061317) regulation of canonical Wnt signaling pathway involved in cardiac muscle cell fate commitment(GO:1901295) negative regulation of canonical Wnt signaling pathway involved in cardiac muscle cell fate commitment(GO:1901296) negative regulation of cardiocyte differentiation(GO:1905208) negative regulation of cardiac muscle cell differentiation(GO:2000726) |
| 0.0 | 0.2 | GO:0015743 | thiosulfate transport(GO:0015709) oxaloacetate transport(GO:0015729) malate transport(GO:0015743) succinate transport(GO:0015744) succinate transmembrane transport(GO:0071422) malate transmembrane transport(GO:0071423) |
| 0.0 | 0.1 | GO:1902024 | L-histidine transmembrane transport(GO:0089709) L-histidine transport(GO:1902024) |
| 0.0 | 0.1 | GO:0017185 | peptidyl-lysine hydroxylation(GO:0017185) |
| 0.0 | 0.1 | GO:0000338 | protein deneddylation(GO:0000338) |
| 0.0 | 0.1 | GO:0000012 | single strand break repair(GO:0000012) |
| 0.0 | 0.7 | GO:0051014 | actin filament severing(GO:0051014) |
| 0.0 | 0.1 | GO:0009120 | deoxyribonucleoside metabolic process(GO:0009120) thymidine metabolic process(GO:0046104) pyrimidine deoxyribonucleoside metabolic process(GO:0046125) |
| 0.0 | 0.1 | GO:0061181 | regulation of chondrocyte development(GO:0061181) |
| 0.0 | 0.2 | GO:0006054 | N-acetylneuraminate metabolic process(GO:0006054) |
| 0.0 | 0.1 | GO:0007288 | sperm axoneme assembly(GO:0007288) |
| 0.0 | 1.3 | GO:0060048 | cardiac muscle contraction(GO:0060048) |
| 0.0 | 0.1 | GO:0070589 | cell wall biogenesis(GO:0042546) cell wall macromolecule biosynthetic process(GO:0044038) cellular component macromolecule biosynthetic process(GO:0070589) |
| 0.0 | 0.2 | GO:0071816 | tail-anchored membrane protein insertion into ER membrane(GO:0071816) |
| 0.0 | 0.2 | GO:0015740 | C4-dicarboxylate transport(GO:0015740) acidic amino acid transport(GO:0015800) aspartate transport(GO:0015810) L-glutamate transport(GO:0015813) malate-aspartate shuttle(GO:0043490) |
| 0.0 | 0.2 | GO:1904263 | positive regulation of TORC1 signaling(GO:1904263) |
| 0.0 | 0.1 | GO:0036268 | swimming(GO:0036268) |
| 0.0 | 0.1 | GO:0042362 | fat-soluble vitamin biosynthetic process(GO:0042362) |
| 0.0 | 0.1 | GO:0006264 | mitochondrial DNA replication(GO:0006264) |
| 0.0 | 0.1 | GO:0042214 | carotene metabolic process(GO:0016119) carotene catabolic process(GO:0016121) terpene metabolic process(GO:0042214) terpene catabolic process(GO:0046247) |
| 0.0 | 0.1 | GO:0006048 | UDP-N-acetylglucosamine biosynthetic process(GO:0006048) |
| 0.0 | 0.0 | GO:0045669 | positive regulation of osteoblast differentiation(GO:0045669) |
| 0.0 | 0.1 | GO:0070584 | mitochondrion morphogenesis(GO:0070584) |
| 0.0 | 0.2 | GO:0002761 | regulation of myeloid leukocyte differentiation(GO:0002761) |
| 0.0 | 0.1 | GO:0060055 | larval development(GO:0002164) larval heart development(GO:0007508) angiogenesis involved in wound healing(GO:0060055) |
| 0.0 | 0.2 | GO:0015671 | oxygen transport(GO:0015671) |
| 0.0 | 0.1 | GO:0006123 | mitochondrial electron transport, cytochrome c to oxygen(GO:0006123) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.6 | GO:0016602 | CCAAT-binding factor complex(GO:0016602) |
| 0.0 | 0.2 | GO:0005790 | smooth endoplasmic reticulum(GO:0005790) |
| 0.0 | 0.3 | GO:0031307 | integral component of mitochondrial outer membrane(GO:0031307) |
| 0.0 | 0.4 | GO:0005682 | U5 snRNP(GO:0005682) |
| 0.0 | 0.9 | GO:0005861 | troponin complex(GO:0005861) |
| 0.0 | 0.1 | GO:0031466 | Cul5-RING ubiquitin ligase complex(GO:0031466) |
| 0.0 | 0.2 | GO:0031838 | haptoglobin-hemoglobin complex(GO:0031838) |
| 0.0 | 0.1 | GO:0030891 | VCB complex(GO:0030891) |
| 0.0 | 0.0 | GO:0034515 | proteasome storage granule(GO:0034515) |
| 0.0 | 0.1 | GO:0071818 | BAT3 complex(GO:0071818) |
| 0.0 | 0.0 | GO:0031310 | integral component of vacuolar membrane(GO:0031166) intrinsic component of vacuolar membrane(GO:0031310) |
| 0.0 | 0.2 | GO:0097038 | perinuclear endoplasmic reticulum(GO:0097038) |
| 0.0 | 0.1 | GO:0031080 | nuclear pore outer ring(GO:0031080) |
| 0.0 | 0.1 | GO:0008180 | COP9 signalosome(GO:0008180) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.3 | GO:0016019 | peptidoglycan receptor activity(GO:0016019) |
| 0.1 | 0.3 | GO:0061656 | SUMO conjugating enzyme activity(GO:0061656) |
| 0.1 | 0.4 | GO:0010859 | calcium-dependent cysteine-type endopeptidase inhibitor activity(GO:0010859) |
| 0.1 | 0.2 | GO:0070513 | death domain binding(GO:0070513) |
| 0.1 | 0.2 | GO:0004377 | GDP-Man:Man3GlcNAc2-PP-Dol alpha-1,2-mannosyltransferase activity(GO:0004377) |
| 0.0 | 0.1 | GO:0016509 | long-chain-3-hydroxyacyl-CoA dehydrogenase activity(GO:0016509) |
| 0.0 | 0.1 | GO:0070815 | peptidyl-lysine 5-dioxygenase activity(GO:0070815) |
| 0.0 | 0.2 | GO:0005219 | ryanodine-sensitive calcium-release channel activity(GO:0005219) calcium-induced calcium release activity(GO:0048763) |
| 0.0 | 0.2 | GO:0015117 | thiosulfate transmembrane transporter activity(GO:0015117) oxaloacetate transmembrane transporter activity(GO:0015131) malate transmembrane transporter activity(GO:0015140) |
| 0.0 | 0.2 | GO:0016316 | phosphatidylinositol-3,4-bisphosphate 4-phosphatase activity(GO:0016316) |
| 0.0 | 0.1 | GO:0000035 | acyl binding(GO:0000035) |
| 0.0 | 0.1 | GO:0003977 | UDP-N-acetylglucosamine diphosphorylase activity(GO:0003977) |
| 0.0 | 0.1 | GO:0004645 | phosphorylase activity(GO:0004645) glycogen phosphorylase activity(GO:0008184) |
| 0.0 | 0.1 | GO:0050699 | WW domain binding(GO:0050699) |
| 0.0 | 0.2 | GO:0080019 | fatty-acyl-CoA reductase (alcohol-forming) activity(GO:0080019) |
| 0.0 | 0.1 | GO:0019784 | NEDD8-specific protease activity(GO:0019784) |
| 0.0 | 0.2 | GO:0004062 | aryl sulfotransferase activity(GO:0004062) |
| 0.0 | 0.1 | GO:0036042 | long-chain fatty acyl-CoA binding(GO:0036042) |
| 0.0 | 0.6 | GO:0048306 | calcium-dependent protein binding(GO:0048306) |
| 0.0 | 0.2 | GO:0031720 | haptoglobin binding(GO:0031720) |
| 0.0 | 0.1 | GO:0070259 | tyrosyl-DNA phosphodiesterase activity(GO:0070259) |
| 0.0 | 0.1 | GO:0033829 | O-fucosylpeptide 3-beta-N-acetylglucosaminyltransferase activity(GO:0033829) |
| 0.0 | 0.1 | GO:0004095 | carnitine O-palmitoyltransferase activity(GO:0004095) O-palmitoyltransferase activity(GO:0016416) |
| 0.0 | 0.1 | GO:0043531 | ADP binding(GO:0043531) |
| 0.0 | 0.1 | GO:0030331 | estrogen receptor binding(GO:0030331) |
| 0.0 | 0.2 | GO:0070567 | cytidylyltransferase activity(GO:0070567) |
| 0.0 | 0.2 | GO:0015183 | L-aspartate transmembrane transporter activity(GO:0015183) |
| 0.0 | 0.1 | GO:0044323 | retinoic acid-responsive element binding(GO:0044323) |
| 0.0 | 0.1 | GO:0015186 | L-glutamine transmembrane transporter activity(GO:0015186) |
| 0.0 | 0.1 | GO:0070577 | lysine-acetylated histone binding(GO:0070577) |
| 0.0 | 0.1 | GO:0003834 | beta-carotene 15,15'-monooxygenase activity(GO:0003834) carotenoid dioxygenase activity(GO:0010436) |
| 0.0 | 0.7 | GO:0005546 | phosphatidylinositol-4,5-bisphosphate binding(GO:0005546) |
| 0.0 | 0.1 | GO:0004142 | diacylglycerol cholinephosphotransferase activity(GO:0004142) ethanolaminephosphotransferase activity(GO:0004307) |
| 0.0 | 0.1 | GO:0032977 | membrane insertase activity(GO:0032977) |
| 0.0 | 0.2 | GO:0070628 | proteasome binding(GO:0070628) |
| 0.0 | 0.0 | GO:0016623 | aldehyde oxidase activity(GO:0004031) oxidoreductase activity, acting on the aldehyde or oxo group of donors, oxygen as acceptor(GO:0016623) |
| 0.0 | 0.2 | GO:0005005 | transmembrane-ephrin receptor activity(GO:0005005) |
| 0.0 | 0.1 | GO:0070888 | E-box binding(GO:0070888) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.3 | PID IL12 STAT4 PATHWAY | IL12 signaling mediated by STAT4 |
| 0.0 | 0.1 | SA FAS SIGNALING | The TNF-type receptor Fas induces apoptosis on ligand binding. |
| 0.0 | 0.3 | PID ECADHERIN KERATINOCYTE PATHWAY | E-cadherin signaling in keratinocytes |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.2 | REACTOME HORMONE SENSITIVE LIPASE HSL MEDIATED TRIACYLGLYCEROL HYDROLYSIS | Genes involved in Hormone-sensitive lipase (HSL)-mediated triacylglycerol hydrolysis |
| 0.0 | 0.3 | REACTOME P130CAS LINKAGE TO MAPK SIGNALING FOR INTEGRINS | Genes involved in p130Cas linkage to MAPK signaling for integrins |
| 0.0 | 0.0 | REACTOME OLFACTORY SIGNALING PATHWAY | Genes involved in Olfactory Signaling Pathway |
| 0.0 | 0.3 | REACTOME GLUCONEOGENESIS | Genes involved in Gluconeogenesis |
| 0.0 | 0.1 | REACTOME SEMA3A PAK DEPENDENT AXON REPULSION | Genes involved in Sema3A PAK dependent Axon repulsion |
| 0.0 | 0.1 | REACTOME ADENYLATE CYCLASE ACTIVATING PATHWAY | Genes involved in Adenylate cyclase activating pathway |
| 0.0 | 0.2 | REACTOME OTHER SEMAPHORIN INTERACTIONS | Genes involved in Other semaphorin interactions |