Project
PRJNA207719: Tissue specific transcriptome profiling
Navigation
Downloads
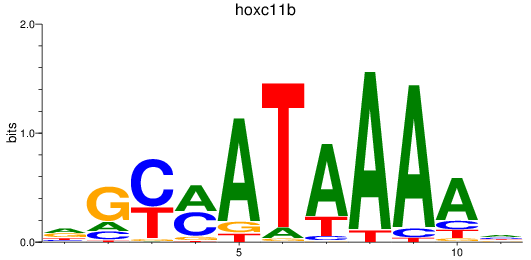
Results for hoxc11b
Z-value: 0.49
Transcription factors associated with hoxc11b
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
hoxc11b
|
ENSDARG00000102631 | homeobox C11b |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| hoxc11b | dr11_v1_chr11_+_2180072_2180072 | -0.31 | 6.1e-01 | Click! |
Activity profile of hoxc11b motif
Sorted Z-values of hoxc11b motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr25_+_29161609 | 0.46 |
ENSDART00000180752
|
pkmb
|
pyruvate kinase M1/2b |
| chr10_+_36650222 | 0.39 |
ENSDART00000126963
|
ucp3
|
uncoupling protein 3 |
| chr7_+_39386982 | 0.32 |
ENSDART00000146702
|
tnni2b.2
|
troponin I type 2b (skeletal, fast), tandem duplicate 2 |
| chr8_+_47633438 | 0.29 |
ENSDART00000139096
|
si:ch211-251b21.1
|
si:ch211-251b21.1 |
| chr14_-_25949951 | 0.27 |
ENSDART00000141304
|
sparc
|
secreted protein, acidic, cysteine-rich (osteonectin) |
| chr23_+_39606108 | 0.26 |
ENSDART00000109464
|
g0s2
|
G0/G1 switch 2 |
| chr2_-_44283554 | 0.23 |
ENSDART00000184684
|
mpz
|
myelin protein zero |
| chr6_-_40697585 | 0.21 |
ENSDART00000113196
|
si:ch211-157b11.14
|
si:ch211-157b11.14 |
| chr14_+_15597049 | 0.19 |
ENSDART00000159732
|
si:dkey-203a12.8
|
si:dkey-203a12.8 |
| chr7_+_46003449 | 0.19 |
ENSDART00000159700
ENSDART00000173625 |
si:ch211-260e23.9
|
si:ch211-260e23.9 |
| chr14_+_15495088 | 0.19 |
ENSDART00000165765
ENSDART00000188577 |
si:dkey-203a12.6
|
si:dkey-203a12.6 |
| chr9_-_41784799 | 0.18 |
ENSDART00000144573
ENSDART00000112542 ENSDART00000190486 |
obsl1b
|
obscurin-like 1b |
| chr6_-_60147517 | 0.17 |
ENSDART00000083453
|
slc32a1
|
solute carrier family 32 (GABA vesicular transporter), member 1 |
| chr3_+_27027781 | 0.17 |
ENSDART00000065495
|
emp2
|
epithelial membrane protein 2 |
| chr12_-_25380028 | 0.16 |
ENSDART00000142674
|
zfp36l2
|
zinc finger protein 36, C3H type-like 2 |
| chr24_-_29822913 | 0.16 |
ENSDART00000160929
|
aglb
|
amylo-alpha-1, 6-glucosidase, 4-alpha-glucanotransferase b |
| chr6_+_35362225 | 0.15 |
ENSDART00000133783
ENSDART00000102483 |
rgs4
|
regulator of G protein signaling 4 |
| chr20_+_40237441 | 0.14 |
ENSDART00000168928
|
si:ch211-199i15.5
|
si:ch211-199i15.5 |
| chr19_+_3840955 | 0.13 |
ENSDART00000172305
|
lsm10
|
LSM10, U7 small nuclear RNA associated |
| chr14_+_30279391 | 0.13 |
ENSDART00000172794
|
fgl1
|
fibrinogen-like 1 |
| chr15_-_25527580 | 0.13 |
ENSDART00000167005
ENSDART00000157498 |
hif1al
|
hypoxia-inducible factor 1, alpha subunit, like |
| chr16_+_13993285 | 0.13 |
ENSDART00000139130
ENSDART00000130353 |
si:dkey-85k15.7
fdps
|
si:dkey-85k15.7 farnesyl diphosphate synthase (farnesyl pyrophosphate synthetase, dimethylallyltranstransferase, geranyltranstransferase) |
| chr5_+_13385837 | 0.13 |
ENSDART00000191190
|
ccl19a.1
|
chemokine (C-C motif) ligand 19a, tandem duplicate 1 |
| chr5_+_70155935 | 0.13 |
ENSDART00000165570
|
rgs3a
|
regulator of G protein signaling 3a |
| chr6_+_49551614 | 0.13 |
ENSDART00000022581
|
rab22a
|
RAB22A, member RAS oncogene family |
| chr22_-_10570749 | 0.13 |
ENSDART00000140736
|
si:dkey-42i9.6
|
si:dkey-42i9.6 |
| chr16_+_45746549 | 0.12 |
ENSDART00000190403
|
paqr6
|
progestin and adipoQ receptor family member VI |
| chr25_+_20089986 | 0.12 |
ENSDART00000143441
ENSDART00000184073 |
tnni4b.2
|
troponin I4b, tandem duplicate 2 |
| chr7_-_38634845 | 0.12 |
ENSDART00000173861
|
c1qtnf4
|
C1q and TNF related 4 |
| chr8_+_11687254 | 0.12 |
ENSDART00000042040
|
atp2a2a
|
ATPase sarcoplasmic/endoplasmic reticulum Ca2+ transporting 2a |
| chr24_+_5237753 | 0.12 |
ENSDART00000106488
ENSDART00000005901 |
plod2
|
procollagen-lysine, 2-oxoglutarate 5-dioxygenase 2 |
| chr24_-_6158933 | 0.12 |
ENSDART00000021609
|
gad2
|
glutamate decarboxylase 2 |
| chr4_+_12292274 | 0.12 |
ENSDART00000061070
ENSDART00000150786 |
mkrn1
|
makorin, ring finger protein, 1 |
| chr14_-_33613794 | 0.11 |
ENSDART00000010022
|
xpnpep2
|
X-prolyl aminopeptidase (aminopeptidase P) 2, membrane-bound |
| chr5_-_48307804 | 0.11 |
ENSDART00000182831
ENSDART00000186920 ENSDART00000183585 |
mef2cb
|
myocyte enhancer factor 2cb |
| chr3_+_52545014 | 0.11 |
ENSDART00000018908
|
slc27a1a
|
solute carrier family 27 (fatty acid transporter), member 1a |
| chr13_-_37465524 | 0.10 |
ENSDART00000100324
ENSDART00000147336 |
ppp2r5eb
|
protein phosphatase 2, regulatory subunit B', epsilon isoform b |
| chr22_-_31517300 | 0.10 |
ENSDART00000164799
|
slc6a6b
|
solute carrier family 6 (neurotransmitter transporter), member 6b |
| chr7_+_34290051 | 0.09 |
ENSDART00000123498
|
fibinb
|
fin bud initiation factor b |
| chr14_-_15171435 | 0.09 |
ENSDART00000159148
ENSDART00000166622 |
si:dkey-77g12.1
|
si:dkey-77g12.1 |
| chr17_-_12389259 | 0.09 |
ENSDART00000185724
|
snap25b
|
synaptosomal-associated protein, 25b |
| chr24_+_39105051 | 0.09 |
ENSDART00000115297
|
mss51
|
MSS51 mitochondrial translational activator |
| chr11_+_25112269 | 0.09 |
ENSDART00000147546
|
ndrg3a
|
ndrg family member 3a |
| chr2_-_48375342 | 0.09 |
ENSDART00000148788
|
per2
|
period circadian clock 2 |
| chr25_+_26798673 | 0.09 |
ENSDART00000157235
|
ca12
|
carbonic anhydrase XII |
| chr22_-_34551568 | 0.08 |
ENSDART00000148147
|
rnf123
|
ring finger protein 123 |
| chr20_-_33961697 | 0.08 |
ENSDART00000061765
|
selp
|
selectin P |
| chr3_+_29714775 | 0.08 |
ENSDART00000041388
|
cacng2a
|
calcium channel, voltage-dependent, gamma subunit 2a |
| chr14_+_49135264 | 0.08 |
ENSDART00000084119
|
si:ch1073-44g3.1
|
si:ch1073-44g3.1 |
| chr3_+_52545400 | 0.08 |
ENSDART00000184183
|
slc27a1a
|
solute carrier family 27 (fatty acid transporter), member 1a |
| chr17_+_52822831 | 0.08 |
ENSDART00000193368
|
meis2a
|
Meis homeobox 2a |
| chr11_-_45152702 | 0.08 |
ENSDART00000168066
|
afmid
|
arylformamidase |
| chr5_-_40510397 | 0.08 |
ENSDART00000146237
ENSDART00000051065 |
fsta
|
follistatin a |
| chr7_-_58952382 | 0.08 |
ENSDART00000167076
|
mrc1a
|
mannose receptor, C type 1a |
| chr6_+_39114345 | 0.08 |
ENSDART00000136835
|
gpr84
|
G protein-coupled receptor 84 |
| chr18_+_26422124 | 0.08 |
ENSDART00000060245
|
ctsh
|
cathepsin H |
| chr19_+_16032383 | 0.07 |
ENSDART00000046530
|
rab42a
|
RAB42, member RAS oncogene family a |
| chr1_+_52481332 | 0.07 |
ENSDART00000074231
|
cldnd1b
|
claudin domain containing 1b |
| chr22_-_5933844 | 0.07 |
ENSDART00000163370
ENSDART00000189331 |
si:rp71-36a1.2
|
si:rp71-36a1.2 |
| chr7_+_30875273 | 0.07 |
ENSDART00000173693
|
apba2b
|
amyloid beta (A4) precursor protein-binding, family A, member 2b |
| chr7_-_30280934 | 0.07 |
ENSDART00000126741
|
serf2
|
small EDRK-rich factor 2 |
| chr12_-_13337033 | 0.07 |
ENSDART00000105903
ENSDART00000139786 |
lsm5
|
LSM5 homolog, U6 small nuclear RNA and mRNA degradation associated |
| chr13_+_4405282 | 0.07 |
ENSDART00000148280
|
prr18
|
proline rich 18 |
| chr25_+_16080181 | 0.07 |
ENSDART00000061753
|
far1
|
fatty acyl CoA reductase 1 |
| chr24_-_17029374 | 0.07 |
ENSDART00000039267
|
ptgdsb.1
|
prostaglandin D2 synthase b, tandem duplicate 1 |
| chr7_+_11543999 | 0.07 |
ENSDART00000173676
|
il16
|
interleukin 16 |
| chr23_+_25354856 | 0.07 |
ENSDART00000109023
ENSDART00000147440 |
fmnl3
|
formin-like 3 |
| chr24_-_32025637 | 0.07 |
ENSDART00000180448
ENSDART00000159034 |
rsu1
|
Ras suppressor protein 1 |
| chr13_-_4018888 | 0.07 |
ENSDART00000058238
|
tjap1
|
tight junction associated protein 1 (peripheral) |
| chr5_+_6796291 | 0.07 |
ENSDART00000166868
ENSDART00000165308 |
me2
|
malic enzyme 2, NAD(+)-dependent, mitochondrial |
| chr14_-_3229384 | 0.07 |
ENSDART00000167247
|
pdgfrb
|
platelet-derived growth factor receptor, beta polypeptide |
| chr2_+_21128391 | 0.07 |
ENSDART00000136814
|
pip4k2ab
|
phosphatidylinositol-5-phosphate 4-kinase, type II, alpha b |
| chr8_-_14121634 | 0.07 |
ENSDART00000184946
|
bgna
|
biglycan a |
| chr2_+_35733335 | 0.06 |
ENSDART00000113489
|
rasal2
|
RAS protein activator like 2 |
| chr23_-_36003282 | 0.06 |
ENSDART00000103150
|
calcoco1a
|
calcium binding and coiled-coil domain 1a |
| chr25_-_34973211 | 0.06 |
ENSDART00000045177
|
cdk10
|
cyclin-dependent kinase 10 |
| chr17_+_15788100 | 0.06 |
ENSDART00000027667
|
rragd
|
ras-related GTP binding D |
| chr21_+_9689103 | 0.06 |
ENSDART00000165132
|
mapk10
|
mitogen-activated protein kinase 10 |
| chr13_+_36585399 | 0.06 |
ENSDART00000030211
|
gmfb
|
glia maturation factor, beta |
| chr21_+_21906671 | 0.06 |
ENSDART00000016916
|
gria4b
|
glutamate receptor, ionotropic, AMPA 4b |
| chr4_-_73136420 | 0.06 |
ENSDART00000160907
|
si:ch73-170d6.3
|
si:ch73-170d6.3 |
| chr3_+_39579393 | 0.06 |
ENSDART00000055170
|
cln3
|
ceroid-lipofuscinosis, neuronal 3 |
| chr7_-_26076970 | 0.06 |
ENSDART00000101120
|
zgc:92664
|
zgc:92664 |
| chr5_-_9073433 | 0.06 |
ENSDART00000099891
|
atp5meb
|
ATP synthase membrane subunit eb |
| chr8_-_25338709 | 0.06 |
ENSDART00000131616
|
atp5pb
|
ATP synthase peripheral stalk-membrane subunit b |
| chr14_-_26425416 | 0.06 |
ENSDART00000088690
|
lman2
|
lectin, mannose-binding 2 |
| chr1_+_8694196 | 0.06 |
ENSDART00000025604
|
zgc:77849
|
zgc:77849 |
| chr16_+_38940758 | 0.05 |
ENSDART00000102482
ENSDART00000136215 |
eny2
|
enhancer of yellow 2 homolog (Drosophila) |
| chr17_+_52823015 | 0.05 |
ENSDART00000160507
ENSDART00000186979 |
meis2a
|
Meis homeobox 2a |
| chr11_-_29737088 | 0.05 |
ENSDART00000159828
|
si:ch211-218g23.6
|
si:ch211-218g23.6 |
| chr19_-_35733401 | 0.05 |
ENSDART00000066712
|
trappc3
|
trafficking protein particle complex 3 |
| chr5_+_24245682 | 0.05 |
ENSDART00000049003
|
atp6v1aa
|
ATPase H+ transporting V1 subunit Aa |
| chr16_+_20934353 | 0.05 |
ENSDART00000052660
|
skap2
|
src kinase associated phosphoprotein 2 |
| chr18_+_910992 | 0.05 |
ENSDART00000161206
ENSDART00000167229 |
pkma
|
pyruvate kinase M1/2a |
| chr18_+_14342326 | 0.05 |
ENSDART00000181013
ENSDART00000138372 |
si:dkey-246g23.2
|
si:dkey-246g23.2 |
| chr25_+_30238021 | 0.05 |
ENSDART00000123220
|
alkbh3
|
alkB homolog 3, alpha-ketoglutarate-dependent dioxygenase |
| chr19_-_28367413 | 0.05 |
ENSDART00000079092
|
si:dkey-261i16.5
|
si:dkey-261i16.5 |
| chr7_-_41420226 | 0.05 |
ENSDART00000074155
|
abcf2b
|
ATP-binding cassette, sub-family F (GCN20), member 2b |
| chr6_+_27667359 | 0.05 |
ENSDART00000159624
ENSDART00000049177 |
rab6ba
|
RAB6B, member RAS oncogene family a |
| chr3_-_9722603 | 0.05 |
ENSDART00000168234
|
crebbpb
|
CREB binding protein b |
| chr20_-_34670236 | 0.05 |
ENSDART00000033325
|
slc25a24
|
solute carrier family 25 (mitochondrial carrier; phosphate carrier), member 24 |
| chr7_-_55051692 | 0.05 |
ENSDART00000170637
|
tpcn2
|
two pore segment channel 2 |
| chr23_-_12014931 | 0.05 |
ENSDART00000134652
|
si:dkey-178k16.1
|
si:dkey-178k16.1 |
| chr20_-_14462995 | 0.05 |
ENSDART00000152418
ENSDART00000044125 |
grcc10
|
gene rich cluster, C10 gene |
| chr24_+_5840258 | 0.05 |
ENSDART00000087034
|
trpc1
|
transient receptor potential cation channel, subfamily C, member 1 |
| chr6_+_27991943 | 0.05 |
ENSDART00000143974
ENSDART00000141354 ENSDART00000088914 ENSDART00000139367 |
amotl2a
|
angiomotin like 2a |
| chr1_+_46194333 | 0.05 |
ENSDART00000010894
|
sox1b
|
SRY (sex determining region Y)-box 1b |
| chr4_-_149334 | 0.05 |
ENSDART00000163280
|
tbk1
|
TANK-binding kinase 1 |
| chr18_-_15373620 | 0.05 |
ENSDART00000031752
|
rfx4
|
regulatory factor X, 4 |
| chr15_-_34567370 | 0.05 |
ENSDART00000099793
|
sostdc1a
|
sclerostin domain containing 1a |
| chr6_-_41138854 | 0.04 |
ENSDART00000128723
ENSDART00000151055 ENSDART00000132484 |
slc6a22.1
|
solute carrier family 6 member 22, tandem duplicate 1 |
| chr2_-_31537521 | 0.04 |
ENSDART00000148244
|
mrc1b
|
mannose receptor, C type 1b |
| chr16_-_49505275 | 0.04 |
ENSDART00000160784
|
satb1b
|
SATB homeobox 1b |
| chr8_+_23731483 | 0.04 |
ENSDART00000099751
|
fance
|
Fanconi anemia, complementation group E |
| chr9_+_2452672 | 0.04 |
ENSDART00000193993
|
chn1
|
chimerin 1 |
| chr11_-_18253111 | 0.04 |
ENSDART00000125984
|
mustn1b
|
musculoskeletal, embryonic nuclear protein 1b |
| chr12_+_10134366 | 0.04 |
ENSDART00000127343
|
smarcd2
|
SWI/SNF related, matrix associated, actin dependent regulator of chromatin, subfamily d, member 2 |
| chr7_-_23745984 | 0.04 |
ENSDART00000048050
|
ITGB1BP2
|
zgc:92429 |
| chr12_+_27117609 | 0.04 |
ENSDART00000076154
|
hoxb8b
|
homeobox B8b |
| chr2_+_31948352 | 0.04 |
ENSDART00000192611
|
ankhb
|
ANKH inorganic pyrophosphate transport regulator b |
| chr25_-_20258508 | 0.04 |
ENSDART00000133860
ENSDART00000006840 ENSDART00000173434 |
dnm1l
|
dynamin 1-like |
| chr10_-_24371312 | 0.04 |
ENSDART00000149362
|
pitpnab
|
phosphatidylinositol transfer protein, alpha b |
| chr21_+_25236297 | 0.04 |
ENSDART00000112783
|
tmem45b
|
transmembrane protein 45B |
| chr3_+_24190207 | 0.04 |
ENSDART00000034762
|
prr15la
|
proline rich 15-like a |
| chr19_-_26736336 | 0.04 |
ENSDART00000109258
ENSDART00000182802 |
csnk2b
|
casein kinase 2, beta polypeptide |
| chr10_-_31562695 | 0.04 |
ENSDART00000186456
|
robo3
|
roundabout, axon guidance receptor, homolog 3 (Drosophila) |
| chr19_+_4916233 | 0.04 |
ENSDART00000159512
|
ppp1r1b
|
protein phosphatase 1, regulatory (inhibitor) subunit 1B |
| chr5_-_28767573 | 0.04 |
ENSDART00000158299
ENSDART00000043466 |
traf2a
|
Tnf receptor-associated factor 2a |
| chr5_-_65662996 | 0.04 |
ENSDART00000147707
|
camsap1b
|
calmodulin regulated spectrin-associated protein 1b |
| chr7_+_36041509 | 0.04 |
ENSDART00000162850
|
irx3a
|
iroquois homeobox 3a |
| chr2_-_32366287 | 0.04 |
ENSDART00000144758
|
ubtfl
|
upstream binding transcription factor, like |
| chr7_+_39634873 | 0.04 |
ENSDART00000114774
|
ptpn5
|
protein tyrosine phosphatase, non-receptor type 5 |
| chr5_-_21888368 | 0.04 |
ENSDART00000020725
|
smarca1
|
SWI/SNF related, matrix associated, actin dependent regulator of chromatin, subfamily a, member 1 |
| chr23_+_36122058 | 0.03 |
ENSDART00000184448
|
hoxc3a
|
homeobox C3a |
| chr14_+_23184517 | 0.03 |
ENSDART00000181410
|
enox2
|
ecto-NOX disulfide-thiol exchanger 2 |
| chr19_+_1370504 | 0.03 |
ENSDART00000158946
|
dgat1a
|
diacylglycerol O-acyltransferase 1a |
| chr11_-_6265574 | 0.03 |
ENSDART00000181974
ENSDART00000104405 |
ccl25b
|
chemokine (C-C motif) ligand 25b |
| chr12_-_20373058 | 0.03 |
ENSDART00000066382
|
aqp8a.1
|
aquaporin 8a, tandem duplicate 1 |
| chr14_+_4276394 | 0.03 |
ENSDART00000038301
|
gnpda2
|
glucosamine-6-phosphate deaminase 2 |
| chr19_-_10243148 | 0.03 |
ENSDART00000148073
|
shisa7b
|
shisa family member 7 |
| chr3_-_19200571 | 0.03 |
ENSDART00000131503
ENSDART00000012335 |
rfx1a
|
regulatory factor X, 1a (influences HLA class II expression) |
| chr1_-_45577980 | 0.03 |
ENSDART00000160961
|
atf7ip
|
activating transcription factor 7 interacting protein |
| chr3_-_13147310 | 0.03 |
ENSDART00000160840
|
prkar1b
|
protein kinase, cAMP-dependent, regulatory, type I, beta |
| chr3_+_54761569 | 0.03 |
ENSDART00000135913
ENSDART00000180983 |
si:ch211-74m13.1
|
si:ch211-74m13.1 |
| chr6_-_53281518 | 0.03 |
ENSDART00000157621
|
rbm5
|
RNA binding motif protein 5 |
| chr8_+_23381892 | 0.03 |
ENSDART00000180950
ENSDART00000063010 ENSDART00000074241 ENSDART00000142783 |
mapre1a
|
microtubule-associated protein, RP/EB family, member 1a |
| chr24_-_8732519 | 0.03 |
ENSDART00000082351
|
tfap2a
|
transcription factor AP-2 alpha |
| chr14_+_30795559 | 0.03 |
ENSDART00000006132
|
cfl1
|
cofilin 1 |
| chr16_-_43679611 | 0.03 |
ENSDART00000123585
|
LO017721.1
|
|
| chr15_+_32643873 | 0.03 |
ENSDART00000189433
|
trpc4b
|
transient receptor potential cation channel, subfamily C, member 4b |
| chr5_-_41307550 | 0.03 |
ENSDART00000143446
|
npr3
|
natriuretic peptide receptor 3 |
| chr10_-_25591194 | 0.03 |
ENSDART00000131640
|
tiam1a
|
T cell lymphoma invasion and metastasis 1a |
| chr21_-_21781158 | 0.03 |
ENSDART00000113734
|
chrdl2
|
chordin-like 2 |
| chr9_-_27442339 | 0.03 |
ENSDART00000138602
|
stxbp5l
|
syntaxin binding protein 5-like |
| chr5_+_45140914 | 0.03 |
ENSDART00000172702
|
smarca2
|
SWI/SNF related, matrix associated, actin dependent regulator of chromatin, subfamily a, member 2 |
| chr8_+_36582728 | 0.03 |
ENSDART00000049230
|
pqbp1
|
polyglutamine binding protein 1 |
| chr10_-_10864331 | 0.03 |
ENSDART00000122657
|
nrarpa
|
NOTCH regulated ankyrin repeat protein a |
| chr16_-_55028740 | 0.03 |
ENSDART00000156368
ENSDART00000161704 |
zgc:114181
|
zgc:114181 |
| chr20_+_19212962 | 0.03 |
ENSDART00000063706
|
fndc4a
|
fibronectin type III domain containing 4a |
| chr6_-_12296170 | 0.03 |
ENSDART00000155685
|
pkp4
|
plakophilin 4 |
| chr15_-_25392589 | 0.03 |
ENSDART00000124205
|
si:dkey-54n8.4
|
si:dkey-54n8.4 |
| chr3_+_24134418 | 0.03 |
ENSDART00000156204
|
si:ch211-246i5.5
|
si:ch211-246i5.5 |
| chr24_+_30392834 | 0.03 |
ENSDART00000162555
|
dpyda.1
|
dihydropyrimidine dehydrogenase a, tandem duplicate 1 |
| chr25_+_28823952 | 0.02 |
ENSDART00000067072
|
nfybb
|
nuclear transcription factor Y, beta b |
| chr4_-_13255700 | 0.02 |
ENSDART00000162277
ENSDART00000026593 |
grip1
|
glutamate receptor interacting protein 1 |
| chr20_-_51831816 | 0.02 |
ENSDART00000060505
|
mia3
|
melanoma inhibitory activity family, member 3 |
| chr16_+_21242491 | 0.02 |
ENSDART00000145886
|
osbpl3b
|
oxysterol binding protein-like 3b |
| chr24_-_40009446 | 0.02 |
ENSDART00000087422
|
aoc1
|
amine oxidase, copper containing 1 |
| chr24_+_11908480 | 0.02 |
ENSDART00000024224
|
fen1
|
flap structure-specific endonuclease 1 |
| chr18_-_15329454 | 0.02 |
ENSDART00000191903
ENSDART00000019818 |
ric8b
|
RIC8 guanine nucleotide exchange factor B |
| chr12_+_1000323 | 0.02 |
ENSDART00000054363
|
si:ch1073-272o11.3
|
si:ch1073-272o11.3 |
| chr2_-_40135942 | 0.02 |
ENSDART00000176951
ENSDART00000098632 ENSDART00000148563 ENSDART00000149895 |
epha4a
|
eph receptor A4a |
| chr1_-_22512063 | 0.02 |
ENSDART00000031546
ENSDART00000190987 |
chrna6
|
cholinergic receptor, nicotinic, alpha 6 |
| chr21_-_20840714 | 0.02 |
ENSDART00000144861
ENSDART00000139430 |
c6
|
complement component 6 |
| chr12_+_8168272 | 0.02 |
ENSDART00000054092
|
arid5b
|
AT-rich interaction domain 5B |
| chr1_-_45039726 | 0.02 |
ENSDART00000186188
|
smu1b
|
SMU1, DNA replication regulator and spliceosomal factor b |
| chr20_-_26491567 | 0.02 |
ENSDART00000147154
|
mthfd1l
|
methylenetetrahydrofolate dehydrogenase (NADP+ dependent) 1 like |
| chr12_+_27022517 | 0.02 |
ENSDART00000152975
|
msl1b
|
male-specific lethal 1 homolog b (Drosophila) |
| chr20_-_4883673 | 0.02 |
ENSDART00000145540
ENSDART00000053877 |
zdhhc14
|
zinc finger, DHHC-type containing 14 |
| chr11_+_30244356 | 0.02 |
ENSDART00000036050
ENSDART00000150080 |
rs1a
|
retinoschisin 1a |
| chr4_+_5741733 | 0.02 |
ENSDART00000110243
|
pou3f2a
|
POU class 3 homeobox 2a |
| chr13_+_15933168 | 0.02 |
ENSDART00000131390
|
fignl1
|
fidgetin-like 1 |
| chr19_+_9455218 | 0.02 |
ENSDART00000139385
|
si:ch211-288g17.3
|
si:ch211-288g17.3 |
| chr24_-_30091937 | 0.02 |
ENSDART00000148249
|
plppr4b
|
phospholipid phosphatase related 4b |
| chr23_-_30431333 | 0.02 |
ENSDART00000146633
|
camta1a
|
calmodulin binding transcription activator 1a |
| chr8_-_10961991 | 0.02 |
ENSDART00000139603
|
trim33
|
tripartite motif containing 33 |
| chr16_-_10547781 | 0.02 |
ENSDART00000166495
|
g6fl
|
g6f-like |
| chr6_-_6448519 | 0.02 |
ENSDART00000180157
ENSDART00000191112 |
si:ch211-194e18.2
|
si:ch211-194e18.2 |
| chr8_-_44298964 | 0.02 |
ENSDART00000098520
|
fzd10
|
frizzled class receptor 10 |
| chr11_-_36009924 | 0.02 |
ENSDART00000189959
ENSDART00000167472 ENSDART00000191211 ENSDART00000191662 ENSDART00000191780 ENSDART00000192622 ENSDART00000179911 |
itpr1b
|
inositol 1,4,5-trisphosphate receptor, type 1b |
| chr13_-_25484659 | 0.02 |
ENSDART00000135321
ENSDART00000022799 |
tial1
|
TIA1 cytotoxic granule-associated RNA binding protein-like 1 |
| chr4_+_20954929 | 0.02 |
ENSDART00000143674
|
nav3
|
neuron navigator 3 |
| chr8_-_12432604 | 0.02 |
ENSDART00000133350
ENSDART00000140699 ENSDART00000101174 |
traf1
|
TNF receptor-associated factor 1 |
| chr4_-_14926106 | 0.02 |
ENSDART00000147629
|
prdm4
|
PR domain containing 4 |
| chr17_+_52822422 | 0.02 |
ENSDART00000158273
ENSDART00000161414 |
meis2a
|
Meis homeobox 2a |
| chr14_-_1958994 | 0.02 |
ENSDART00000161783
|
pcdh2g5
|
protocadherin 2 gamma 5 |
| chr5_-_65037525 | 0.02 |
ENSDART00000158856
|
anxa1b
|
annexin A1b |
| chr23_+_43718115 | 0.02 |
ENSDART00000149266
ENSDART00000149503 |
anapc10
|
anaphase promoting complex subunit 10 |
| chr11_+_21586335 | 0.02 |
ENSDART00000091182
|
FAM72B
|
zgc:101564 |
| chr2_-_43196595 | 0.01 |
ENSDART00000141087
|
crema
|
cAMP responsive element modulator a |
Network of associatons between targets according to the STRING database.
First level regulatory network of hoxc11b
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.2 | GO:0003093 | renal system process involved in regulation of blood volume(GO:0001977) renal system process involved in regulation of systemic arterial blood pressure(GO:0003071) regulation of glomerular filtration(GO:0003093) |
| 0.0 | 0.4 | GO:1990845 | adaptive thermogenesis(GO:1990845) |
| 0.0 | 0.1 | GO:0009449 | gamma-aminobutyric acid biosynthetic process(GO:0009449) |
| 0.0 | 0.2 | GO:0071072 | heat generation(GO:0031649) regulation of heat generation(GO:0031650) positive regulation of heat generation(GO:0031652) regulation of phospholipid biosynthetic process(GO:0071071) negative regulation of phospholipid biosynthetic process(GO:0071072) |
| 0.0 | 0.1 | GO:0017185 | peptidyl-lysine hydroxylation(GO:0017185) |
| 0.0 | 0.1 | GO:0008591 | regulation of Wnt signaling pathway, calcium modulating pathway(GO:0008591) negative regulation of non-canonical Wnt signaling pathway(GO:2000051) |
| 0.0 | 0.2 | GO:0098700 | neurotransmitter loading into synaptic vesicle(GO:0098700) |
| 0.0 | 0.1 | GO:0045337 | farnesyl diphosphate biosynthetic process(GO:0045337) |
| 0.0 | 0.3 | GO:0014823 | response to activity(GO:0014823) |
| 0.0 | 0.1 | GO:0014909 | smooth muscle cell migration(GO:0014909) |
| 0.0 | 0.1 | GO:0042753 | positive regulation of circadian rhythm(GO:0042753) |
| 0.0 | 0.1 | GO:0097241 | hematopoietic stem cell migration to bone marrow(GO:0097241) |
| 0.0 | 0.0 | GO:2000471 | regulation of hematopoietic stem cell migration(GO:2000471) |
| 0.0 | 0.0 | GO:0006043 | glucosamine metabolic process(GO:0006041) glucosamine catabolic process(GO:0006043) |
| 0.0 | 0.0 | GO:0021557 | oculomotor nerve development(GO:0021557) |
| 0.0 | 0.1 | GO:0016572 | histone phosphorylation(GO:0016572) |
| 0.0 | 0.0 | GO:0030043 | actin filament fragmentation(GO:0030043) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.3 | GO:0032983 | kainate selective glutamate receptor complex(GO:0032983) |
| 0.0 | 0.1 | GO:0016589 | NURF complex(GO:0016589) |
| 0.0 | 0.1 | GO:0070032 | synaptobrevin 2-SNAP-25-syntaxin-1a-complexin I complex(GO:0070032) |
| 0.0 | 0.2 | GO:0043209 | myelin sheath(GO:0043209) |
| 0.0 | 0.1 | GO:1990131 | EGO complex(GO:0034448) Gtr1-Gtr2 GTPase complex(GO:1990131) |
| 0.0 | 0.2 | GO:0030285 | integral component of synaptic vesicle membrane(GO:0030285) intrinsic component of synaptic vesicle membrane(GO:0098563) |
| 0.0 | 0.1 | GO:0070390 | transcription export complex 2(GO:0070390) |
| 0.0 | 0.1 | GO:1990726 | Lsm1-7-Pat1 complex(GO:1990726) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.4 | GO:0017077 | oxidative phosphorylation uncoupler activity(GO:0017077) |
| 0.1 | 0.5 | GO:0004743 | pyruvate kinase activity(GO:0004743) |
| 0.1 | 0.3 | GO:0015277 | kainate selective glutamate receptor activity(GO:0015277) |
| 0.0 | 0.1 | GO:0004337 | dimethylallyltranstransferase activity(GO:0004161) geranyltranstransferase activity(GO:0004337) |
| 0.0 | 0.2 | GO:0004135 | glycogen debranching enzyme activity(GO:0004133) 4-alpha-glucanotransferase activity(GO:0004134) amylo-alpha-1,6-glucosidase activity(GO:0004135) |
| 0.0 | 0.1 | GO:0070815 | peptidyl-lysine 5-dioxygenase activity(GO:0070815) |
| 0.0 | 0.1 | GO:0004351 | glutamate decarboxylase activity(GO:0004351) |
| 0.0 | 0.2 | GO:0031957 | very long-chain fatty acid-CoA ligase activity(GO:0031957) |
| 0.0 | 0.1 | GO:0004061 | arylformamidase activity(GO:0004061) |
| 0.0 | 0.1 | GO:0042609 | CD4 receptor binding(GO:0042609) |
| 0.0 | 0.1 | GO:0004470 | malic enzyme activity(GO:0004470) malate dehydrogenase (decarboxylating) (NAD+) activity(GO:0004471) malate dehydrogenase (decarboxylating) (NADP+) activity(GO:0004473) |
| 0.0 | 0.1 | GO:0005017 | platelet-derived growth factor-activated receptor activity(GO:0005017) platelet-derived growth factor binding(GO:0048407) |
| 0.0 | 0.2 | GO:0015185 | gamma-aminobutyric acid transmembrane transporter activity(GO:0015185) |
| 0.0 | 0.0 | GO:0004342 | glucosamine-6-phosphate deaminase activity(GO:0004342) |
| 0.0 | 0.1 | GO:0001222 | transcription corepressor binding(GO:0001222) |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.3 | REACTOME GABA SYNTHESIS RELEASE REUPTAKE AND DEGRADATION | Genes involved in GABA synthesis, release, reuptake and degradation |
| 0.0 | 0.1 | REACTOME SLBP DEPENDENT PROCESSING OF REPLICATION DEPENDENT HISTONE PRE MRNAS | Genes involved in SLBP Dependent Processing of Replication-Dependent Histone Pre-mRNAs |