Project
PRJNA207719: Tissue specific transcriptome profiling
Navigation
Downloads
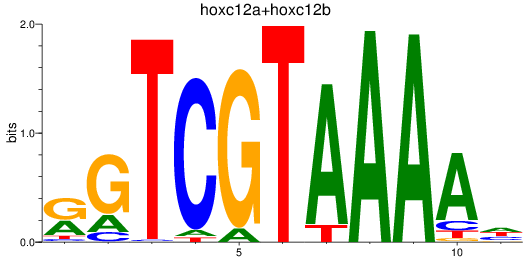
Results for hoxc12a+hoxc12b
Z-value: 1.13
Transcription factors associated with hoxc12a+hoxc12b
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
hoxc12a
|
ENSDARG00000070352 | homeobox C12a |
|
hoxc12b
|
ENSDARG00000103133 | homeobox C12b |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| hoxc12b | dr11_v1_chr11_+_2172335_2172335 | 0.92 | 2.9e-02 | Click! |
| hoxc12a | dr11_v1_chr23_+_36063599_36063599 | -0.52 | 3.7e-01 | Click! |
Activity profile of hoxc12a+hoxc12b motif
Sorted Z-values of hoxc12a+hoxc12b motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr14_+_35691889 | 1.21 |
ENSDART00000074685
|
glrbb
|
glycine receptor, beta b |
| chr21_-_19018455 | 1.04 |
ENSDART00000080256
|
nefma
|
neurofilament, medium polypeptide a |
| chr23_-_4855122 | 0.98 |
ENSDART00000133701
|
slc6a1a
|
solute carrier family 6 (neurotransmitter transporter), member 1a |
| chr24_-_40009446 | 0.97 |
ENSDART00000087422
|
aoc1
|
amine oxidase, copper containing 1 |
| chr21_+_13861589 | 0.94 |
ENSDART00000015629
ENSDART00000171306 |
stxbp1a
|
syntaxin binding protein 1a |
| chr25_-_11057753 | 0.94 |
ENSDART00000186551
|
sv2bb
|
synaptic vesicle glycoprotein 2Bb |
| chr8_-_14049404 | 0.94 |
ENSDART00000093117
|
atp2b3a
|
ATPase plasma membrane Ca2+ transporting 3a |
| chr21_-_4032650 | 0.94 |
ENSDART00000151648
|
ntng2b
|
netrin g2b |
| chr1_-_8101495 | 0.81 |
ENSDART00000161938
|
si:dkeyp-9d4.3
|
si:dkeyp-9d4.3 |
| chr1_+_36437585 | 0.79 |
ENSDART00000189182
|
pou4f2
|
POU class 4 homeobox 2 |
| chr20_-_47422469 | 0.72 |
ENSDART00000021341
|
kif3ca
|
kinesin family member 3Ca |
| chr23_-_12015139 | 0.70 |
ENSDART00000110627
ENSDART00000193988 ENSDART00000184528 |
si:dkey-178k16.1
|
si:dkey-178k16.1 |
| chr24_-_38374744 | 0.69 |
ENSDART00000007208
|
lrrc4bb
|
leucine rich repeat containing 4Bb |
| chr19_-_10243148 | 0.69 |
ENSDART00000148073
|
shisa7b
|
shisa family member 7 |
| chr10_+_21776911 | 0.68 |
ENSDART00000163077
ENSDART00000186093 |
pcdh1g22
|
protocadherin 1 gamma 22 |
| chr25_+_21176937 | 0.68 |
ENSDART00000163751
ENSDART00000188930 ENSDART00000187155 |
erc1a
|
ELKS/RAB6-interacting/CAST family member 1a |
| chr3_-_27868183 | 0.68 |
ENSDART00000185812
|
abat
|
4-aminobutyrate aminotransferase |
| chr4_+_20318127 | 0.67 |
ENSDART00000028856
ENSDART00000132909 |
cacna1c
|
calcium channel, voltage-dependent, L type, alpha 1C subunit |
| chr1_-_26782573 | 0.66 |
ENSDART00000090611
|
sh3gl2a
|
SH3 domain containing GRB2 like 2a, endophilin A1 |
| chr8_-_19649617 | 0.64 |
ENSDART00000189033
|
fam78bb
|
family with sequence similarity 78, member B b |
| chr6_+_49412754 | 0.63 |
ENSDART00000027398
|
kcna2a
|
potassium voltage-gated channel, shaker-related subfamily, member 2a |
| chr15_-_19250543 | 0.61 |
ENSDART00000092705
ENSDART00000138895 |
igsf9ba
|
immunoglobulin superfamily, member 9Ba |
| chr24_-_6898302 | 0.58 |
ENSDART00000158646
|
dpp6a
|
dipeptidyl-peptidase 6a |
| chr25_-_7999756 | 0.57 |
ENSDART00000159908
|
camk1db
|
calcium/calmodulin-dependent protein kinase 1Db |
| chr21_+_20715020 | 0.57 |
ENSDART00000015224
|
gadd45gb.1
|
growth arrest and DNA-damage-inducible, gamma b, tandem duplicate 1 |
| chr23_-_12014931 | 0.55 |
ENSDART00000134652
|
si:dkey-178k16.1
|
si:dkey-178k16.1 |
| chr7_-_43840418 | 0.55 |
ENSDART00000174592
|
cdh11
|
cadherin 11, type 2, OB-cadherin (osteoblast) |
| chr5_+_36611128 | 0.54 |
ENSDART00000097684
|
nova1
|
neuro-oncological ventral antigen 1 |
| chr7_-_23996133 | 0.54 |
ENSDART00000173761
|
si:dkey-183c6.8
|
si:dkey-183c6.8 |
| chr23_-_28025943 | 0.53 |
ENSDART00000181146
|
sp5l
|
Sp5 transcription factor-like |
| chr4_-_13156971 | 0.53 |
ENSDART00000182164
|
grip1
|
glutamate receptor interacting protein 1 |
| chr10_-_31805923 | 0.53 |
ENSDART00000077785
|
vps26bl
|
vacuolar protein sorting 26 homolog B, like |
| chr7_+_20966434 | 0.53 |
ENSDART00000185570
|
efnb3b
|
ephrin-B3b |
| chr9_+_22634073 | 0.52 |
ENSDART00000181822
|
etv5a
|
ets variant 5a |
| chr13_-_24447332 | 0.51 |
ENSDART00000043004
|
slc1a4
|
solute carrier family 1 (glutamate/neutral amino acid transporter), member 4 |
| chr17_-_23727978 | 0.49 |
ENSDART00000079600
|
minpp1a
|
multiple inositol-polyphosphate phosphatase 1a |
| chr10_+_15967643 | 0.49 |
ENSDART00000136709
|
apbb1
|
amyloid beta (A4) precursor protein-binding, family B, member 1 (Fe65) |
| chr7_+_27898091 | 0.49 |
ENSDART00000186451
ENSDART00000173510 |
tub
|
tubby bipartite transcription factor |
| chr11_+_42730639 | 0.47 |
ENSDART00000165297
|
zgc:194981
|
zgc:194981 |
| chr6_-_53334259 | 0.46 |
ENSDART00000172465
|
gnb1b
|
guanine nucleotide binding protein (G protein), beta polypeptide 1b |
| chr18_-_2433011 | 0.45 |
ENSDART00000181922
ENSDART00000193276 |
CR769778.1
|
|
| chr20_-_44576949 | 0.45 |
ENSDART00000148639
|
ubxn2a
|
UBX domain protein 2A |
| chr5_-_31856681 | 0.44 |
ENSDART00000187817
|
pkn3
|
protein kinase N3 |
| chr1_-_22834824 | 0.44 |
ENSDART00000043556
|
ldb2b
|
LIM domain binding 2b |
| chr25_+_7670683 | 0.44 |
ENSDART00000040275
|
kcnj11l
|
potassium inwardly-rectifying channel, subfamily J, member 11, like |
| chr15_-_39820491 | 0.44 |
ENSDART00000097134
|
robo1
|
roundabout, axon guidance receptor, homolog 1 (Drosophila) |
| chr10_+_21660447 | 0.43 |
ENSDART00000164519
|
pcdh1g3
|
protocadherin 1 gamma 3 |
| chr5_-_21065094 | 0.42 |
ENSDART00000143785
|
si:dkey-13n15.2
|
si:dkey-13n15.2 |
| chr14_-_48348973 | 0.42 |
ENSDART00000185822
|
CABZ01080056.1
|
|
| chr5_-_51819027 | 0.42 |
ENSDART00000164267
|
homer1b
|
homer scaffolding protein 1b |
| chr18_+_31280984 | 0.41 |
ENSDART00000170285
ENSDART00000150608 ENSDART00000159720 |
def8
|
differentially expressed in FDCP 8 homolog (mouse) |
| chr14_+_32022272 | 0.41 |
ENSDART00000105760
|
zic6
|
zic family member 6 |
| chr23_-_39849155 | 0.39 |
ENSDART00000115330
|
ppp1r14c
|
protein phosphatase 1, regulatory (inhibitor) subunit 14C |
| chr15_+_44093286 | 0.38 |
ENSDART00000114352
|
zgc:112998
|
zgc:112998 |
| chr12_-_48552716 | 0.38 |
ENSDART00000063438
|
LO017829.1
|
Danio rerio testis expressed 47 (tex47), mRNA. |
| chr11_+_43751263 | 0.38 |
ENSDART00000163843
|
zgc:153431
|
zgc:153431 |
| chr7_-_32833153 | 0.38 |
ENSDART00000099871
ENSDART00000099872 |
slc17a6b
|
solute carrier family 17 (vesicular glutamate transporter), member 6b |
| chr6_-_53281518 | 0.37 |
ENSDART00000157621
|
rbm5
|
RNA binding motif protein 5 |
| chr16_+_5184402 | 0.36 |
ENSDART00000156685
|
soga3a
|
SOGA family member 3a |
| chr2_-_39759059 | 0.36 |
ENSDART00000007333
|
slc25a36a
|
solute carrier family 25 (pyrimidine nucleotide carrier ), member 36a |
| chr15_-_14552101 | 0.35 |
ENSDART00000171169
|
numbl
|
numb homolog (Drosophila)-like |
| chr22_-_31517300 | 0.34 |
ENSDART00000164799
|
slc6a6b
|
solute carrier family 6 (neurotransmitter transporter), member 6b |
| chr25_-_18002937 | 0.34 |
ENSDART00000149696
|
cep290
|
centrosomal protein 290 |
| chr1_-_7894255 | 0.33 |
ENSDART00000167126
ENSDART00000145460 |
radil
|
Ras association and DIL domains |
| chr20_+_48803248 | 0.33 |
ENSDART00000164006
|
nkx2.4b
|
NK2 homeobox 4b |
| chr3_+_24190207 | 0.33 |
ENSDART00000034762
|
prr15la
|
proline rich 15-like a |
| chr6_+_2174082 | 0.32 |
ENSDART00000073936
|
acvr1bb
|
activin A receptor type 1Bb |
| chr7_+_59739164 | 0.32 |
ENSDART00000110412
|
gpr78b
|
G protein-coupled receptor 78b |
| chr14_-_31060082 | 0.32 |
ENSDART00000111601
ENSDART00000161113 |
mbnl3
|
muscleblind-like splicing regulator 3 |
| chr6_-_28943056 | 0.31 |
ENSDART00000065138
|
tbc1d23
|
TBC1 domain family, member 23 |
| chr23_-_19434199 | 0.31 |
ENSDART00000132543
|
klhdc8b
|
kelch domain containing 8B |
| chr4_-_23082985 | 0.31 |
ENSDART00000133644
|
magi2a
|
membrane associated guanylate kinase, WW and PDZ domain containing 2a |
| chr15_-_6863150 | 0.30 |
ENSDART00000153815
|
meis3
|
myeloid ecotropic viral integration site 3 |
| chr10_+_3049636 | 0.30 |
ENSDART00000081794
ENSDART00000183167 ENSDART00000191634 ENSDART00000183514 |
rasgrf2a
|
Ras protein-specific guanine nucleotide-releasing factor 2a |
| chr21_-_4539899 | 0.30 |
ENSDART00000112460
|
dolk
|
dolichol kinase |
| chr2_-_18830722 | 0.30 |
ENSDART00000165330
ENSDART00000165698 |
pbx1a
|
pre-B-cell leukemia homeobox 1a |
| chrM_+_3803 | 0.28 |
ENSDART00000093596
|
mt-nd1
|
NADH dehydrogenase 1, mitochondrial |
| chr4_-_6567355 | 0.27 |
ENSDART00000134820
ENSDART00000142087 |
foxp2
|
forkhead box P2 |
| chr9_-_10145795 | 0.27 |
ENSDART00000004745
ENSDART00000143295 |
hnmt
|
histamine N-methyltransferase |
| chr9_-_38399432 | 0.27 |
ENSDART00000148268
|
znf142
|
zinc finger protein 142 |
| chr14_+_45883687 | 0.27 |
ENSDART00000114790
|
flrt1b
|
fibronectin leucine rich transmembrane protein 1b |
| chr13_-_9875538 | 0.26 |
ENSDART00000041609
|
tm9sf3
|
transmembrane 9 superfamily member 3 |
| chr5_-_31857345 | 0.25 |
ENSDART00000112546
|
pkn3
|
protein kinase N3 |
| chr13_+_39182099 | 0.25 |
ENSDART00000131434
|
fam135a
|
family with sequence similarity 135, member A |
| chr16_-_6205790 | 0.25 |
ENSDART00000038495
|
ctnnb1
|
catenin (cadherin-associated protein), beta 1 |
| chr6_-_14038804 | 0.25 |
ENSDART00000184606
ENSDART00000184609 |
etv5b
|
ets variant 5b |
| chr1_-_22803147 | 0.24 |
ENSDART00000086867
|
tapt1b
|
transmembrane anterior posterior transformation 1b |
| chr15_+_5132439 | 0.24 |
ENSDART00000010350
|
pgm2l1
|
phosphoglucomutase 2-like 1 |
| chr17_+_49500820 | 0.23 |
ENSDART00000170306
|
AREL1
|
apoptosis resistant E3 ubiquitin protein ligase 1 |
| chr6_-_28222592 | 0.23 |
ENSDART00000131126
|
bcl6a
|
B-cell CLL/lymphoma 6a (zinc finger protein 51) |
| chr23_+_23658474 | 0.23 |
ENSDART00000162838
|
agrn
|
agrin |
| chr5_-_32489796 | 0.23 |
ENSDART00000168870
|
gpr107
|
G protein-coupled receptor 107 |
| chr6_-_13709591 | 0.22 |
ENSDART00000151771
|
chpfb
|
chondroitin polymerizing factor b |
| chr6_+_49551614 | 0.21 |
ENSDART00000022581
|
rab22a
|
RAB22A, member RAS oncogene family |
| chr16_+_38659475 | 0.21 |
ENSDART00000023238
|
emc2
|
ER membrane protein complex subunit 2 |
| chr8_+_26293673 | 0.21 |
ENSDART00000144977
|
mgll
|
monoglyceride lipase |
| chr17_+_23255365 | 0.21 |
ENSDART00000180277
|
AL935174.5
|
|
| chr11_-_26832685 | 0.21 |
ENSDART00000153519
|
iqsec1b
|
IQ motif and Sec7 domain 1b |
| chr15_-_20412286 | 0.21 |
ENSDART00000008589
|
chp2
|
calcineurin-like EF-hand protein 2 |
| chr24_+_21540842 | 0.21 |
ENSDART00000091529
|
wasf3b
|
WAS protein family, member 3b |
| chr5_+_17624463 | 0.20 |
ENSDART00000183869
ENSDART00000081064 |
fbrsl1
|
fibrosin-like 1 |
| chr14_-_36863432 | 0.19 |
ENSDART00000158052
|
rnf130
|
ring finger protein 130 |
| chr3_+_36284986 | 0.19 |
ENSDART00000059533
|
wipi1
|
WD repeat domain, phosphoinositide interacting 1 |
| chr10_-_15910974 | 0.19 |
ENSDART00000148169
|
pip5k1ba
|
phosphatidylinositol-4-phosphate 5-kinase, type I, beta a |
| chr25_-_18002498 | 0.19 |
ENSDART00000158688
|
cep290
|
centrosomal protein 290 |
| chr15_-_23942861 | 0.19 |
ENSDART00000002824
|
appbp2
|
amyloid beta precursor protein (cytoplasmic tail) binding protein 2 |
| chr24_-_26715174 | 0.18 |
ENSDART00000079726
|
pld1b
|
phospholipase D1b |
| chr11_-_37717709 | 0.18 |
ENSDART00000177280
|
yod1
|
YOD1 deubiquitinase |
| chr2_-_22966076 | 0.18 |
ENSDART00000143412
ENSDART00000146014 ENSDART00000183443 ENSDART00000191056 ENSDART00000183539 |
sap130b
|
Sin3A-associated protein b |
| chr7_+_29167744 | 0.18 |
ENSDART00000076345
|
slc38a8b
|
solute carrier family 38, member 8b |
| chr20_-_23656516 | 0.17 |
ENSDART00000149735
|
cbr4
|
carbonyl reductase 4 |
| chr20_-_23657304 | 0.17 |
ENSDART00000016913
|
cbr4
|
carbonyl reductase 4 |
| chr15_-_28262632 | 0.17 |
ENSDART00000134601
ENSDART00000175022 |
prpf8
|
pre-mRNA processing factor 8 |
| chr10_+_31222433 | 0.17 |
ENSDART00000185080
|
tmem218
|
transmembrane protein 218 |
| chr20_+_38837238 | 0.16 |
ENSDART00000061334
|
ift172
|
intraflagellar transport 172 |
| chr4_-_38259998 | 0.16 |
ENSDART00000172389
|
znf1093
|
zinc finger protein 1093 |
| chr19_+_26718074 | 0.16 |
ENSDART00000134455
|
zgc:100906
|
zgc:100906 |
| chr2_+_54696042 | 0.15 |
ENSDART00000074270
|
ankrd12
|
ankyrin repeat domain 12 |
| chr15_+_29116063 | 0.15 |
ENSDART00000016112
ENSDART00000153609 ENSDART00000155630 |
capns1b
|
calpain, small subunit 1 b |
| chr18_-_21685055 | 0.15 |
ENSDART00000019861
|
mbtps1
|
membrane-bound transcription factor peptidase, site 1 |
| chr6_+_40775800 | 0.14 |
ENSDART00000085090
|
si:ch211-157b11.8
|
si:ch211-157b11.8 |
| chr24_-_31223232 | 0.14 |
ENSDART00000164155
|
alg14
|
ALG14, UDP-N-acetylglucosaminyltransferase subunit |
| chr13_-_18119696 | 0.14 |
ENSDART00000148125
|
washc2c
|
WASH complex subunit 2C |
| chr2_-_37134169 | 0.14 |
ENSDART00000146123
ENSDART00000146533 ENSDART00000040427 |
elavl1a
|
ELAV like RNA binding protein 1a |
| chr7_+_30240791 | 0.13 |
ENSDART00000109243
|
sema4bb
|
sema domain, immunoglobulin domain (Ig), transmembrane domain (TM) and short cytoplasmic domain, (semaphorin) 4Bb |
| chr6_-_54816567 | 0.13 |
ENSDART00000150079
|
tnni1b
|
troponin I type 1b (skeletal, slow) |
| chr16_-_31452416 | 0.13 |
ENSDART00000140880
ENSDART00000008297 ENSDART00000147373 |
csnk2a1
|
casein kinase 2, alpha 1 polypeptide |
| chr19_+_26640096 | 0.12 |
ENSDART00000067793
|
ints3
|
integrator complex subunit 3 |
| chr17_-_32865788 | 0.12 |
ENSDART00000077476
|
prox1a
|
prospero homeobox 1a |
| chr22_+_17286841 | 0.12 |
ENSDART00000133176
|
FAM163A
|
si:dkey-171o17.8 |
| chr6_+_13207139 | 0.12 |
ENSDART00000185601
ENSDART00000182182 |
ino80db
|
INO80 complex subunit Db |
| chr2_+_30103285 | 0.12 |
ENSDART00000019417
|
dnajb6a
|
DnaJ (Hsp40) homolog, subfamily B, member 6a |
| chr2_-_55797318 | 0.12 |
ENSDART00000158147
|
calr3b
|
calreticulin 3b |
| chr6_-_34838397 | 0.11 |
ENSDART00000060169
ENSDART00000169605 |
mier1a
|
mesoderm induction early response 1a, transcriptional regulator |
| chr14_-_26425416 | 0.11 |
ENSDART00000088690
|
lman2
|
lectin, mannose-binding 2 |
| chr11_+_6881001 | 0.11 |
ENSDART00000170331
|
klhl26
|
kelch-like family member 26 |
| chr11_-_18557929 | 0.11 |
ENSDART00000110882
ENSDART00000181381 ENSDART00000189312 |
dido1
|
death inducer-obliterator 1 |
| chr5_-_69499486 | 0.10 |
ENSDART00000023983
ENSDART00000180293 |
psat1
|
phosphoserine aminotransferase 1 |
| chr1_+_36612660 | 0.10 |
ENSDART00000190784
|
ednraa
|
endothelin receptor type Aa |
| chr16_+_5898878 | 0.09 |
ENSDART00000180930
|
ulk4
|
unc-51 like kinase 4 |
| chr3_-_40254634 | 0.09 |
ENSDART00000154562
|
top3a
|
DNA topoisomerase III alpha |
| chr14_-_32866351 | 0.09 |
ENSDART00000166133
ENSDART00000172545 |
ube2a
|
ubiquitin-conjugating enzyme E2A (RAD6 homolog) |
| chr13_+_3880882 | 0.09 |
ENSDART00000169595
|
timm23b
|
translocase of inner mitochondrial membrane 23 homolog b (yeast) |
| chr2_+_23039041 | 0.09 |
ENSDART00000056921
|
csnk1g2a
|
casein kinase 1, gamma 2a |
| chr4_-_30712588 | 0.08 |
ENSDART00000142393
|
si:dkey-16p19.5
|
si:dkey-16p19.5 |
| chr5_-_54395488 | 0.08 |
ENSDART00000160781
|
zmynd19
|
zinc finger, MYND-type containing 19 |
| chr22_+_30137374 | 0.08 |
ENSDART00000187808
|
add3a
|
adducin 3 (gamma) a |
| chr9_+_44034790 | 0.08 |
ENSDART00000166110
ENSDART00000176954 |
itga4
|
integrin alpha 4 |
| chr12_-_17199381 | 0.08 |
ENSDART00000193292
|
lipf
|
lipase, gastric |
| chr2_+_8261109 | 0.07 |
ENSDART00000024677
|
dia1a
|
deleted in autism 1a |
| chr6_+_27992886 | 0.07 |
ENSDART00000160354
|
amotl2a
|
angiomotin like 2a |
| chr4_-_28958601 | 0.07 |
ENSDART00000111294
|
zgc:174315
|
zgc:174315 |
| chr2_+_31330358 | 0.07 |
ENSDART00000178066
|
clul1
|
clusterin-like 1 (retinal) |
| chr2_+_55199721 | 0.06 |
ENSDART00000016143
|
zmp:0000000521
|
zmp:0000000521 |
| chr4_-_13995766 | 0.06 |
ENSDART00000147955
|
prickle1b
|
prickle homolog 1b |
| chr13_-_36050303 | 0.06 |
ENSDART00000134955
ENSDART00000139087 |
lgmn
|
legumain |
| chr4_-_73520581 | 0.06 |
ENSDART00000171513
|
si:ch73-266f23.1
|
si:ch73-266f23.1 |
| chr12_+_46696867 | 0.06 |
ENSDART00000152928
ENSDART00000153445 ENSDART00000123357 ENSDART00000152880 ENSDART00000123834 ENSDART00000174765 ENSDART00000189923 |
exoc7
|
exocyst complex component 7 |
| chr6_-_9236309 | 0.06 |
ENSDART00000160397
ENSDART00000164603 |
iqcb1
|
IQ motif containing B1 |
| chr2_-_50966124 | 0.06 |
ENSDART00000170470
|
ghrhra
|
growth hormone releasing hormone receptor a |
| chr8_+_52442785 | 0.05 |
ENSDART00000189958
|
zgc:77112
|
zgc:77112 |
| chr6_+_3730843 | 0.05 |
ENSDART00000019630
|
FO704755.1
|
|
| chr13_-_3351708 | 0.05 |
ENSDART00000042875
|
si:ch73-193i2.2
|
si:ch73-193i2.2 |
| chr10_+_26612321 | 0.04 |
ENSDART00000134322
|
fhl1b
|
four and a half LIM domains 1b |
| chr7_-_30624435 | 0.04 |
ENSDART00000173828
|
rnf111
|
ring finger protein 111 |
| chr3_-_18737126 | 0.04 |
ENSDART00000055767
|
e4f1
|
E4F transcription factor 1 |
| chr5_+_32490238 | 0.04 |
ENSDART00000191839
|
ndor1
|
NADPH dependent diflavin oxidoreductase 1 |
| chr12_-_11560794 | 0.03 |
ENSDART00000149098
ENSDART00000169975 |
plekha1b
|
pleckstrin homology domain containing, family A (phosphoinositide binding specific) member 1b |
| chr3_-_48612078 | 0.03 |
ENSDART00000169923
|
ndel1b
|
nudE neurodevelopment protein 1-like 1b |
| chr16_-_47426482 | 0.03 |
ENSDART00000148631
ENSDART00000149723 |
sept7b
|
septin 7b |
| chr13_+_8892784 | 0.03 |
ENSDART00000075054
ENSDART00000143705 |
thada
|
thyroid adenoma associated |
| chr18_+_18000887 | 0.02 |
ENSDART00000147797
|
si:ch211-212o1.2
|
si:ch211-212o1.2 |
| chr8_-_65189 | 0.02 |
ENSDART00000168412
|
hsd17b4
|
hydroxysteroid (17-beta) dehydrogenase 4 |
| chr4_-_18434924 | 0.02 |
ENSDART00000190271
|
socs2
|
suppressor of cytokine signaling 2 |
| chr14_+_2487672 | 0.02 |
ENSDART00000170629
ENSDART00000123063 |
fgf18a
|
fibroblast growth factor 18a |
| chr20_-_19530751 | 0.02 |
ENSDART00000148574
|
eif2b4
|
eukaryotic translation initiation factor 2B, subunit 4 delta |
| chr12_-_10038870 | 0.01 |
ENSDART00000152250
|
ngfrb
|
nerve growth factor receptor b |
| chr19_+_30884960 | 0.01 |
ENSDART00000140603
ENSDART00000183224 ENSDART00000135484 ENSDART00000139599 |
yars
|
tyrosyl-tRNA synthetase |
| chr15_-_2754056 | 0.01 |
ENSDART00000129380
|
ppp5c
|
protein phosphatase 5, catalytic subunit |
| chr21_+_4540127 | 0.01 |
ENSDART00000043431
|
nup188
|
nucleoporin 188 |
| chr13_-_25196758 | 0.01 |
ENSDART00000184722
|
adka
|
adenosine kinase a |
| chr20_+_34671386 | 0.01 |
ENSDART00000152836
ENSDART00000138226 |
elp3
|
elongator acetyltransferase complex subunit 3 |
| chr22_+_2403068 | 0.01 |
ENSDART00000132925
ENSDART00000132569 |
zgc:112977
|
zgc:112977 |
| chr5_-_24542726 | 0.01 |
ENSDART00000182975
|
trmt2a
|
tRNA methyltransferase 2 homolog A |
| chr25_-_37121335 | 0.01 |
ENSDART00000017805
|
nfat5a
|
nuclear factor of activated T cells 5a |
| chr19_+_5315987 | 0.01 |
ENSDART00000145749
|
si:dkeyp-113d7.1
|
si:dkeyp-113d7.1 |
Network of associatons between targets according to the STRING database.
First level regulatory network of hoxc12a+hoxc12b
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 1.0 | GO:0046677 | response to antibiotic(GO:0046677) |
| 0.2 | 1.1 | GO:0060052 | neurofilament cytoskeleton organization(GO:0060052) |
| 0.1 | 0.7 | GO:0009450 | gamma-aminobutyric acid catabolic process(GO:0009450) |
| 0.1 | 0.7 | GO:0014896 | muscle hypertrophy(GO:0014896) |
| 0.1 | 0.4 | GO:1990519 | pyrimidine nucleotide transport(GO:0006864) mitochondrial pyrimidine nucleotide import(GO:1990519) |
| 0.1 | 0.3 | GO:1990403 | embryonic brain development(GO:1990403) |
| 0.1 | 0.3 | GO:0001692 | histamine metabolic process(GO:0001692) |
| 0.1 | 0.5 | GO:0042769 | DNA damage response, detection of DNA damage(GO:0042769) |
| 0.1 | 0.2 | GO:2000055 | positive regulation of Wnt signaling pathway involved in dorsal/ventral axis specification(GO:2000055) |
| 0.1 | 1.2 | GO:0060012 | synaptic transmission, glycinergic(GO:0060012) |
| 0.1 | 0.2 | GO:0045724 | positive regulation of cilium assembly(GO:0045724) |
| 0.1 | 0.2 | GO:1902571 | regulation of serine-type peptidase activity(GO:1902571) |
| 0.1 | 0.6 | GO:0000185 | activation of MAPKKK activity(GO:0000185) |
| 0.1 | 0.7 | GO:0099558 | maintenance of presynaptic active zone structure(GO:0048790) maintenance of synapse structure(GO:0099558) |
| 0.1 | 0.3 | GO:1901166 | neural crest cell migration involved in autonomic nervous system development(GO:1901166) |
| 0.1 | 0.4 | GO:2001256 | regulation of store-operated calcium entry(GO:2001256) |
| 0.1 | 0.4 | GO:0003262 | endocardial progenitor cell migration to the midline involved in heart field formation(GO:0003262) |
| 0.1 | 0.6 | GO:0030431 | sleep(GO:0030431) |
| 0.1 | 0.5 | GO:0048172 | regulation of short-term neuronal synaptic plasticity(GO:0048172) |
| 0.0 | 0.3 | GO:0048795 | swim bladder morphogenesis(GO:0048795) |
| 0.0 | 0.3 | GO:0051290 | protein heterotetramerization(GO:0051290) |
| 0.0 | 0.5 | GO:0060217 | hemangioblast cell differentiation(GO:0060217) |
| 0.0 | 0.4 | GO:0098700 | neurotransmitter loading into synaptic vesicle(GO:0098700) |
| 0.0 | 0.2 | GO:0032534 | regulation of microvillus organization(GO:0032530) regulation of microvillus assembly(GO:0032534) |
| 0.0 | 0.2 | GO:1990167 | protein K27-linked deubiquitination(GO:1990167) |
| 0.0 | 0.5 | GO:1902624 | positive regulation of neutrophil migration(GO:1902624) |
| 0.0 | 0.5 | GO:0099638 | neurotransmitter receptor transport, endosome to postsynaptic membrane(GO:0098887) endosome to plasma membrane protein transport(GO:0099638) neurotransmitter receptor transport, endosome to plasma membrane(GO:0099639) |
| 0.0 | 0.9 | GO:0070831 | basement membrane assembly(GO:0070831) |
| 0.0 | 0.1 | GO:0006564 | L-serine biosynthetic process(GO:0006564) |
| 0.0 | 0.2 | GO:0045050 | protein insertion into ER membrane by stop-transfer membrane-anchor sequence(GO:0045050) |
| 0.0 | 0.4 | GO:0090110 | cargo loading into COPII-coated vesicle(GO:0090110) |
| 0.0 | 0.2 | GO:0000244 | spliceosomal tri-snRNP complex assembly(GO:0000244) |
| 0.0 | 0.4 | GO:0031468 | nuclear envelope reassembly(GO:0031468) |
| 0.0 | 0.5 | GO:0021654 | rhombomere boundary formation(GO:0021654) |
| 0.0 | 0.9 | GO:0006904 | vesicle docking involved in exocytosis(GO:0006904) |
| 0.0 | 0.6 | GO:1901379 | regulation of potassium ion transmembrane transport(GO:1901379) |
| 0.0 | 0.3 | GO:0030878 | thyroid gland development(GO:0030878) |
| 0.0 | 1.2 | GO:0030866 | cortical actin cytoskeleton organization(GO:0030866) |
| 0.0 | 0.5 | GO:0036342 | post-anal tail morphogenesis(GO:0036342) |
| 0.0 | 2.3 | GO:0006836 | neurotransmitter transport(GO:0006836) |
| 0.0 | 0.2 | GO:0021979 | hypothalamus cell differentiation(GO:0021979) |
| 0.0 | 0.1 | GO:0035176 | social behavior(GO:0035176) intraspecies interaction between organisms(GO:0051703) |
| 0.0 | 0.2 | GO:0034497 | protein localization to pre-autophagosomal structure(GO:0034497) |
| 0.0 | 0.6 | GO:0016339 | calcium-dependent cell-cell adhesion via plasma membrane cell adhesion molecules(GO:0016339) |
| 0.0 | 0.1 | GO:0045668 | negative regulation of osteoblast differentiation(GO:0045668) |
| 0.0 | 0.1 | GO:0006488 | dolichol-linked oligosaccharide biosynthetic process(GO:0006488) |
| 0.0 | 0.2 | GO:2000601 | positive regulation of actin nucleation(GO:0051127) positive regulation of Arp2/3 complex-mediated actin nucleation(GO:2000601) |
| 0.0 | 0.2 | GO:0030206 | chondroitin sulfate biosynthetic process(GO:0030206) |
| 0.0 | 1.2 | GO:0000381 | regulation of alternative mRNA splicing, via spliceosome(GO:0000381) |
| 0.0 | 0.2 | GO:0021508 | floor plate formation(GO:0021508) floor plate morphogenesis(GO:0033505) |
| 0.0 | 0.4 | GO:0045746 | negative regulation of Notch signaling pathway(GO:0045746) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 1.0 | GO:0005883 | neurofilament(GO:0005883) |
| 0.1 | 0.3 | GO:0071203 | WASH complex(GO:0071203) |
| 0.0 | 0.6 | GO:0044224 | juxtaparanode region of axon(GO:0044224) |
| 0.0 | 0.5 | GO:0034451 | centriolar satellite(GO:0034451) |
| 0.0 | 0.5 | GO:0030904 | retromer complex(GO:0030904) |
| 0.0 | 0.9 | GO:0043256 | basal lamina(GO:0005605) laminin complex(GO:0043256) |
| 0.0 | 0.2 | GO:0005915 | cell-cell adherens junction(GO:0005913) zonula adherens(GO:0005915) |
| 0.0 | 0.5 | GO:0032591 | dendritic spine membrane(GO:0032591) |
| 0.0 | 0.4 | GO:0030285 | integral component of synaptic vesicle membrane(GO:0030285) intrinsic component of synaptic vesicle membrane(GO:0098563) |
| 0.0 | 0.2 | GO:0034045 | pre-autophagosomal structure membrane(GO:0034045) |
| 0.0 | 0.7 | GO:0048788 | cytoskeleton of presynaptic active zone(GO:0048788) |
| 0.0 | 0.3 | GO:0048179 | activin receptor complex(GO:0048179) |
| 0.0 | 0.1 | GO:0070876 | SOSS complex(GO:0070876) |
| 0.0 | 0.4 | GO:0030127 | COPII vesicle coat(GO:0030127) |
| 0.0 | 0.2 | GO:0072546 | ER membrane protein complex(GO:0072546) |
| 0.0 | 0.7 | GO:0032154 | cleavage furrow(GO:0032154) cell surface furrow(GO:0097610) |
| 0.0 | 0.1 | GO:0033503 | HULC complex(GO:0033503) |
| 0.0 | 0.9 | GO:0030672 | synaptic vesicle membrane(GO:0030672) exocytic vesicle membrane(GO:0099501) |
| 0.0 | 0.2 | GO:0031209 | SCAR complex(GO:0031209) |
| 0.0 | 0.9 | GO:0030141 | secretory granule(GO:0030141) |
| 0.0 | 0.6 | GO:0005788 | endoplasmic reticulum lumen(GO:0005788) |
| 0.0 | 0.1 | GO:0005956 | protein kinase CK2 complex(GO:0005956) |
| 0.0 | 0.6 | GO:0016342 | catenin complex(GO:0016342) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 1.0 | GO:0008131 | primary amine oxidase activity(GO:0008131) |
| 0.1 | 1.2 | GO:0016934 | extracellular-glycine-gated ion channel activity(GO:0016933) extracellular-glycine-gated chloride channel activity(GO:0016934) |
| 0.1 | 0.5 | GO:0034416 | bisphosphoglycerate phosphatase activity(GO:0034416) |
| 0.1 | 0.4 | GO:0015218 | pyrimidine nucleotide transmembrane transporter activity(GO:0015218) |
| 0.1 | 0.4 | GO:0008046 | axon guidance receptor activity(GO:0008046) |
| 0.1 | 0.4 | GO:0035256 | G-protein coupled glutamate receptor binding(GO:0035256) |
| 0.1 | 0.3 | GO:0031697 | beta-1 adrenergic receptor binding(GO:0031697) type II activin receptor binding(GO:0070699) |
| 0.1 | 1.0 | GO:0005332 | gamma-aminobutyric acid:sodium symporter activity(GO:0005332) |
| 0.1 | 0.4 | GO:0030274 | LIM domain binding(GO:0030274) |
| 0.1 | 0.5 | GO:0004563 | beta-N-acetylhexosaminidase activity(GO:0004563) |
| 0.1 | 0.2 | GO:0030623 | U5 snRNA binding(GO:0030623) |
| 0.1 | 0.3 | GO:0016361 | activin receptor activity, type I(GO:0016361) |
| 0.0 | 0.5 | GO:0035259 | glucocorticoid receptor binding(GO:0035259) |
| 0.0 | 0.4 | GO:0004865 | protein serine/threonine phosphatase inhibitor activity(GO:0004865) |
| 0.0 | 0.7 | GO:0098882 | structural constituent of presynaptic active zone(GO:0098882) |
| 0.0 | 0.9 | GO:0005388 | calcium-transporting ATPase activity(GO:0005388) |
| 0.0 | 0.6 | GO:0008239 | dipeptidyl-peptidase activity(GO:0008239) |
| 0.0 | 0.4 | GO:0015272 | ATP-activated inward rectifier potassium channel activity(GO:0015272) |
| 0.0 | 0.5 | GO:0031682 | G-protein gamma-subunit binding(GO:0031682) |
| 0.0 | 0.4 | GO:0005326 | neurotransmitter transporter activity(GO:0005326) |
| 0.0 | 0.2 | GO:0045294 | alpha-catenin binding(GO:0045294) |
| 0.0 | 0.8 | GO:0008483 | transaminase activity(GO:0008483) |
| 0.0 | 0.2 | GO:0004630 | phospholipase D activity(GO:0004630) |
| 0.0 | 0.7 | GO:0004697 | protein kinase C activity(GO:0004697) calcium-dependent protein kinase C activity(GO:0004698) |
| 0.0 | 0.5 | GO:0046875 | ephrin receptor binding(GO:0046875) |
| 0.0 | 0.2 | GO:0047238 | glucuronosyl-N-acetylgalactosaminyl-proteoglycan 4-beta-N-acetylgalactosaminyltransferase activity(GO:0047238) |
| 0.0 | 0.2 | GO:0032977 | membrane insertase activity(GO:0032977) |
| 0.0 | 0.9 | GO:0017075 | syntaxin-1 binding(GO:0017075) |
| 0.0 | 0.2 | GO:0071933 | Arp2/3 complex binding(GO:0071933) |
| 0.0 | 0.7 | GO:0008331 | high voltage-gated calcium channel activity(GO:0008331) |
| 0.0 | 0.4 | GO:0050136 | NADH dehydrogenase (ubiquinone) activity(GO:0008137) NADH dehydrogenase (quinone) activity(GO:0050136) |
| 0.0 | 0.5 | GO:0001540 | beta-amyloid binding(GO:0001540) |
| 0.0 | 0.3 | GO:0048038 | quinone binding(GO:0048038) |
| 0.0 | 0.2 | GO:0016868 | intramolecular transferase activity, phosphotransferases(GO:0016868) |
| 0.0 | 0.2 | GO:0032050 | clathrin heavy chain binding(GO:0032050) |
| 0.0 | 0.2 | GO:0047372 | acylglycerol lipase activity(GO:0047372) |
| 0.0 | 0.3 | GO:0005343 | organic acid:sodium symporter activity(GO:0005343) |
| 0.0 | 0.6 | GO:0005251 | delayed rectifier potassium channel activity(GO:0005251) |
| 0.0 | 0.1 | GO:0004962 | endothelin receptor activity(GO:0004962) |
| 0.0 | 0.0 | GO:0032184 | SUMO polymer binding(GO:0032184) |
| 0.0 | 0.6 | GO:0004683 | calmodulin-dependent protein kinase activity(GO:0004683) |
| 0.0 | 0.5 | GO:0004869 | cysteine-type endopeptidase inhibitor activity(GO:0004869) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 1.3 | PID ERA GENOMIC PATHWAY | Validated nuclear estrogen receptor alpha network |
| 0.0 | 0.2 | PID BETA CATENIN DEG PATHWAY | Degradation of beta catenin |
| 0.0 | 0.4 | PID AJDISS 2PATHWAY | Posttranslational regulation of adherens junction stability and dissassembly |
| 0.0 | 0.4 | PID ILK PATHWAY | Integrin-linked kinase signaling |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.2 | REACTOME APOPTOTIC CLEAVAGE OF CELL ADHESION PROTEINS | Genes involved in Apoptotic cleavage of cell adhesion proteins |
| 0.0 | 0.7 | REACTOME GABA SYNTHESIS RELEASE REUPTAKE AND DEGRADATION | Genes involved in GABA synthesis, release, reuptake and degradation |
| 0.0 | 0.2 | REACTOME HORMONE SENSITIVE LIPASE HSL MEDIATED TRIACYLGLYCEROL HYDROLYSIS | Genes involved in Hormone-sensitive lipase (HSL)-mediated triacylglycerol hydrolysis |
| 0.0 | 0.4 | REACTOME ACTIVATION OF RAC | Genes involved in Activation of Rac |
| 0.0 | 0.6 | REACTOME ADHERENS JUNCTIONS INTERACTIONS | Genes involved in Adherens junctions interactions |
| 0.0 | 0.5 | REACTOME AMINO ACID TRANSPORT ACROSS THE PLASMA MEMBRANE | Genes involved in Amino acid transport across the plasma membrane |
| 0.0 | 0.3 | REACTOME SYNTHESIS OF SUBSTRATES IN N GLYCAN BIOSYTHESIS | Genes involved in Synthesis of substrates in N-glycan biosythesis |
| 0.0 | 0.2 | REACTOME HS GAG BIOSYNTHESIS | Genes involved in HS-GAG biosynthesis |
| 0.0 | 0.1 | REACTOME ACTIVATION OF CHAPERONES BY ATF6 ALPHA | Genes involved in Activation of Chaperones by ATF6-alpha |
| 0.0 | 0.5 | REACTOME LOSS OF NLP FROM MITOTIC CENTROSOMES | Genes involved in Loss of Nlp from mitotic centrosomes |
| 0.0 | 0.1 | REACTOME AMINO ACID SYNTHESIS AND INTERCONVERSION TRANSAMINATION | Genes involved in Amino acid synthesis and interconversion (transamination) |