Project
PRJNA207719: Tissue specific transcriptome profiling
Navigation
Downloads
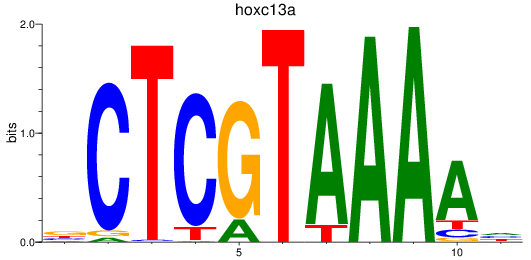
Results for hoxc13a
Z-value: 0.76
Transcription factors associated with hoxc13a
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
hoxc13a
|
ENSDARG00000070353 | homeobox C13a |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| hoxc13a | dr11_v1_chr23_+_36052944_36052944 | -0.19 | 7.6e-01 | Click! |
Activity profile of hoxc13a motif
Sorted Z-values of hoxc13a motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr25_+_31277415 | 0.59 |
ENSDART00000036275
|
tnni2a.4
|
troponin I type 2a (skeletal, fast), tandem duplicate 4 |
| chr15_-_23647078 | 0.57 |
ENSDART00000059366
|
ckmb
|
creatine kinase, muscle b |
| chr25_+_31276842 | 0.49 |
ENSDART00000187238
|
tnni2a.4
|
troponin I type 2a (skeletal, fast), tandem duplicate 4 |
| chr24_-_6158933 | 0.48 |
ENSDART00000021609
|
gad2
|
glutamate decarboxylase 2 |
| chr15_-_25518084 | 0.42 |
ENSDART00000158594
|
hif1al
|
hypoxia-inducible factor 1, alpha subunit, like |
| chr3_+_59851537 | 0.39 |
ENSDART00000180997
|
CU693479.1
|
|
| chr25_+_19947298 | 0.31 |
ENSDART00000067648
|
kcna6a
|
potassium voltage-gated channel, shaker-related, subfamily, member 6 a |
| chr19_+_4916233 | 0.31 |
ENSDART00000159512
|
ppp1r1b
|
protein phosphatase 1, regulatory (inhibitor) subunit 1B |
| chr9_+_38967998 | 0.29 |
ENSDART00000135581
|
map2
|
microtubule-associated protein 2 |
| chr3_+_23691847 | 0.29 |
ENSDART00000078453
|
hoxb7a
|
homeobox B7a |
| chr14_-_3031810 | 0.28 |
ENSDART00000090213
|
ndst1a
|
N-deacetylase/N-sulfotransferase (heparan glucosaminyl) 1a |
| chr3_-_48612078 | 0.25 |
ENSDART00000169923
|
ndel1b
|
nudE neurodevelopment protein 1-like 1b |
| chr1_+_18965750 | 0.25 |
ENSDART00000132379
|
limch1a
|
LIM and calponin homology domains 1a |
| chr1_+_20069535 | 0.24 |
ENSDART00000088653
|
prss12
|
protease, serine, 12 (neurotrypsin, motopsin) |
| chr14_-_3032016 | 0.24 |
ENSDART00000183461
ENSDART00000183035 |
ndst1a
|
N-deacetylase/N-sulfotransferase (heparan glucosaminyl) 1a |
| chr22_+_18477934 | 0.22 |
ENSDART00000132684
|
cilp2
|
cartilage intermediate layer protein 2 |
| chr21_-_25756119 | 0.20 |
ENSDART00000002341
|
cldnc
|
claudin c |
| chr19_-_28367413 | 0.19 |
ENSDART00000079092
|
si:dkey-261i16.5
|
si:dkey-261i16.5 |
| chr6_+_49412754 | 0.18 |
ENSDART00000027398
|
kcna2a
|
potassium voltage-gated channel, shaker-related subfamily, member 2a |
| chr23_-_21471022 | 0.17 |
ENSDART00000104206
|
her4.2
|
hairy-related 4, tandem duplicate 2 |
| chr4_-_2727491 | 0.17 |
ENSDART00000141760
ENSDART00000039083 ENSDART00000134442 |
slco1c1
|
solute carrier organic anion transporter family, member 1C1 |
| chr1_-_40994259 | 0.17 |
ENSDART00000101562
|
adra2c
|
adrenoceptor alpha 2C |
| chr23_+_28582865 | 0.16 |
ENSDART00000020296
|
l1cama
|
L1 cell adhesion molecule, paralog a |
| chr20_-_27190393 | 0.16 |
ENSDART00000149024
|
btbd7
|
BTB (POZ) domain containing 7 |
| chr12_+_20700961 | 0.15 |
ENSDART00000016099
|
si:ch211-119c20.2
|
si:ch211-119c20.2 |
| chr21_+_11503212 | 0.14 |
ENSDART00000146701
|
si:dkey-184p9.7
|
si:dkey-184p9.7 |
| chr13_-_24447332 | 0.14 |
ENSDART00000043004
|
slc1a4
|
solute carrier family 1 (glutamate/neutral amino acid transporter), member 4 |
| chr7_-_26518086 | 0.13 |
ENSDART00000058913
|
eif4a1a
|
eukaryotic translation initiation factor 4A1A |
| chr10_-_34867401 | 0.13 |
ENSDART00000145545
|
dclk1a
|
doublecortin-like kinase 1a |
| chr8_-_28449782 | 0.13 |
ENSDART00000062702
|
cebpb
|
CCAAT/enhancer binding protein (C/EBP), beta |
| chr13_+_8892784 | 0.12 |
ENSDART00000075054
ENSDART00000143705 |
thada
|
thyroid adenoma associated |
| chr16_+_36906693 | 0.11 |
ENSDART00000160645
|
si:ch73-215d9.1
|
si:ch73-215d9.1 |
| chr21_-_21148623 | 0.11 |
ENSDART00000184364
|
ank1b
|
ankyrin 1, erythrocytic b |
| chr13_-_1349922 | 0.11 |
ENSDART00000140970
|
si:ch73-52p7.1
|
si:ch73-52p7.1 |
| chr6_-_40098641 | 0.11 |
ENSDART00000017402
|
ip6k2b
|
inositol hexakisphosphate kinase 2b |
| chr4_-_16545085 | 0.11 |
ENSDART00000033188
|
btg1
|
B-cell translocation gene 1, anti-proliferative |
| chr4_-_23083187 | 0.10 |
ENSDART00000009768
|
magi2a
|
membrane associated guanylate kinase, WW and PDZ domain containing 2a |
| chr9_-_25443094 | 0.10 |
ENSDART00000105492
|
acvr2aa
|
activin A receptor type 2Aa |
| chr16_-_17541890 | 0.10 |
ENSDART00000131328
|
clcn1b
|
chloride channel, voltage-sensitive 1b |
| chr21_+_42226113 | 0.10 |
ENSDART00000170362
|
GABRB2 (1 of many)
|
gamma-aminobutyric acid type A receptor beta2 subunit |
| chr5_-_71838520 | 0.09 |
ENSDART00000174396
|
CU927890.1
|
|
| chr11_-_33857911 | 0.09 |
ENSDART00000165370
|
nxph2b
|
neurexophilin 2b |
| chr1_-_22803147 | 0.09 |
ENSDART00000086867
|
tapt1b
|
transmembrane anterior posterior transformation 1b |
| chr25_-_15214161 | 0.09 |
ENSDART00000031499
|
wt1a
|
wilms tumor 1a |
| chr8_+_14792830 | 0.09 |
ENSDART00000139972
|
cacna1ea
|
calcium channel, voltage-dependent, R type, alpha 1E subunit a |
| chr14_+_36497250 | 0.09 |
ENSDART00000184727
|
TENM3
|
si:dkey-237h12.3 |
| chr16_+_22618620 | 0.09 |
ENSDART00000185728
ENSDART00000041625 |
chrnb2b
|
cholinergic receptor, nicotinic, beta 2b |
| chr21_-_35419486 | 0.08 |
ENSDART00000138529
|
si:dkeyp-23e4.3
|
si:dkeyp-23e4.3 |
| chr6_+_13779857 | 0.08 |
ENSDART00000154793
|
tmem198b
|
transmembrane protein 198b |
| chr13_-_10261383 | 0.08 |
ENSDART00000080808
|
six3a
|
SIX homeobox 3a |
| chr14_+_23811808 | 0.08 |
ENSDART00000014411
|
kctd16a
|
potassium channel tetramerization domain containing 16a |
| chr21_-_37790727 | 0.08 |
ENSDART00000162907
|
gabrb4
|
gamma-aminobutyric acid (GABA) A receptor, beta 4 |
| chr9_-_1965727 | 0.08 |
ENSDART00000082354
|
hoxd9a
|
homeobox D9a |
| chr4_-_23082985 | 0.08 |
ENSDART00000133644
|
magi2a
|
membrane associated guanylate kinase, WW and PDZ domain containing 2a |
| chr12_-_4028079 | 0.07 |
ENSDART00000128676
|
si:ch211-180a12.2
|
si:ch211-180a12.2 |
| chr17_-_3291369 | 0.07 |
ENSDART00000181840
|
CABZ01007222.1
|
|
| chr16_+_38159758 | 0.07 |
ENSDART00000058666
ENSDART00000112165 |
pi4kb
|
phosphatidylinositol 4-kinase, catalytic, beta |
| chr16_-_16761164 | 0.07 |
ENSDART00000135872
|
si:dkey-27n14.1
|
si:dkey-27n14.1 |
| chr22_+_12361317 | 0.07 |
ENSDART00000189963
ENSDART00000159614 |
r3hdm1
|
R3H domain containing 1 |
| chr12_-_3828854 | 0.07 |
ENSDART00000158529
|
taok2b
|
TAO kinase 2b |
| chr13_-_2440622 | 0.07 |
ENSDART00000102767
ENSDART00000172616 |
fbxo9
|
F-box protein 9 |
| chr18_+_17151916 | 0.07 |
ENSDART00000175488
|
trhr2
|
thyrotropin releasing hormone receptor 2 |
| chr2_-_20323901 | 0.06 |
ENSDART00000125531
|
plppr5a
|
phospholipid phosphatase related 5a |
| chr25_-_31739309 | 0.06 |
ENSDART00000098896
|
acot19
|
acyl-CoA thioesterase 19 |
| chr22_+_30184039 | 0.06 |
ENSDART00000049075
|
add3a
|
adducin 3 (gamma) a |
| chr22_+_24214665 | 0.06 |
ENSDART00000163980
ENSDART00000167996 |
glrx2
|
glutaredoxin 2 |
| chr19_+_7506380 | 0.06 |
ENSDART00000182837
|
prcc
|
papillary renal cell carcinoma (translocation-associated) |
| chr25_-_1720736 | 0.06 |
ENSDART00000097256
|
SLC6A13
|
solute carrier family 6 member 13 |
| chr22_+_24215007 | 0.06 |
ENSDART00000162227
|
glrx2
|
glutaredoxin 2 |
| chr9_-_1959917 | 0.06 |
ENSDART00000082359
|
hoxd3a
|
homeobox D3a |
| chr22_+_10676981 | 0.06 |
ENSDART00000138016
|
hyal2b
|
hyaluronoglucosaminidase 2b |
| chr7_-_46777876 | 0.06 |
ENSDART00000193954
|
tshz3b
|
teashirt zinc finger homeobox 3b |
| chr25_-_13871118 | 0.05 |
ENSDART00000160866
|
cry2
|
cryptochrome circadian clock 2 |
| chr19_+_20201593 | 0.05 |
ENSDART00000163026
|
igf2bp3
|
insulin-like growth factor 2 mRNA binding protein 3 |
| chr7_-_31941330 | 0.05 |
ENSDART00000144682
|
bdnf
|
brain-derived neurotrophic factor |
| chr9_-_12574473 | 0.05 |
ENSDART00000191372
ENSDART00000193667 |
igf2bp2a
|
insulin-like growth factor 2 mRNA binding protein 2a |
| chr10_+_42589707 | 0.05 |
ENSDART00000075269
|
fgfr1b
|
fibroblast growth factor receptor 1b |
| chr1_-_25144439 | 0.05 |
ENSDART00000132355
|
fbxw7
|
F-box and WD repeat domain containing 7 |
| chr2_+_38881165 | 0.05 |
ENSDART00000141850
|
carmil3
|
capping protein regulator and myosin 1 linker 3 |
| chr14_+_16083818 | 0.05 |
ENSDART00000168462
|
rnf103
|
ring finger protein 103 |
| chr5_-_31875645 | 0.05 |
ENSDART00000098160
|
tmem119b
|
transmembrane protein 119b |
| chr9_+_45428041 | 0.05 |
ENSDART00000193087
|
adarb1b
|
adenosine deaminase, RNA-specific, B1b |
| chr14_-_26425416 | 0.05 |
ENSDART00000088690
|
lman2
|
lectin, mannose-binding 2 |
| chr2_-_37353098 | 0.05 |
ENSDART00000056522
|
skila
|
SKI-like proto-oncogene a |
| chr18_+_14633974 | 0.05 |
ENSDART00000133834
|
vps9d1
|
VPS9 domain containing 1 |
| chr2_-_39759059 | 0.04 |
ENSDART00000007333
|
slc25a36a
|
solute carrier family 25 (pyrimidine nucleotide carrier ), member 36a |
| chr7_-_23873173 | 0.04 |
ENSDART00000101408
|
zgc:162171
|
zgc:162171 |
| chr15_+_27387555 | 0.04 |
ENSDART00000018603
|
tbx4
|
T-box 4 |
| chr22_-_16997887 | 0.04 |
ENSDART00000138233
|
nfia
|
nuclear factor I/A |
| chr18_+_34225520 | 0.04 |
ENSDART00000126115
|
v2rl1
|
vomeronasal 2 receptor, l1 |
| chr3_-_7656059 | 0.04 |
ENSDART00000170917
|
junbb
|
JunB proto-oncogene, AP-1 transcription factor subunit b |
| chr23_+_41912151 | 0.04 |
ENSDART00000191115
|
podn
|
podocan |
| chr3_+_35005062 | 0.04 |
ENSDART00000181163
|
prkcbb
|
protein kinase C, beta b |
| chr6_+_55285578 | 0.04 |
ENSDART00000180183
|
zgc:109913
|
zgc:109913 |
| chr18_+_48428713 | 0.04 |
ENSDART00000076861
|
fli1a
|
Fli-1 proto-oncogene, ETS transcription factor a |
| chr22_-_3299100 | 0.03 |
ENSDART00000160305
|
si:zfos-943e10.1
|
si:zfos-943e10.1 |
| chr22_-_16997475 | 0.03 |
ENSDART00000090237
|
nfia
|
nuclear factor I/A |
| chr10_+_42589391 | 0.03 |
ENSDART00000067689
ENSDART00000075259 |
fgfr1b
|
fibroblast growth factor receptor 1b |
| chr11_-_16093018 | 0.03 |
ENSDART00000139309
ENSDART00000139819 |
SPATA1
|
si:dkey-205k8.5 |
| chr2_+_56891858 | 0.03 |
ENSDART00000159912
|
zgc:85843
|
zgc:85843 |
| chr20_-_26421112 | 0.03 |
ENSDART00000183767
ENSDART00000182330 |
akap12b
|
A kinase (PRKA) anchor protein 12b |
| chr20_-_26420939 | 0.03 |
ENSDART00000110883
|
akap12b
|
A kinase (PRKA) anchor protein 12b |
| chr24_-_26282672 | 0.03 |
ENSDART00000176663
|
and1
|
actinodin1 |
| chr6_+_24661736 | 0.03 |
ENSDART00000169283
|
znf644b
|
zinc finger protein 644b |
| chr5_+_2815021 | 0.03 |
ENSDART00000020472
|
hpda
|
4-hydroxyphenylpyruvate dioxygenase a |
| chr2_+_24700922 | 0.03 |
ENSDART00000170467
|
pik3r2
|
phosphoinositide-3-kinase, regulatory subunit 2 (beta) |
| chr7_+_29890292 | 0.03 |
ENSDART00000170403
ENSDART00000168600 |
tln2a
|
talin 2a |
| chr16_-_26296477 | 0.03 |
ENSDART00000157553
|
erfl1
|
Ets2 repressor factor like 1 |
| chr20_+_51006362 | 0.02 |
ENSDART00000028084
|
mpp5b
|
membrane protein, palmitoylated 5b (MAGUK p55 subfamily member 5) |
| chr19_+_43017931 | 0.02 |
ENSDART00000132213
|
nkain1
|
sodium/potassium transporting ATPase interacting 1 |
| chr17_-_31221655 | 0.02 |
ENSDART00000145010
ENSDART00000137834 ENSDART00000132858 |
pcmtl
|
l-isoaspartyl protein carboxyl methyltransferase, like |
| chr17_+_42274825 | 0.02 |
ENSDART00000020156
|
pax1a
|
paired box 1a |
| chr4_+_37406676 | 0.02 |
ENSDART00000130981
|
si:ch73-134f24.1
|
si:ch73-134f24.1 |
| chr6_-_19664848 | 0.02 |
ENSDART00000159749
|
usp43a
|
ubiquitin specific peptidase 43a |
| chr4_+_73672430 | 0.02 |
ENSDART00000174310
ENSDART00000150505 |
si:dkey-262g12.7
|
si:dkey-262g12.7 |
| chr24_+_1294176 | 0.02 |
ENSDART00000106637
|
si:ch73-134f24.1
|
si:ch73-134f24.1 |
| chr6_-_19665114 | 0.01 |
ENSDART00000168985
|
usp43a
|
ubiquitin specific peptidase 43a |
| chr1_-_22370660 | 0.01 |
ENSDART00000127506
|
si:ch73-380n15.2
|
si:ch73-380n15.2 |
| chr12_+_22560067 | 0.01 |
ENSDART00000172066
|
polr2a
|
polymerase (RNA) II (DNA directed) polypeptide A |
| chr18_-_2433011 | 0.01 |
ENSDART00000181922
ENSDART00000193276 |
CR769778.1
|
|
| chr7_+_5905091 | 0.01 |
ENSDART00000167099
|
CU459186.3
|
Histone H3.2 |
| chr8_+_18044257 | 0.00 |
ENSDART00000029255
|
glis1b
|
GLIS family zinc finger 1b |
| chr21_-_21537452 | 0.00 |
ENSDART00000142549
|
or133-7
|
odorant receptor, family H, subfamily 133, member 7 |
| chr24_+_25032340 | 0.00 |
ENSDART00000005845
|
mtmr6
|
myotubularin related protein 6 |
| chr9_+_22364997 | 0.00 |
ENSDART00000188054
ENSDART00000046116 |
crygs3
|
crystallin, gamma S3 |
Network of associatons between targets according to the STRING database.
First level regulatory network of hoxc13a
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.5 | GO:0009449 | gamma-aminobutyric acid biosynthetic process(GO:0009449) |
| 0.0 | 0.5 | GO:0015014 | heparan sulfate proteoglycan biosynthetic process, polysaccharide chain biosynthetic process(GO:0015014) |
| 0.0 | 0.6 | GO:0042396 | phosphagen metabolic process(GO:0006599) phosphocreatine metabolic process(GO:0006603) phosphagen biosynthetic process(GO:0042396) phosphocreatine biosynthetic process(GO:0046314) |
| 0.0 | 0.3 | GO:0060052 | neurofilament cytoskeleton organization(GO:0060052) |
| 0.0 | 0.2 | GO:0071881 | adenylate cyclase-inhibiting adrenergic receptor signaling pathway(GO:0071881) |
| 0.0 | 0.1 | GO:0045724 | positive regulation of cilium assembly(GO:0045724) |
| 0.0 | 0.1 | GO:0072314 | visceral serous pericardium development(GO:0061032) glomerular visceral epithelial cell fate commitment(GO:0072149) glomerular epithelial cell fate commitment(GO:0072314) |
| 0.0 | 0.4 | GO:0014823 | response to activity(GO:0014823) |
| 0.0 | 0.2 | GO:0007045 | cell-substrate adherens junction assembly(GO:0007045) focal adhesion assembly(GO:0048041) regulation of focal adhesion assembly(GO:0051893) regulation of cell-substrate junction assembly(GO:0090109) |
| 0.0 | 0.1 | GO:1904969 | bleb assembly(GO:0032060) slow muscle cell migration(GO:1904969) |
| 0.0 | 0.1 | GO:0031645 | negative regulation of myelination(GO:0031642) negative regulation of neurological system process(GO:0031645) |
| 0.0 | 0.0 | GO:0033690 | positive regulation of osteoblast proliferation(GO:0033690) |
| 0.0 | 0.8 | GO:0003009 | skeletal muscle contraction(GO:0003009) |
| 0.0 | 0.2 | GO:0030431 | sleep(GO:0030431) |
| 0.0 | 0.0 | GO:0006864 | pyrimidine nucleotide transport(GO:0006864) mitochondrial pyrimidine nucleotide import(GO:1990519) |
| 0.0 | 0.1 | GO:0044030 | regulation of DNA methylation(GO:0044030) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.2 | GO:0016460 | myosin II complex(GO:0016460) |
| 0.0 | 0.2 | GO:0043083 | synaptic cleft(GO:0043083) |
| 0.0 | 0.4 | GO:1990907 | beta-catenin-TCF complex(GO:1990907) |
| 0.0 | 0.8 | GO:0005861 | troponin complex(GO:0005861) |
| 0.0 | 0.2 | GO:0044224 | juxtaparanode region of axon(GO:0044224) |
| 0.0 | 0.2 | GO:0044295 | axonal growth cone(GO:0044295) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.5 | GO:0004351 | glutamate decarboxylase activity(GO:0004351) |
| 0.1 | 0.5 | GO:0015016 | [heparan sulfate]-glucosamine N-sulfotransferase activity(GO:0015016) |
| 0.1 | 0.2 | GO:0032028 | myosin head/neck binding(GO:0032028) myosin II head/neck binding(GO:0032034) myosin II heavy chain binding(GO:0032038) |
| 0.0 | 0.2 | GO:0031697 | beta-1 adrenergic receptor binding(GO:0031697) type II activin receptor binding(GO:0070699) |
| 0.0 | 0.6 | GO:0004111 | creatine kinase activity(GO:0004111) phosphotransferase activity, nitrogenous group as acceptor(GO:0016775) |
| 0.0 | 0.2 | GO:0008046 | axon guidance receptor activity(GO:0008046) |
| 0.0 | 0.2 | GO:0004938 | alpha2-adrenergic receptor activity(GO:0004938) |
| 0.0 | 0.1 | GO:0004362 | glutathione-disulfide reductase activity(GO:0004362) |
| 0.0 | 0.0 | GO:0015218 | pyrimidine nucleotide transmembrane transporter activity(GO:0015218) |
| 0.0 | 0.1 | GO:0033906 | hyaluronoglucuronidase activity(GO:0033906) |
| 0.0 | 0.1 | GO:0004997 | thyrotropin-releasing hormone receptor activity(GO:0004997) |
| 0.0 | 0.5 | GO:0005251 | delayed rectifier potassium channel activity(GO:0005251) |
| 0.0 | 0.1 | GO:0004430 | 1-phosphatidylinositol 4-kinase activity(GO:0004430) |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.5 | REACTOME GABA SYNTHESIS RELEASE REUPTAKE AND DEGRADATION | Genes involved in GABA synthesis, release, reuptake and degradation |
| 0.0 | 0.2 | REACTOME TRANSPORT OF ORGANIC ANIONS | Genes involved in Transport of organic anions |
| 0.0 | 0.3 | REACTOME DARPP 32 EVENTS | Genes involved in DARPP-32 events |