Project
PRJNA207719: Tissue specific transcriptome profiling
Navigation
Downloads
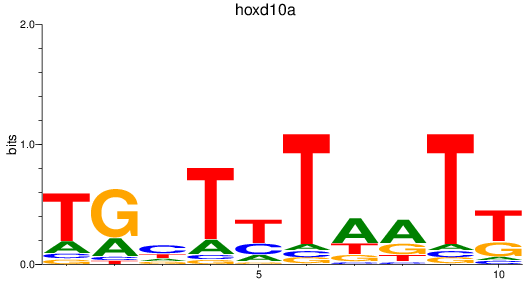
Results for hoxd10a
Z-value: 0.60
Transcription factors associated with hoxd10a
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
hoxd10a
|
ENSDARG00000057859 | homeobox D10a |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| hoxd10a | dr11_v1_chr9_-_1970071_1970071 | 0.26 | 6.7e-01 | Click! |
Activity profile of hoxd10a motif
Sorted Z-values of hoxd10a motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr14_+_38786298 | 0.41 |
ENSDART00000164440
|
si:ch211-195b11.3
|
si:ch211-195b11.3 |
| chr6_+_13201358 | 0.39 |
ENSDART00000190290
|
CT009620.1
|
|
| chr10_-_17484762 | 0.38 |
ENSDART00000137905
ENSDART00000007961 |
nt5c2l1
|
5'-nucleotidase, cytosolic II, like 1 |
| chr5_+_22459087 | 0.37 |
ENSDART00000134781
|
BX546499.1
|
|
| chr21_-_3796461 | 0.36 |
ENSDART00000021976
|
nsa2
|
NSA2 ribosome biogenesis homolog (S. cerevisiae) |
| chr16_-_45178430 | 0.36 |
ENSDART00000165186
|
si:dkey-33i11.9
|
si:dkey-33i11.9 |
| chr7_-_22981796 | 0.35 |
ENSDART00000167565
|
si:dkey-171c9.3
|
si:dkey-171c9.3 |
| chr1_-_52431220 | 0.35 |
ENSDART00000111256
|
zgc:194101
|
zgc:194101 |
| chr6_+_12316438 | 0.32 |
ENSDART00000156854
|
si:dkey-276j7.2
|
si:dkey-276j7.2 |
| chr7_+_22982201 | 0.30 |
ENSDART00000134116
|
ccnb3
|
cyclin B3 |
| chr1_-_18811517 | 0.30 |
ENSDART00000142026
|
si:dkey-167i21.2
|
si:dkey-167i21.2 |
| chr21_+_15295685 | 0.28 |
ENSDART00000135603
|
BX901878.1
|
|
| chr16_-_42186093 | 0.28 |
ENSDART00000076030
|
fbl
|
fibrillarin |
| chr21_-_41025340 | 0.28 |
ENSDART00000148231
|
plac8l1
|
PLAC8-like 1 |
| chr18_-_20458412 | 0.26 |
ENSDART00000012241
|
kif23
|
kinesin family member 23 |
| chr22_+_19366866 | 0.26 |
ENSDART00000137301
|
si:dkey-21e2.12
|
si:dkey-21e2.12 |
| chr13_-_31017960 | 0.24 |
ENSDART00000145287
|
wdfy4
|
WDFY family member 4 |
| chr8_+_36142734 | 0.22 |
ENSDART00000159361
ENSDART00000161194 |
mhc2b
|
major histocompatibility complex class II integral membrane beta chain gene |
| chr23_+_28322986 | 0.22 |
ENSDART00000134710
|
birc5b
|
baculoviral IAP repeat containing 5b |
| chr22_-_38819603 | 0.20 |
ENSDART00000104437
|
si:ch211-262h13.5
|
si:ch211-262h13.5 |
| chr1_-_37377509 | 0.19 |
ENSDART00000113542
|
tnip2
|
TNFAIP3 interacting protein 2 |
| chr25_+_34915762 | 0.19 |
ENSDART00000191776
|
sntb2
|
syntrophin, beta 2 |
| chr8_+_36354125 | 0.19 |
ENSDART00000109439
|
si:zfos-367g9.1
|
si:zfos-367g9.1 |
| chr14_+_26439227 | 0.19 |
ENSDART00000054183
|
gpr137
|
G protein-coupled receptor 137 |
| chr7_+_22981909 | 0.18 |
ENSDART00000122449
|
ccnb3
|
cyclin B3 |
| chr21_+_3796620 | 0.18 |
ENSDART00000099535
ENSDART00000144515 |
spout1
|
SPOUT domain containing methyltransferase 1 |
| chr4_-_16824556 | 0.18 |
ENSDART00000165289
ENSDART00000185839 |
gys2
|
glycogen synthase 2 |
| chr18_+_15644559 | 0.17 |
ENSDART00000061794
|
nr1h4
|
nuclear receptor subfamily 1, group H, member 4 |
| chr7_+_4694924 | 0.17 |
ENSDART00000144873
|
si:ch211-225k7.3
|
si:ch211-225k7.3 |
| chr11_-_31039533 | 0.17 |
ENSDART00000127355
|
ier2b
|
immediate early response 2b |
| chr8_-_15292197 | 0.17 |
ENSDART00000140867
|
spata6
|
spermatogenesis associated 6 |
| chr25_-_3217115 | 0.15 |
ENSDART00000032390
|
gtf2h1
|
general transcription factor IIH, polypeptide 1 |
| chr19_-_35450661 | 0.15 |
ENSDART00000113574
ENSDART00000136895 |
anln
|
anillin, actin binding protein |
| chr2_-_1486023 | 0.14 |
ENSDART00000113774
|
si:ch211-188c16.1
|
si:ch211-188c16.1 |
| chr9_-_2594410 | 0.14 |
ENSDART00000188306
ENSDART00000164276 |
sp9
|
sp9 transcription factor |
| chr2_-_42628028 | 0.14 |
ENSDART00000179866
|
myo10
|
myosin X |
| chr3_-_29899172 | 0.14 |
ENSDART00000140518
ENSDART00000020311 |
rpl27
|
ribosomal protein L27 |
| chr6_-_39521832 | 0.14 |
ENSDART00000065038
|
atf1
|
activating transcription factor 1 |
| chr18_+_31056645 | 0.13 |
ENSDART00000159316
|
mvda
|
mevalonate (diphospho) decarboxylase a |
| chr9_+_45605410 | 0.13 |
ENSDART00000136444
ENSDART00000007189 ENSDART00000158713 ENSDART00000182671 |
traf3ip1
|
TNF receptor-associated factor 3 interacting protein 1 |
| chr19_+_4051535 | 0.13 |
ENSDART00000167084
|
btr24
|
bloodthirsty-related gene family, member 24 |
| chr13_+_27232848 | 0.12 |
ENSDART00000138043
|
rin2
|
Ras and Rab interactor 2 |
| chr25_+_34915576 | 0.12 |
ENSDART00000073441
|
sntb2
|
syntrophin, beta 2 |
| chr1_-_19764038 | 0.12 |
ENSDART00000005733
|
tma16
|
translation machinery associated 16 homolog |
| chr17_+_42274825 | 0.12 |
ENSDART00000020156
|
pax1a
|
paired box 1a |
| chr5_+_35458346 | 0.12 |
ENSDART00000141239
|
erlin2
|
ER lipid raft associated 2 |
| chr21_+_3796196 | 0.11 |
ENSDART00000146754
|
spout1
|
SPOUT domain containing methyltransferase 1 |
| chr10_-_20453995 | 0.11 |
ENSDART00000168541
ENSDART00000164072 |
si:ch211-113d22.2
|
si:ch211-113d22.2 |
| chr7_+_5007310 | 0.11 |
ENSDART00000146083
|
si:ch211-272h9.3
|
si:ch211-272h9.3 |
| chr23_+_36074798 | 0.11 |
ENSDART00000133760
ENSDART00000103146 |
hoxc11a
|
homeobox C11a |
| chr9_-_3519253 | 0.11 |
ENSDART00000169586
|
dcaf17
|
ddb1 and cul4 associated factor 17 |
| chr10_-_3138403 | 0.11 |
ENSDART00000183365
|
ube2l3a
|
ubiquitin-conjugating enzyme E2L 3a |
| chr3_-_26184018 | 0.11 |
ENSDART00000191604
|
si:ch211-11k18.4
|
si:ch211-11k18.4 |
| chr7_-_3850090 | 0.10 |
ENSDART00000113403
ENSDART00000137683 ENSDART00000049926 |
si:dkey-28d5.12
si:dkey-88n24.7
|
si:dkey-28d5.12 si:dkey-88n24.7 |
| chr19_-_3056235 | 0.10 |
ENSDART00000137020
|
bop1
|
block of proliferation 1 |
| chr5_-_23805387 | 0.10 |
ENSDART00000133805
|
gbgt1l4
|
globoside alpha-1,3-N-acetylgalactosaminyltransferase 1, like 4 |
| chr7_+_4219234 | 0.10 |
ENSDART00000187891
ENSDART00000173246 ENSDART00000172808 |
si:ch211-63p21.4
|
si:ch211-63p21.4 |
| chr6_+_29386728 | 0.10 |
ENSDART00000104303
|
actl6a
|
actin-like 6A |
| chr13_+_27232694 | 0.10 |
ENSDART00000131128
|
rin2
|
Ras and Rab interactor 2 |
| chr2_-_23349116 | 0.10 |
ENSDART00000099690
|
fam129ab
|
family with sequence similarity 129, member Ab |
| chr5_+_25317061 | 0.10 |
ENSDART00000170097
|
trpm6
|
transient receptor potential cation channel, subfamily M, member 6 |
| chr1_-_40911332 | 0.10 |
ENSDART00000027463
|
hmx4
|
H6 family homeobox 4 |
| chr5_+_6672870 | 0.09 |
ENSDART00000126598
|
pxna
|
paxillin a |
| chr19_+_47299212 | 0.09 |
ENSDART00000158262
|
tpmt.1
|
thiopurine S-methyltransferase, tandem duplicate 1 |
| chr2_-_23411368 | 0.09 |
ENSDART00000159495
|
si:ch73-129a22.11
|
si:ch73-129a22.11 |
| chr12_+_30788912 | 0.09 |
ENSDART00000160422
|
aldh18a1
|
aldehyde dehydrogenase 18 family, member A1 |
| chr20_+_18993452 | 0.09 |
ENSDART00000025509
|
tdh
|
L-threonine dehydrogenase |
| chr25_+_13620555 | 0.08 |
ENSDART00000163642
|
si:ch211-172l8.4
|
si:ch211-172l8.4 |
| chr13_-_50108337 | 0.08 |
ENSDART00000133308
|
nid1a
|
nidogen 1a |
| chr4_-_67589158 | 0.08 |
ENSDART00000184361
|
BX088712.2
|
|
| chr7_+_4854690 | 0.08 |
ENSDART00000131645
|
si:dkey-28d5.7
|
si:dkey-28d5.7 |
| chr7_+_4194306 | 0.08 |
ENSDART00000190454
|
si:dkey-28d5.5
|
si:dkey-28d5.5 |
| chr11_-_42752884 | 0.08 |
ENSDART00000186025
|
si:ch73-106k19.5
|
si:ch73-106k19.5 |
| chr18_+_7543347 | 0.08 |
ENSDART00000103467
|
arf5
|
ADP-ribosylation factor 5 |
| chr22_+_22302614 | 0.08 |
ENSDART00000049434
|
scamp4
|
secretory carrier membrane protein 4 |
| chr9_+_19489304 | 0.08 |
ENSDART00000151920
|
si:ch211-140m22.7
|
si:ch211-140m22.7 |
| chr7_+_4592867 | 0.08 |
ENSDART00000192614
|
si:dkey-83f18.15
|
si:dkey-83f18.15 |
| chr17_-_28707898 | 0.08 |
ENSDART00000135752
ENSDART00000061853 |
ap4s1
|
adaptor-related protein complex 4, sigma 1 subunit |
| chr11_+_37265692 | 0.07 |
ENSDART00000184691
|
il17rc
|
interleukin 17 receptor C |
| chr9_+_3519191 | 0.07 |
ENSDART00000008606
|
mettl8
|
methyltransferase like 8 |
| chr6_-_25165693 | 0.07 |
ENSDART00000167259
|
znf326
|
zinc finger protein 326 |
| chr9_+_21151138 | 0.07 |
ENSDART00000133903
|
hao2
|
hydroxyacid oxidase 2 (long chain) |
| chr9_-_22272181 | 0.07 |
ENSDART00000113174
|
crygm2d7
|
crystallin, gamma M2d7 |
| chr25_-_12412704 | 0.07 |
ENSDART00000168275
|
det1
|
DET1, COP1 ubiquitin ligase partner |
| chr9_-_19489264 | 0.06 |
ENSDART00000122894
|
wdr4
|
WD repeat domain 4 |
| chr7_+_4938463 | 0.06 |
ENSDART00000143722
|
si:dkey-28d5.14
|
si:dkey-28d5.14 |
| chr16_-_16152199 | 0.06 |
ENSDART00000012718
|
fabp11b
|
fatty acid binding protein 11b |
| chr20_+_13894123 | 0.06 |
ENSDART00000007744
|
slc30a1a
|
solute carrier family 30 (zinc transporter), member 1a |
| chr3_-_43954343 | 0.06 |
ENSDART00000157580
|
ubfd1
|
ubiquitin family domain containing 1 |
| chr5_+_50912729 | 0.06 |
ENSDART00000190837
|
col4a3bpa
|
collagen, type IV, alpha 3 (Goodpasture antigen) binding protein a |
| chr7_+_17973269 | 0.05 |
ENSDART00000079969
|
kcnk7
|
potassium channel, subfamily K, member 7 |
| chr11_-_39118882 | 0.05 |
ENSDART00000113185
ENSDART00000156526 |
ap5b1
|
adaptor-related protein complex 5, beta 1 subunit |
| chr9_-_3519717 | 0.05 |
ENSDART00000145043
|
dcaf17
|
ddb1 and cul4 associated factor 17 |
| chr15_-_568645 | 0.05 |
ENSDART00000156744
|
cbln18
|
cerebellin 18 |
| chr20_-_25445237 | 0.05 |
ENSDART00000145034
ENSDART00000018049 |
mrpl19
|
mitochondrial ribosomal protein L19 |
| chr21_+_5192016 | 0.05 |
ENSDART00000139288
|
si:dkey-121h17.7
|
si:dkey-121h17.7 |
| chr2_+_10280645 | 0.04 |
ENSDART00000063996
|
gadd45aa
|
growth arrest and DNA-damage-inducible, alpha, a |
| chr24_+_17069420 | 0.04 |
ENSDART00000014787
|
pip4k2aa
|
phosphatidylinositol-5-phosphate 4-kinase, type II, alpha a |
| chr11_-_27953135 | 0.04 |
ENSDART00000168338
|
ece1
|
endothelin converting enzyme 1 |
| chr14_+_20941038 | 0.04 |
ENSDART00000182539
|
zgc:66433
|
zgc:66433 |
| chr24_+_15655069 | 0.04 |
ENSDART00000042943
ENSDART00000181296 |
fbxo15
|
F-box protein 15 |
| chr11_+_14104417 | 0.04 |
ENSDART00000059752
ENSDART00000186575 |
med16
|
mediator complex subunit 16 |
| chr8_+_23142946 | 0.04 |
ENSDART00000152933
|
si:ch211-196c10.13
|
si:ch211-196c10.13 |
| chr24_-_24038800 | 0.04 |
ENSDART00000080549
|
lyz
|
lysozyme |
| chr16_+_54829293 | 0.04 |
ENSDART00000024729
|
pabpc1a
|
poly(A) binding protein, cytoplasmic 1a |
| chr13_+_24717880 | 0.04 |
ENSDART00000147713
|
cfap43
|
cilia and flagella associated protein 43 |
| chr16_+_813780 | 0.04 |
ENSDART00000162474
ENSDART00000161774 |
irx1a
|
iroquois homeobox 1a |
| chr19_-_10324573 | 0.03 |
ENSDART00000171795
|
u2af2b
|
U2 small nuclear RNA auxiliary factor 2b |
| chr10_-_22491353 | 0.03 |
ENSDART00000180783
ENSDART00000159564 |
epob
|
erythropoietin b |
| chr3_-_11828206 | 0.03 |
ENSDART00000018159
|
si:ch211-262e15.1
|
si:ch211-262e15.1 |
| chr19_-_10324182 | 0.03 |
ENSDART00000151352
ENSDART00000151162 ENSDART00000023571 |
u2af2b
|
U2 small nuclear RNA auxiliary factor 2b |
| chr23_-_17451264 | 0.03 |
ENSDART00000190748
|
tpd52l2b
|
tumor protein D52-like 2b |
| chr3_-_25107802 | 0.03 |
ENSDART00000055432
|
rbx1
|
ring-box 1, E3 ubiquitin protein ligase |
| chr20_-_30931139 | 0.03 |
ENSDART00000006778
ENSDART00000146376 |
acat2
|
acetyl-CoA acetyltransferase 2 |
| chr18_-_2222128 | 0.02 |
ENSDART00000171402
|
pigb
|
phosphatidylinositol glycan anchor biosynthesis, class B |
| chr25_-_2723902 | 0.02 |
ENSDART00000143721
ENSDART00000175224 |
adpgk
|
ADP-dependent glucokinase |
| chr25_-_2723682 | 0.02 |
ENSDART00000113382
|
adpgk
|
ADP-dependent glucokinase |
| chr2_+_21356242 | 0.02 |
ENSDART00000145670
ENSDART00000146600 |
ctdsplb
|
CTD (carboxy-terminal domain, RNA polymerase II, polypeptide A) small phosphatase-like b |
| chr16_-_26296477 | 0.02 |
ENSDART00000157553
|
erfl1
|
Ets2 repressor factor like 1 |
| chr10_-_2788668 | 0.02 |
ENSDART00000131749
ENSDART00000124356 ENSDART00000085031 |
ash2l
|
ash2 (absent, small, or homeotic)-like (Drosophila) |
| chr17_-_19344999 | 0.02 |
ENSDART00000138315
|
gsc
|
goosecoid |
| chr7_+_4194055 | 0.02 |
ENSDART00000105073
|
si:dkey-28d5.14
|
si:dkey-28d5.14 |
| chr19_+_13994563 | 0.02 |
ENSDART00000164696
|
tmem222b
|
transmembrane protein 222b |
| chr16_-_41059919 | 0.01 |
ENSDART00000188803
|
si:dkey-201i6.8
|
si:dkey-201i6.8 |
| chr24_+_15655233 | 0.01 |
ENSDART00000143160
|
fbxo15
|
F-box protein 15 |
| chr1_+_32341734 | 0.01 |
ENSDART00000135078
|
pudp
|
pseudouridine 5'-phosphatase |
| chr17_-_15201031 | 0.01 |
ENSDART00000124426
|
socs4
|
suppressor of cytokine signaling 4 |
| chr18_+_7363242 | 0.01 |
ENSDART00000133375
|
si:dkey-30c15.17
|
si:dkey-30c15.17 |
| chr17_+_31621788 | 0.01 |
ENSDART00000111629
ENSDART00000157148 |
cinp
|
cyclin-dependent kinase 2 interacting protein |
| chr5_-_24270989 | 0.01 |
ENSDART00000146251
|
si:ch211-137i24.12
|
si:ch211-137i24.12 |
| chr23_+_12852655 | 0.01 |
ENSDART00000183015
|
smc1al
|
structural maintenance of chromosomes 1A, like |
Network of associatons between targets according to the STRING database.
First level regulatory network of hoxd10a
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.2 | GO:0000212 | meiotic spindle organization(GO:0000212) |
| 0.1 | 0.3 | GO:0051661 | maintenance of centrosome location(GO:0051661) |
| 0.1 | 0.2 | GO:0034134 | toll-like receptor 2 signaling pathway(GO:0034134) toll-like receptor 3 signaling pathway(GO:0034138) |
| 0.0 | 0.2 | GO:0008356 | asymmetric cell division(GO:0008356) |
| 0.0 | 0.3 | GO:0040016 | embryonic cleavage(GO:0040016) |
| 0.0 | 0.1 | GO:0019287 | isopentenyl diphosphate biosynthetic process, mevalonate pathway(GO:0019287) |
| 0.0 | 0.3 | GO:0031126 | snoRNA 3'-end processing(GO:0031126) |
| 0.0 | 0.1 | GO:0030326 | embryonic limb morphogenesis(GO:0030326) |
| 0.0 | 0.1 | GO:1990359 | detoxification of zinc ion(GO:0010312) stress response to zinc ion(GO:1990359) |
| 0.0 | 0.1 | GO:0090660 | regulation of microtubule polymerization(GO:0031113) cerebrospinal fluid circulation(GO:0090660) |
| 0.0 | 0.2 | GO:1904263 | positive regulation of TORC1 signaling(GO:1904263) |
| 0.0 | 0.1 | GO:0036265 | RNA (guanine-N7)-methylation(GO:0036265) |
| 0.0 | 0.5 | GO:0000460 | maturation of 5.8S rRNA(GO:0000460) |
| 0.0 | 0.5 | GO:0010389 | regulation of G2/M transition of mitotic cell cycle(GO:0010389) |
| 0.0 | 0.2 | GO:0050728 | negative regulation of inflammatory response(GO:0050728) |
| 0.0 | 0.1 | GO:0006567 | threonine metabolic process(GO:0006566) threonine catabolic process(GO:0006567) |
| 0.0 | 0.2 | GO:0070816 | phosphorylation of RNA polymerase II C-terminal domain(GO:0070816) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.3 | GO:0031428 | box C/D snoRNP complex(GO:0031428) |
| 0.0 | 0.1 | GO:0098556 | cytoplasmic side of rough endoplasmic reticulum membrane(GO:0098556) |
| 0.0 | 0.3 | GO:0031616 | spindle pole centrosome(GO:0031616) |
| 0.0 | 0.1 | GO:0070545 | PeBoW complex(GO:0070545) |
| 0.0 | 0.2 | GO:0097224 | sperm connecting piece(GO:0097224) |
| 0.0 | 0.2 | GO:0042613 | MHC class II protein complex(GO:0042613) |
| 0.0 | 0.2 | GO:0000439 | core TFIIH complex(GO:0000439) |
| 0.0 | 0.2 | GO:0005826 | actomyosin contractile ring(GO:0005826) |
| 0.0 | 0.1 | GO:0044609 | DBIRD complex(GO:0044609) |
| 0.0 | 0.4 | GO:0030687 | preribosome, large subunit precursor(GO:0030687) |
| 0.0 | 0.3 | GO:0090665 | dystrophin-associated glycoprotein complex(GO:0016010) glycoprotein complex(GO:0090665) |
| 0.0 | 0.1 | GO:1990589 | ATF4-CREB1 transcription factor complex(GO:1990589) |
| 0.0 | 0.5 | GO:0000307 | cyclin-dependent protein kinase holoenzyme complex(GO:0000307) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.2 | GO:0032027 | myosin light chain binding(GO:0032027) |
| 0.0 | 0.2 | GO:0004373 | glycogen (starch) synthase activity(GO:0004373) |
| 0.0 | 0.1 | GO:0008119 | thiopurine S-methyltransferase activity(GO:0008119) |
| 0.0 | 0.3 | GO:0035198 | miRNA binding(GO:0035198) |
| 0.0 | 0.2 | GO:0032052 | bile acid binding(GO:0032052) |
| 0.0 | 0.1 | GO:0016427 | tRNA (cytosine) methyltransferase activity(GO:0016427) |
| 0.0 | 0.4 | GO:0008253 | 5'-nucleotidase activity(GO:0008253) |
| 0.0 | 0.1 | GO:0016774 | phosphotransferase activity, carboxyl group as acceptor(GO:0016774) |
| 0.0 | 0.1 | GO:0008743 | L-threonine 3-dehydrogenase activity(GO:0008743) |
| 0.0 | 0.2 | GO:0043028 | cysteine-type endopeptidase inhibitor activity involved in apoptotic process(GO:0043027) cysteine-type endopeptidase regulator activity involved in apoptotic process(GO:0043028) |
| 0.0 | 0.3 | GO:0008649 | rRNA methyltransferase activity(GO:0008649) |
| 0.0 | 0.1 | GO:1990756 | protein binding, bridging involved in substrate recognition for ubiquitination(GO:1990756) |
| 0.0 | 0.2 | GO:0070530 | K63-linked polyubiquitin binding(GO:0070530) |
| 0.0 | 0.0 | GO:0005128 | erythropoietin receptor binding(GO:0005128) |
| 0.0 | 0.0 | GO:0016308 | 1-phosphatidylinositol-4-phosphate 5-kinase activity(GO:0016308) |
| 0.0 | 0.4 | GO:0051018 | protein kinase A binding(GO:0051018) |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.3 | REACTOME KINESINS | Genes involved in Kinesins |