Project
PRJNA207719: Tissue specific transcriptome profiling
Navigation
Downloads
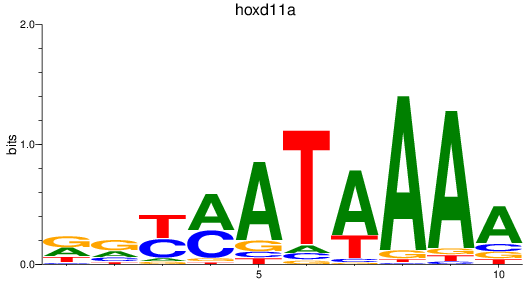
Results for hoxd11a
Z-value: 0.78
Transcription factors associated with hoxd11a
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
hoxd11a
|
ENSDARG00000059267 | homeobox D11a |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| hoxd11a | dr11_v1_chr9_-_1978090_1978090 | -0.50 | 3.9e-01 | Click! |
Activity profile of hoxd11a motif
Sorted Z-values of hoxd11a motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr5_+_44805028 | 0.78 |
ENSDART00000141198
|
ctsla
|
cathepsin La |
| chr5_+_44805269 | 0.69 |
ENSDART00000136965
|
ctsla
|
cathepsin La |
| chr12_-_20373058 | 0.66 |
ENSDART00000066382
|
aqp8a.1
|
aquaporin 8a, tandem duplicate 1 |
| chr15_+_20403903 | 0.56 |
ENSDART00000134182
|
cox7a1
|
cytochrome c oxidase subunit VIIa polypeptide 1 (muscle) |
| chr8_-_2616326 | 0.52 |
ENSDART00000027214
|
slc25a25a
|
solute carrier family 25 (mitochondrial carrier; phosphate carrier), member 25a |
| chr13_-_36034582 | 0.50 |
ENSDART00000133565
|
si:dkey-157l19.2
|
si:dkey-157l19.2 |
| chr7_-_16562200 | 0.48 |
ENSDART00000169093
ENSDART00000173491 |
csrp3
|
cysteine and glycine-rich protein 3 (cardiac LIM protein) |
| chr4_+_7391110 | 0.47 |
ENSDART00000160708
ENSDART00000187823 |
tnni4a
|
troponin I4a |
| chr23_+_7379728 | 0.42 |
ENSDART00000012194
|
gata5
|
GATA binding protein 5 |
| chr3_+_19299309 | 0.42 |
ENSDART00000046297
ENSDART00000146955 |
ldlra
|
low density lipoprotein receptor a |
| chr13_-_319996 | 0.38 |
ENSDART00000148675
|
ddx21
|
DEAD (Asp-Glu-Ala-Asp) box helicase 21 |
| chr1_-_14332283 | 0.36 |
ENSDART00000090025
|
wfs1a
|
Wolfram syndrome 1a (wolframin) |
| chr16_-_45917322 | 0.36 |
ENSDART00000060822
|
afp4
|
antifreeze protein type IV |
| chr23_+_39606108 | 0.34 |
ENSDART00000109464
|
g0s2
|
G0/G1 switch 2 |
| chr11_+_24313931 | 0.32 |
ENSDART00000017599
ENSDART00000166045 |
rem1
|
RAS (RAD and GEM)-like GTP-binding 1 |
| chr22_+_16308806 | 0.31 |
ENSDART00000162685
|
lrrc39
|
leucine rich repeat containing 39 |
| chr11_-_18253111 | 0.31 |
ENSDART00000125984
|
mustn1b
|
musculoskeletal, embryonic nuclear protein 1b |
| chr7_+_13639473 | 0.31 |
ENSDART00000157870
ENSDART00000032899 |
clpxa
|
caseinolytic mitochondrial matrix peptidase chaperone subunit a |
| chr25_+_29161609 | 0.31 |
ENSDART00000180752
|
pkmb
|
pyruvate kinase M1/2b |
| chr15_+_14856307 | 0.31 |
ENSDART00000167213
|
diabloa
|
diablo, IAP-binding mitochondrial protein a |
| chr9_-_41784799 | 0.30 |
ENSDART00000144573
ENSDART00000112542 ENSDART00000190486 |
obsl1b
|
obscurin-like 1b |
| chr17_-_2690083 | 0.30 |
ENSDART00000135374
|
ptpn21
|
protein tyrosine phosphatase, non-receptor type 21 |
| chr23_+_18722715 | 0.30 |
ENSDART00000137438
|
myh7bb
|
myosin, heavy chain 7B, cardiac muscle, beta b |
| chr17_-_15498275 | 0.30 |
ENSDART00000156905
ENSDART00000080661 |
si:ch211-266g18.10
|
si:ch211-266g18.10 |
| chr8_+_28467893 | 0.28 |
ENSDART00000189724
|
slc52a3
|
solute carrier family 52 (riboflavin transporter), member 3 |
| chr14_+_15495088 | 0.28 |
ENSDART00000165765
ENSDART00000188577 |
si:dkey-203a12.6
|
si:dkey-203a12.6 |
| chr2_+_52847049 | 0.28 |
ENSDART00000121980
|
creb3l3b
|
cAMP responsive element binding protein 3-like 3b |
| chr1_+_17900306 | 0.28 |
ENSDART00000089480
|
cyp4v8
|
cytochrome P450, family 4, subfamily V, polypeptide 8 |
| chr2_+_20406399 | 0.27 |
ENSDART00000006817
ENSDART00000137848 |
palmda
|
palmdelphin a |
| chr16_+_13993285 | 0.27 |
ENSDART00000139130
ENSDART00000130353 |
si:dkey-85k15.7
fdps
|
si:dkey-85k15.7 farnesyl diphosphate synthase (farnesyl pyrophosphate synthetase, dimethylallyltranstransferase, geranyltranstransferase) |
| chr23_+_2666944 | 0.27 |
ENSDART00000192861
|
CABZ01057928.1
|
|
| chr16_-_28658341 | 0.25 |
ENSDART00000148456
|
abcb4
|
ATP-binding cassette, sub-family B (MDR/TAP), member 4 |
| chr8_+_40477264 | 0.25 |
ENSDART00000085559
|
gck
|
glucokinase (hexokinase 4) |
| chr15_-_25527580 | 0.24 |
ENSDART00000167005
ENSDART00000157498 |
hif1al
|
hypoxia-inducible factor 1, alpha subunit, like |
| chr8_-_38159805 | 0.24 |
ENSDART00000112331
ENSDART00000180006 |
adgra2
|
adhesion G protein-coupled receptor A2 |
| chr17_+_17764979 | 0.24 |
ENSDART00000105013
|
alkbh1
|
alkB homolog 1, histone H2A dioxygenase |
| chr24_+_39105051 | 0.24 |
ENSDART00000115297
|
mss51
|
MSS51 mitochondrial translational activator |
| chr13_-_25719628 | 0.23 |
ENSDART00000135383
|
si:dkey-192p21.6
|
si:dkey-192p21.6 |
| chr23_+_8797143 | 0.23 |
ENSDART00000132992
|
sox18
|
SRY (sex determining region Y)-box 18 |
| chr14_+_32918172 | 0.22 |
ENSDART00000182867
|
lnx2b
|
ligand of numb-protein X 2b |
| chr10_+_26612321 | 0.22 |
ENSDART00000134322
|
fhl1b
|
four and a half LIM domains 1b |
| chr10_-_13116337 | 0.22 |
ENSDART00000164568
|
musk
|
muscle, skeletal, receptor tyrosine kinase |
| chr24_+_5237753 | 0.22 |
ENSDART00000106488
ENSDART00000005901 |
plod2
|
procollagen-lysine, 2-oxoglutarate 5-dioxygenase 2 |
| chr21_-_30648106 | 0.21 |
ENSDART00000160800
ENSDART00000177022 |
phka1b
|
phosphorylase kinase, alpha 1b (muscle) |
| chr3_+_54761569 | 0.21 |
ENSDART00000135913
ENSDART00000180983 |
si:ch211-74m13.1
|
si:ch211-74m13.1 |
| chr10_+_35526528 | 0.21 |
ENSDART00000184110
|
phldb2a
|
pleckstrin homology-like domain, family B, member 2a |
| chr20_-_33961697 | 0.21 |
ENSDART00000061765
|
selp
|
selectin P |
| chr1_-_9249943 | 0.21 |
ENSDART00000055011
|
zgc:136472
|
zgc:136472 |
| chr23_+_44349252 | 0.21 |
ENSDART00000097644
|
ephb4b
|
eph receptor B4b |
| chr22_+_8979955 | 0.21 |
ENSDART00000144005
|
si:ch211-213a13.1
|
si:ch211-213a13.1 |
| chr6_+_14301214 | 0.20 |
ENSDART00000129491
|
tmem182b
|
transmembrane protein 182b |
| chr8_+_41037541 | 0.20 |
ENSDART00000129344
|
gpat2
|
glycerol-3-phosphate acyltransferase 2, mitochondrial |
| chr15_+_29024895 | 0.20 |
ENSDART00000141164
ENSDART00000144126 |
si:ch211-137a8.2
|
si:ch211-137a8.2 |
| chr3_+_23726148 | 0.20 |
ENSDART00000174580
|
hoxb3a
|
homeobox B3a |
| chr22_-_4644484 | 0.19 |
ENSDART00000167748
|
fbn2b
|
fibrillin 2b |
| chr3_+_49074008 | 0.19 |
ENSDART00000168864
|
zgc:112146
|
zgc:112146 |
| chr23_+_27068225 | 0.18 |
ENSDART00000054238
|
mipa
|
major intrinsic protein of lens fiber a |
| chr24_+_21540842 | 0.18 |
ENSDART00000091529
|
wasf3b
|
WAS protein family, member 3b |
| chr24_+_7782313 | 0.18 |
ENSDART00000111090
|
ptprh
|
protein tyrosine phosphatase, receptor type, h |
| chr7_-_24112484 | 0.16 |
ENSDART00000111923
|
ajuba
|
ajuba LIM protein |
| chr14_+_15597049 | 0.16 |
ENSDART00000159732
|
si:dkey-203a12.8
|
si:dkey-203a12.8 |
| chr21_+_11969603 | 0.16 |
ENSDART00000142247
ENSDART00000140652 |
mlnl
|
motilin-like |
| chr22_-_32360464 | 0.16 |
ENSDART00000148886
|
pcbp4
|
poly(rC) binding protein 4 |
| chr25_+_3549584 | 0.16 |
ENSDART00000165913
|
ccdc77
|
coiled-coil domain containing 77 |
| chr17_+_15788100 | 0.15 |
ENSDART00000027667
|
rragd
|
ras-related GTP binding D |
| chr15_+_11381532 | 0.15 |
ENSDART00000124172
|
si:ch73-321d9.2
|
si:ch73-321d9.2 |
| chr21_-_11970199 | 0.15 |
ENSDART00000114524
|
nop56
|
NOP56 ribonucleoprotein homolog |
| chr3_+_13482542 | 0.15 |
ENSDART00000161287
|
CABZ01079251.1
|
|
| chr21_+_39197628 | 0.14 |
ENSDART00000113607
|
cpdb
|
carboxypeptidase D, b |
| chr11_-_25257045 | 0.14 |
ENSDART00000130477
|
snai1a
|
snail family zinc finger 1a |
| chr20_+_52546186 | 0.14 |
ENSDART00000110777
ENSDART00000153377 ENSDART00000153013 ENSDART00000042704 |
eef1db
|
eukaryotic translation elongation factor 1 delta b (guanine nucleotide exchange protein) |
| chr20_+_25586099 | 0.14 |
ENSDART00000063122
ENSDART00000134047 |
cyp2p10
|
cytochrome P450, family 2, subfamily P, polypeptide 10 |
| chr11_-_38899816 | 0.13 |
ENSDART00000141515
|
pimr127
|
Pim proto-oncogene, serine/threonine kinase, related 127 |
| chr8_-_43834442 | 0.13 |
ENSDART00000191927
|
adgrd1
|
adhesion G protein-coupled receptor D1 |
| chr13_-_31349284 | 0.13 |
ENSDART00000145887
|
rrp12
|
ribosomal RNA processing 12 homolog |
| chr2_-_31537521 | 0.13 |
ENSDART00000148244
|
mrc1b
|
mannose receptor, C type 1b |
| chr8_+_30709685 | 0.13 |
ENSDART00000133989
|
upb1
|
ureidopropionase, beta |
| chr5_-_23715861 | 0.13 |
ENSDART00000019992
|
gbgt1l1
|
globoside alpha-1,3-N-acetylgalactosaminyltransferase 1, like 1 |
| chr16_-_10547781 | 0.13 |
ENSDART00000166495
|
g6fl
|
g6f-like |
| chr7_+_66822229 | 0.13 |
ENSDART00000112109
|
lyve1a
|
lymphatic vessel endothelial hyaluronic receptor 1a |
| chr24_+_1042594 | 0.13 |
ENSDART00000109117
|
si:dkey-192l18.9
|
si:dkey-192l18.9 |
| chr8_+_23521974 | 0.12 |
ENSDART00000188130
ENSDART00000129378 |
sema3gb
|
sema domain, immunoglobulin domain (Ig), short basic domain, secreted, (semaphorin) 3Gb |
| chr9_+_30275139 | 0.12 |
ENSDART00000131781
|
rpgra
|
retinitis pigmentosa GTPase regulator a |
| chr7_-_71384391 | 0.12 |
ENSDART00000112841
|
ccdc149a
|
coiled-coil domain containing 149a |
| chr5_+_67371650 | 0.12 |
ENSDART00000142156
|
sebox
|
SEBOX homeobox |
| chr12_-_11258404 | 0.12 |
ENSDART00000149229
|
si:ch73-30l9.1
|
si:ch73-30l9.1 |
| chr6_-_40697585 | 0.12 |
ENSDART00000113196
|
si:ch211-157b11.14
|
si:ch211-157b11.14 |
| chr22_-_30999000 | 0.12 |
ENSDART00000109841
|
ssuh2.3
|
ssu-2 homolog, tandem duplicate 3 |
| chr3_+_23710839 | 0.12 |
ENSDART00000151584
|
hoxb4a
|
homeobox B4a |
| chr14_+_36497250 | 0.11 |
ENSDART00000184727
|
TENM3
|
si:dkey-237h12.3 |
| chr2_+_5927255 | 0.11 |
ENSDART00000152866
|
si:ch211-168b3.2
|
si:ch211-168b3.2 |
| chr22_-_5933844 | 0.11 |
ENSDART00000163370
ENSDART00000189331 |
si:rp71-36a1.2
|
si:rp71-36a1.2 |
| chr7_-_38689562 | 0.11 |
ENSDART00000167209
|
aplnr2
|
apelin receptor 2 |
| chr24_+_25258904 | 0.11 |
ENSDART00000155714
|
gabrr3b
|
gamma-aminobutyric acid (GABA) A receptor, rho 3b |
| chr19_+_19761966 | 0.11 |
ENSDART00000163697
|
hoxa3a
|
homeobox A3a |
| chr16_-_28268201 | 0.11 |
ENSDART00000121671
ENSDART00000141911 |
si:dkey-12j5.1
|
si:dkey-12j5.1 |
| chr11_+_41981959 | 0.11 |
ENSDART00000055707
|
her15.1
|
hairy and enhancer of split-related 15, tandem duplicate 1 |
| chr4_+_14899720 | 0.11 |
ENSDART00000187456
|
akr1b1
|
aldo-keto reductase family 1, member B1 (aldose reductase) |
| chr9_-_34986827 | 0.11 |
ENSDART00000137862
|
si:ch211-160b11.4
|
si:ch211-160b11.4 |
| chr25_+_16356083 | 0.11 |
ENSDART00000125925
ENSDART00000125444 |
tead1a
|
TEA domain family member 1a |
| chr9_-_23990416 | 0.11 |
ENSDART00000113176
|
col6a3
|
collagen, type VI, alpha 3 |
| chr21_+_20396858 | 0.10 |
ENSDART00000003299
ENSDART00000146615 |
zgc:103482
|
zgc:103482 |
| chr3_+_52545014 | 0.10 |
ENSDART00000018908
|
slc27a1a
|
solute carrier family 27 (fatty acid transporter), member 1a |
| chr10_+_2587234 | 0.10 |
ENSDART00000126937
|
wu:fb59d01
|
wu:fb59d01 |
| chr11_-_25257595 | 0.10 |
ENSDART00000123567
|
snai1a
|
snail family zinc finger 1a |
| chr14_+_80685 | 0.10 |
ENSDART00000188443
|
stag3
|
stromal antigen 3 |
| chr12_+_27117609 | 0.10 |
ENSDART00000076154
|
hoxb8b
|
homeobox B8b |
| chr25_+_15933411 | 0.10 |
ENSDART00000191581
|
ppfibp2b
|
PTPRF interacting protein, binding protein 2b (liprin beta 2) |
| chr21_-_33032715 | 0.10 |
ENSDART00000065349
ENSDART00000191502 ENSDART00000149386 |
slc34a1b
|
solute carrier family 34 (type II sodium/phosphate cotransporter), member 1b |
| chr3_+_52545400 | 0.10 |
ENSDART00000184183
|
slc27a1a
|
solute carrier family 27 (fatty acid transporter), member 1a |
| chr24_+_30392834 | 0.10 |
ENSDART00000162555
|
dpyda.1
|
dihydropyrimidine dehydrogenase a, tandem duplicate 1 |
| chr10_-_42923385 | 0.10 |
ENSDART00000076731
|
ACOT12
|
acyl-CoA thioesterase 12 |
| chr12_+_34854562 | 0.10 |
ENSDART00000130366
|
si:dkey-21c1.4
|
si:dkey-21c1.4 |
| chr22_+_1953096 | 0.10 |
ENSDART00000166630
ENSDART00000186234 |
si:dkey-15h8.16
|
si:dkey-15h8.16 |
| chr10_+_41199660 | 0.10 |
ENSDART00000125314
|
adrb3b
|
adrenoceptor beta 3b |
| chr2_-_32637592 | 0.09 |
ENSDART00000136353
|
si:dkeyp-73d8.8
|
si:dkeyp-73d8.8 |
| chr3_-_55121125 | 0.09 |
ENSDART00000125092
|
hbae1
|
hemoglobin, alpha embryonic 1 |
| chr20_-_51831816 | 0.09 |
ENSDART00000060505
|
mia3
|
melanoma inhibitory activity family, member 3 |
| chr24_-_32025637 | 0.09 |
ENSDART00000180448
ENSDART00000159034 |
rsu1
|
Ras suppressor protein 1 |
| chr2_-_43196595 | 0.09 |
ENSDART00000141087
|
crema
|
cAMP responsive element modulator a |
| chr15_+_31344472 | 0.09 |
ENSDART00000146695
ENSDART00000159182 ENSDART00000060125 |
or107-1
|
odorant receptor, family D, subfamily 107, member 1 |
| chr10_-_44788554 | 0.09 |
ENSDART00000166965
|
tacr1b
|
tachykinin receptor 1b |
| chr20_-_48701593 | 0.09 |
ENSDART00000132835
|
pax1b
|
paired box 1b |
| chr7_-_44957503 | 0.09 |
ENSDART00000165077
|
cdh16
|
cadherin 16, KSP-cadherin |
| chr11_-_1392468 | 0.09 |
ENSDART00000004423
|
iars
|
isoleucyl-tRNA synthetase |
| chr18_-_5875433 | 0.09 |
ENSDART00000151727
|
nob1
|
NIN1/RPN12 binding protein 1 homolog (S. cerevisiae) |
| chr11_+_37049347 | 0.09 |
ENSDART00000109235
|
bicd2
|
bicaudal D homolog 2 (Drosophila) |
| chr12_-_18872717 | 0.09 |
ENSDART00000126300
|
shisa8b
|
shisa family member 8b |
| chr17_-_23727978 | 0.09 |
ENSDART00000079600
|
minpp1a
|
multiple inositol-polyphosphate phosphatase 1a |
| chr25_-_11378623 | 0.09 |
ENSDART00000166586
|
enc2
|
ectodermal-neural cortex 2 |
| chr12_+_19191787 | 0.09 |
ENSDART00000152892
|
slc16a8
|
solute carrier family 16 (monocarboxylate transporter), member 8 |
| chr7_+_18075504 | 0.08 |
ENSDART00000173689
|
si:ch73-40a2.1
|
si:ch73-40a2.1 |
| chr15_-_16946124 | 0.08 |
ENSDART00000154923
|
hip1
|
huntingtin interacting protein 1 |
| chr13_-_37474989 | 0.08 |
ENSDART00000114136
|
wdr89
|
WD repeat domain 89 |
| chr8_-_35960987 | 0.08 |
ENSDART00000160503
|
slc15a4
|
solute carrier family 15 (oligopeptide transporter), member 4 |
| chr19_+_7810028 | 0.08 |
ENSDART00000081592
ENSDART00000140719 |
aqp10b
|
aquaporin 10b |
| chr12_-_18872927 | 0.08 |
ENSDART00000187717
|
shisa8b
|
shisa family member 8b |
| chr2_-_40196547 | 0.08 |
ENSDART00000168098
|
ccl34a.3
|
chemokine (C-C motif) ligand 34a, duplicate 3 |
| chr17_-_50355512 | 0.07 |
ENSDART00000041685
|
otofb
|
otoferlin b |
| chr21_-_19918286 | 0.07 |
ENSDART00000180816
|
ppp1r3b
|
protein phosphatase 1, regulatory subunit 3B |
| chr17_+_14965570 | 0.07 |
ENSDART00000066604
|
gpr137c
|
G protein-coupled receptor 137c |
| chr6_+_28208973 | 0.07 |
ENSDART00000171216
ENSDART00000171377 ENSDART00000167389 ENSDART00000166988 |
LSM2 (1 of many)
|
si:ch73-14h10.2 |
| chr18_+_30508729 | 0.07 |
ENSDART00000185140
|
cox4i1
|
cytochrome c oxidase subunit IV isoform 1 |
| chr12_+_43585527 | 0.07 |
ENSDART00000161590
|
clrn3
|
clarin 3 |
| chr7_-_7692992 | 0.07 |
ENSDART00000192619
|
aadat
|
aminoadipate aminotransferase |
| chr9_+_54290896 | 0.07 |
ENSDART00000149175
|
pou4f3
|
POU class 4 homeobox 3 |
| chr11_+_42556395 | 0.07 |
ENSDART00000039206
|
rps23
|
ribosomal protein S23 |
| chr15_+_44053244 | 0.07 |
ENSDART00000059550
|
lrrc51
|
leucine rich repeat containing 51 |
| chr9_+_41080029 | 0.07 |
ENSDART00000141179
ENSDART00000019289 |
zgc:136439
|
zgc:136439 |
| chr20_-_51831657 | 0.06 |
ENSDART00000165076
|
mia3
|
melanoma inhibitory activity family, member 3 |
| chr22_-_14115292 | 0.06 |
ENSDART00000105717
ENSDART00000165670 |
aox5
|
aldehyde oxidase 5 |
| chr5_-_37881345 | 0.06 |
ENSDART00000084819
|
arhgap35b
|
Rho GTPase activating protein 35b |
| chr24_+_40529346 | 0.06 |
ENSDART00000168548
|
CU929259.1
|
|
| chr15_+_29727799 | 0.06 |
ENSDART00000182006
|
zgc:153372
|
zgc:153372 |
| chr22_-_24818066 | 0.06 |
ENSDART00000143443
|
vtg6
|
vitellogenin 6 |
| chr23_+_36052944 | 0.06 |
ENSDART00000103149
|
hoxc13a
|
homeobox C13a |
| chr20_-_2713120 | 0.06 |
ENSDART00000138753
ENSDART00000104606 |
rars2
|
arginyl-tRNA synthetase 2, mitochondrial (putative) |
| chr1_+_54683655 | 0.06 |
ENSDART00000132785
|
knop1
|
lysine-rich nucleolar protein 1 |
| chr22_-_9736050 | 0.06 |
ENSDART00000152919
|
si:dkey-286j17.4
|
si:dkey-286j17.4 |
| chr1_+_44826593 | 0.06 |
ENSDART00000162200
|
STX3
|
zgc:165520 |
| chr19_-_37154436 | 0.06 |
ENSDART00000103155
|
cx39.4
|
connexin 39.4 |
| chr22_+_2207502 | 0.05 |
ENSDART00000169162
|
si:dkeyp-79b7.12
|
si:dkeyp-79b7.12 |
| chr7_-_7692723 | 0.05 |
ENSDART00000183352
|
aadat
|
aminoadipate aminotransferase |
| chr8_-_25338709 | 0.05 |
ENSDART00000131616
|
atp5pb
|
ATP synthase peripheral stalk-membrane subunit b |
| chr13_-_25745089 | 0.05 |
ENSDART00000189333
ENSDART00000147420 |
sgpl1
|
sphingosine-1-phosphate lyase 1 |
| chr22_-_13165186 | 0.05 |
ENSDART00000105762
|
ahr2
|
aryl hydrocarbon receptor 2 |
| chr23_+_24085531 | 0.05 |
ENSDART00000139710
|
ttll10
|
tubulin tyrosine ligase-like family, member 10 |
| chr17_+_8799661 | 0.05 |
ENSDART00000105326
|
tonsl
|
tonsoku-like, DNA repair protein |
| chr24_+_13635108 | 0.05 |
ENSDART00000183008
|
trpa1b
|
transient receptor potential cation channel, subfamily A, member 1b |
| chr3_-_24456451 | 0.05 |
ENSDART00000024480
ENSDART00000156814 |
baiap2l2a
|
BAI1-associated protein 2-like 2a |
| chr10_+_36178713 | 0.05 |
ENSDART00000140816
|
or108-3
|
odorant receptor, family D, subfamily 108, member 3 |
| chr9_+_21977383 | 0.05 |
ENSDART00000135032
|
si:dkey-57a22.11
|
si:dkey-57a22.11 |
| chr23_-_43718067 | 0.05 |
ENSDART00000015777
|
abce1
|
ATP-binding cassette, sub-family E (OABP), member 1 |
| chr17_+_23554932 | 0.05 |
ENSDART00000135814
|
pank1a
|
pantothenate kinase 1a |
| chr14_+_34490445 | 0.05 |
ENSDART00000132193
ENSDART00000148044 |
wnt8a
|
wingless-type MMTV integration site family, member 8a |
| chr20_-_49889111 | 0.04 |
ENSDART00000058858
|
kif13bb
|
kinesin family member 13Bb |
| chr1_+_961607 | 0.04 |
ENSDART00000184660
|
n6amt1
|
N-6 adenine-specific DNA methyltransferase 1 |
| chr5_+_11290851 | 0.04 |
ENSDART00000180408
|
CABZ01077124.1
|
|
| chr2_+_19578079 | 0.04 |
ENSDART00000144413
|
pimr50
|
Pim proto-oncogene, serine/threonine kinase, related 50 |
| chr14_+_3071110 | 0.04 |
ENSDART00000162445
|
hmgxb3
|
HMG box domain containing 3 |
| chr4_-_16334362 | 0.04 |
ENSDART00000101461
|
epyc
|
epiphycan |
| chr6_-_42388608 | 0.04 |
ENSDART00000049425
|
sec61a1l
|
Sec61 translocon alpha 1 subunit, like |
| chr22_+_2632961 | 0.04 |
ENSDART00000106278
|
znf1173
|
zinc finger protein 1173 |
| chr21_+_3775551 | 0.04 |
ENSDART00000111699
|
tor1
|
torsin family 1 |
| chr21_+_27278120 | 0.04 |
ENSDART00000193882
|
si:dkey-175m17.7
|
si:dkey-175m17.7 |
| chr9_-_23152092 | 0.04 |
ENSDART00000180155
ENSDART00000186935 |
lypd6b
|
LY6/PLAUR domain containing 6B |
| chr10_-_42882215 | 0.04 |
ENSDART00000180580
ENSDART00000187374 |
FP236542.1
|
|
| chr15_-_17024779 | 0.04 |
ENSDART00000154719
|
hip1
|
huntingtin interacting protein 1 |
| chr5_-_48307804 | 0.04 |
ENSDART00000182831
ENSDART00000186920 ENSDART00000183585 |
mef2cb
|
myocyte enhancer factor 2cb |
| chr3_-_44412 | 0.04 |
ENSDART00000191690
|
zgc:110249
|
zgc:110249 |
| chr17_-_23412705 | 0.04 |
ENSDART00000126995
|
si:ch211-149k12.3
|
si:ch211-149k12.3 |
| chr21_+_2102829 | 0.04 |
ENSDART00000157922
|
AL627305.1
|
|
| chr11_+_30244356 | 0.04 |
ENSDART00000036050
ENSDART00000150080 |
rs1a
|
retinoschisin 1a |
| chr5_-_57289872 | 0.03 |
ENSDART00000189893
ENSDART00000050957 |
fer
|
fer (fps/fes related) tyrosine kinase |
| chr10_-_3332362 | 0.03 |
ENSDART00000007577
ENSDART00000055140 |
tor4aa
|
torsin family 4, member Aa |
| chr23_+_25354856 | 0.03 |
ENSDART00000109023
ENSDART00000147440 |
fmnl3
|
formin-like 3 |
| chr1_-_20593778 | 0.03 |
ENSDART00000124770
|
ugt8
|
UDP glycosyltransferase 8 |
| chr10_+_20628698 | 0.03 |
ENSDART00000064666
|
prnpb
|
prion protein b |
| chr10_+_5234327 | 0.03 |
ENSDART00000133927
ENSDART00000063120 |
sptlc1
|
serine palmitoyltransferase, long chain base subunit 1 |
| chr25_-_31739309 | 0.03 |
ENSDART00000098896
|
acot19
|
acyl-CoA thioesterase 19 |
Network of associatons between targets according to the STRING database.
First level regulatory network of hoxd11a
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.5 | GO:0035994 | response to muscle stretch(GO:0035994) |
| 0.1 | 0.4 | GO:0048618 | post-embryonic foregut morphogenesis(GO:0048618) |
| 0.1 | 0.6 | GO:0097250 | mitochondrial respiratory chain supercomplex assembly(GO:0097250) |
| 0.1 | 0.3 | GO:0010430 | fatty acid omega-oxidation(GO:0010430) |
| 0.1 | 0.2 | GO:0022009 | central nervous system vasculogenesis(GO:0022009) |
| 0.1 | 0.4 | GO:0032367 | intracellular cholesterol transport(GO:0032367) |
| 0.1 | 0.2 | GO:2000793 | cell proliferation involved in heart valve morphogenesis(GO:0003249) regulation of cell proliferation involved in heart valve morphogenesis(GO:0003250) endocardial cell development(GO:0060958) cell proliferation involved in heart valve development(GO:2000793) |
| 0.1 | 0.3 | GO:1901841 | regulation of high voltage-gated calcium channel activity(GO:1901841) negative regulation of high voltage-gated calcium channel activity(GO:1901842) |
| 0.1 | 0.3 | GO:0035988 | chondrocyte proliferation(GO:0035988) |
| 0.1 | 0.3 | GO:0008635 | activation of cysteine-type endopeptidase activity involved in apoptotic process by cytochrome c(GO:0008635) |
| 0.1 | 0.2 | GO:0060148 | negative regulation of hippo signaling(GO:0035331) positive regulation of posttranscriptional gene silencing(GO:0060148) positive regulation of gene silencing by miRNA(GO:2000637) |
| 0.1 | 0.2 | GO:0017185 | peptidyl-lysine hydroxylation(GO:0017185) |
| 0.0 | 0.1 | GO:0006212 | uracil catabolic process(GO:0006212) uracil metabolic process(GO:0019860) |
| 0.0 | 0.2 | GO:0035513 | oxidative RNA demethylation(GO:0035513) |
| 0.0 | 0.3 | GO:0032218 | riboflavin transport(GO:0032218) |
| 0.0 | 0.3 | GO:0045337 | farnesyl diphosphate biosynthetic process(GO:0045337) |
| 0.0 | 0.2 | GO:0031650 | heat generation(GO:0031649) regulation of heat generation(GO:0031650) positive regulation of heat generation(GO:0031652) regulation of phospholipid biosynthetic process(GO:0071071) negative regulation of phospholipid biosynthetic process(GO:0071072) |
| 0.0 | 0.2 | GO:0097241 | hematopoietic stem cell migration to bone marrow(GO:0097241) |
| 0.0 | 0.2 | GO:0033119 | negative regulation of RNA splicing(GO:0033119) negative regulation of mRNA splicing, via spliceosome(GO:0048025) |
| 0.0 | 0.2 | GO:0032024 | positive regulation of insulin secretion(GO:0032024) |
| 0.0 | 0.1 | GO:0042220 | response to cocaine(GO:0042220) cellular response to alkaloid(GO:0071312) cellular response to cocaine(GO:0071314) |
| 0.0 | 0.3 | GO:0006833 | water transport(GO:0006833) |
| 0.0 | 0.2 | GO:1990511 | piRNA biosynthetic process(GO:1990511) |
| 0.0 | 0.2 | GO:0071340 | skeletal muscle acetylcholine-gated channel clustering(GO:0071340) |
| 0.0 | 0.1 | GO:0019482 | beta-alanine metabolic process(GO:0019482) beta-alanine biosynthetic process(GO:0019483) |
| 0.0 | 0.1 | GO:0042776 | mitochondrial ATP synthesis coupled proton transport(GO:0042776) |
| 0.0 | 0.1 | GO:0005981 | regulation of glycogen catabolic process(GO:0005981) |
| 0.0 | 0.1 | GO:0018872 | arsonoacetate metabolic process(GO:0018872) |
| 0.0 | 0.1 | GO:0002025 | vasodilation by norepinephrine-epinephrine involved in regulation of systemic arterial blood pressure(GO:0002025) |
| 0.0 | 0.1 | GO:0060017 | parathyroid gland development(GO:0060017) |
| 0.0 | 0.2 | GO:0048172 | regulation of short-term neuronal synaptic plasticity(GO:0048172) |
| 0.0 | 0.4 | GO:0014823 | response to activity(GO:0014823) |
| 0.0 | 0.1 | GO:0044341 | sodium-dependent phosphate transport(GO:0044341) |
| 0.0 | 0.0 | GO:0050968 | chemosensory behavior(GO:0007635) detection of chemical stimulus involved in sensory perception of pain(GO:0050968) |
| 0.0 | 0.0 | GO:0039015 | spinal cord anterior/posterior patterning(GO:0021512) cell proliferation involved in pronephros development(GO:0039015) cell proliferation involved in kidney development(GO:0072111) |
| 0.0 | 0.6 | GO:0030968 | endoplasmic reticulum unfolded protein response(GO:0030968) |
| 0.0 | 0.2 | GO:0051127 | positive regulation of actin nucleation(GO:0051127) positive regulation of Arp2/3 complex-mediated actin nucleation(GO:2000601) |
| 0.0 | 0.2 | GO:1990798 | pancreas regeneration(GO:1990798) |
| 0.0 | 0.4 | GO:0060030 | dorsal convergence(GO:0060030) |
| 0.0 | 0.3 | GO:0055008 | cardiac muscle tissue morphogenesis(GO:0055008) |
| 0.0 | 0.2 | GO:0008078 | mesodermal cell migration(GO:0008078) |
| 0.0 | 0.1 | GO:0045948 | positive regulation of translational initiation(GO:0045948) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.2 | GO:1990909 | Wnt signalosome(GO:1990909) |
| 0.0 | 0.1 | GO:1990131 | EGO complex(GO:0034448) Gtr1-Gtr2 GTPase complex(GO:1990131) |
| 0.0 | 0.1 | GO:0098890 | extrinsic component of postsynaptic membrane(GO:0098890) |
| 0.0 | 0.1 | GO:0031428 | box C/D snoRNP complex(GO:0031428) |
| 0.0 | 0.1 | GO:0005853 | eukaryotic translation elongation factor 1 complex(GO:0005853) |
| 0.0 | 0.2 | GO:0005964 | phosphorylase kinase complex(GO:0005964) |
| 0.0 | 0.0 | GO:0097189 | apoptotic body(GO:0097189) |
| 0.0 | 0.3 | GO:0031430 | M band(GO:0031430) |
| 0.0 | 0.1 | GO:0097225 | sperm midpiece(GO:0097225) |
| 0.0 | 0.3 | GO:0033017 | sarcoplasmic reticulum membrane(GO:0033017) |
| 0.0 | 0.2 | GO:0031209 | SCAR complex(GO:0031209) |
| 0.0 | 0.2 | GO:1990907 | beta-catenin-TCF complex(GO:1990907) |
| 0.0 | 0.5 | GO:0005861 | troponin complex(GO:0005861) |
| 0.0 | 0.2 | GO:0034992 | microtubule organizing center attachment site(GO:0034992) LINC complex(GO:0034993) |
| 0.0 | 0.2 | GO:0032591 | dendritic spine membrane(GO:0032591) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.4 | GO:0030228 | lipoprotein particle receptor activity(GO:0030228) |
| 0.1 | 0.8 | GO:0015250 | water channel activity(GO:0015250) |
| 0.1 | 0.3 | GO:0004161 | dimethylallyltranstransferase activity(GO:0004161) geranyltranstransferase activity(GO:0004337) |
| 0.1 | 0.3 | GO:0015562 | protein-transmembrane transporting ATPase activity(GO:0015462) efflux transmembrane transporter activity(GO:0015562) |
| 0.1 | 0.2 | GO:0070815 | peptidyl-lysine 5-dioxygenase activity(GO:0070815) |
| 0.1 | 0.2 | GO:0051059 | NF-kappaB binding(GO:0051059) |
| 0.1 | 0.2 | GO:0035516 | oxidative DNA demethylase activity(GO:0035516) |
| 0.1 | 0.3 | GO:0004743 | pyruvate kinase activity(GO:0004743) |
| 0.0 | 0.3 | GO:0032217 | riboflavin transporter activity(GO:0032217) |
| 0.0 | 0.2 | GO:0004396 | glucokinase activity(GO:0004340) hexokinase activity(GO:0004396) glucose binding(GO:0005536) fructokinase activity(GO:0008865) mannokinase activity(GO:0019158) |
| 0.0 | 0.1 | GO:0016496 | substance P receptor activity(GO:0016496) |
| 0.0 | 0.2 | GO:0031957 | very long-chain fatty acid-CoA ligase activity(GO:0031957) |
| 0.0 | 0.2 | GO:0004366 | glycerol-3-phosphate O-acyltransferase activity(GO:0004366) |
| 0.0 | 0.2 | GO:0038064 | protein tyrosine kinase collagen receptor activity(GO:0038062) collagen receptor activity(GO:0038064) |
| 0.0 | 0.1 | GO:0051380 | norepinephrine binding(GO:0051380) |
| 0.0 | 0.6 | GO:0015002 | cytochrome-c oxidase activity(GO:0004129) heme-copper terminal oxidase activity(GO:0015002) oxidoreductase activity, acting on a heme group of donors(GO:0016675) oxidoreductase activity, acting on a heme group of donors, oxygen as acceptor(GO:0016676) |
| 0.0 | 0.1 | GO:0060182 | apelin receptor activity(GO:0060182) |
| 0.0 | 0.1 | GO:0016623 | aldehyde oxidase activity(GO:0004031) oxidoreductase activity, acting on the aldehyde or oxo group of donors, oxygen as acceptor(GO:0016623) |
| 0.0 | 0.1 | GO:0034416 | bisphosphoglycerate phosphatase activity(GO:0034416) |
| 0.0 | 0.1 | GO:0030792 | arsenite methyltransferase activity(GO:0030791) methylarsonite methyltransferase activity(GO:0030792) |
| 0.0 | 0.2 | GO:0071933 | Arp2/3 complex binding(GO:0071933) |
| 0.0 | 0.1 | GO:0002061 | uracil binding(GO:0002058) pyrimidine nucleobase binding(GO:0002061) dihydropyrimidine dehydrogenase (NADP+) activity(GO:0017113) |
| 0.0 | 0.1 | GO:0016212 | kynurenine-oxoglutarate transaminase activity(GO:0016212) kynurenine aminotransferase activity(GO:0036137) |
| 0.0 | 0.5 | GO:0008307 | structural constituent of muscle(GO:0008307) |
| 0.0 | 0.1 | GO:0005436 | sodium:phosphate symporter activity(GO:0005436) |
| 0.0 | 0.3 | GO:0035497 | cAMP response element binding(GO:0035497) |
| 0.0 | 0.1 | GO:0008117 | sphinganine-1-phosphate aldolase activity(GO:0008117) |
| 0.0 | 0.3 | GO:0005246 | calcium channel regulator activity(GO:0005246) |
| 0.0 | 0.1 | GO:0034452 | dynactin binding(GO:0034452) |
| 0.0 | 0.1 | GO:0043325 | phosphatidylinositol-3,4-bisphosphate binding(GO:0043325) |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | REACTOME PYRIMIDINE CATABOLISM | Genes involved in Pyrimidine catabolism |
| 0.0 | 0.2 | REACTOME REGULATION OF GENE EXPRESSION IN BETA CELLS | Genes involved in Regulation of gene expression in beta cells |
| 0.0 | 0.3 | REACTOME CHOLESTEROL BIOSYNTHESIS | Genes involved in Cholesterol biosynthesis |