Project
PRJNA207719: Tissue specific transcriptome profiling
Navigation
Downloads
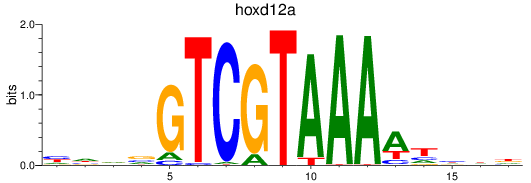
Results for hoxd12a
Z-value: 0.50
Transcription factors associated with hoxd12a
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
hoxd12a
|
ENSDARG00000059263 | homeobox D12a |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| hoxd12a | dr11_v1_chr9_-_1984604_1984604 | 0.75 | 1.5e-01 | Click! |
Activity profile of hoxd12a motif
Sorted Z-values of hoxd12a motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr9_-_39005317 | 0.38 |
ENSDART00000014207
|
myl1
|
myosin, light chain 1, alkali; skeletal, fast |
| chr24_-_17029374 | 0.27 |
ENSDART00000039267
|
ptgdsb.1
|
prostaglandin D2 synthase b, tandem duplicate 1 |
| chr14_-_34026316 | 0.25 |
ENSDART00000186062
|
cyfip2
|
cytoplasmic FMR1 interacting protein 2 |
| chr10_-_20357013 | 0.23 |
ENSDART00000080143
|
sfrp1b
|
secreted frizzled-related protein 1b |
| chr1_+_31019165 | 0.18 |
ENSDART00000102210
|
itga6b
|
integrin, alpha 6b |
| chr14_-_25985698 | 0.17 |
ENSDART00000172909
ENSDART00000123053 |
atox1
|
antioxidant 1 copper chaperone |
| chr1_+_45056371 | 0.15 |
ENSDART00000073689
ENSDART00000167309 |
btr01
|
bloodthirsty-related gene family, member 1 |
| chr11_+_2416064 | 0.15 |
ENSDART00000067117
|
ube2v1
|
ubiquitin-conjugating enzyme E2 variant 1 |
| chr1_-_26782573 | 0.15 |
ENSDART00000090611
|
sh3gl2a
|
SH3 domain containing GRB2 like 2a, endophilin A1 |
| chr15_+_21744680 | 0.14 |
ENSDART00000016860
|
ppp2r1bb
|
protein phosphatase 2, regulatory subunit A, beta b |
| chr8_-_12432604 | 0.14 |
ENSDART00000133350
ENSDART00000140699 ENSDART00000101174 |
traf1
|
TNF receptor-associated factor 1 |
| chr16_-_31754102 | 0.14 |
ENSDART00000185043
|
ptpn6
|
protein tyrosine phosphatase, non-receptor type 6 |
| chr5_+_58372164 | 0.13 |
ENSDART00000057910
|
nrgna
|
neurogranin (protein kinase C substrate, RC3) a |
| chr22_+_10440991 | 0.13 |
ENSDART00000064805
|
cenpp
|
centromere protein P |
| chr15_+_29116063 | 0.13 |
ENSDART00000016112
ENSDART00000153609 ENSDART00000155630 |
capns1b
|
calpain, small subunit 1 b |
| chr1_-_38171648 | 0.13 |
ENSDART00000137451
ENSDART00000047159 |
hmgb2a
|
high mobility group box 2a |
| chr1_+_49568335 | 0.12 |
ENSDART00000142957
|
col17a1a
|
collagen, type XVII, alpha 1a |
| chr8_-_14049404 | 0.12 |
ENSDART00000093117
|
atp2b3a
|
ATPase plasma membrane Ca2+ transporting 3a |
| chr7_+_24205034 | 0.12 |
ENSDART00000018580
|
nedd8
|
neural precursor cell expressed, developmentally down-regulated 8 |
| chr18_+_8812549 | 0.11 |
ENSDART00000017619
|
impdh1a
|
IMP (inosine 5'-monophosphate) dehydrogenase 1a |
| chr6_+_49551614 | 0.11 |
ENSDART00000022581
|
rab22a
|
RAB22A, member RAS oncogene family |
| chr13_+_35856463 | 0.11 |
ENSDART00000171056
ENSDART00000017202 |
kcnk1b
|
potassium channel, subfamily K, member 1b |
| chr5_-_69499486 | 0.10 |
ENSDART00000023983
ENSDART00000180293 |
psat1
|
phosphoserine aminotransferase 1 |
| chr24_+_25913162 | 0.10 |
ENSDART00000143099
ENSDART00000184814 |
map3k15
|
mitogen-activated protein kinase kinase kinase 15 |
| chr21_+_26720803 | 0.10 |
ENSDART00000053797
|
slc3a2b
|
solute carrier family 3 (amino acid transporter heavy chain), member 2b |
| chr3_+_34140507 | 0.10 |
ENSDART00000131802
|
si:dkey-204f11.64
|
si:dkey-204f11.64 |
| chr7_+_20524064 | 0.10 |
ENSDART00000052917
|
slc3a2a
|
solute carrier family 3 (amino acid transporter heavy chain), member 2a |
| chr25_+_20077225 | 0.10 |
ENSDART00000136543
|
tnni4b.1
|
troponin I4b, tandem duplicate 1 |
| chr9_-_9242777 | 0.10 |
ENSDART00000021191
|
u2af1
|
U2 small nuclear RNA auxiliary factor 1 |
| chr6_-_2154137 | 0.09 |
ENSDART00000162656
|
tgm5l
|
transglutaminase 5, like |
| chr24_-_26632171 | 0.09 |
ENSDART00000008374
ENSDART00000017384 |
tnikb
|
TRAF2 and NCK interacting kinase b |
| chr15_-_28904371 | 0.09 |
ENSDART00000155154
|
eml2
|
echinoderm microtubule associated protein like 2 |
| chr20_+_405811 | 0.09 |
ENSDART00000149311
|
gpr63
|
G protein-coupled receptor 63 |
| chr11_-_21303946 | 0.09 |
ENSDART00000185786
|
RASSF5
|
si:dkey-85p17.3 |
| chr13_-_37631092 | 0.09 |
ENSDART00000108855
|
si:dkey-188i13.7
|
si:dkey-188i13.7 |
| chr13_+_35745572 | 0.09 |
ENSDART00000159690
|
gpr75
|
G protein-coupled receptor 75 |
| chr12_-_10038870 | 0.09 |
ENSDART00000152250
|
ngfrb
|
nerve growth factor receptor b |
| chr21_-_40174647 | 0.09 |
ENSDART00000183738
ENSDART00000076840 ENSDART00000145109 |
slco2b1
|
solute carrier organic anion transporter family, member 2B1 |
| chr16_-_10316359 | 0.09 |
ENSDART00000104025
|
flot1b
|
flotillin 1b |
| chr19_-_35400819 | 0.09 |
ENSDART00000148080
|
rnf19b
|
ring finger protein 19B |
| chr23_-_27505825 | 0.09 |
ENSDART00000137229
ENSDART00000013797 |
asb8
|
ankyrin repeat and SOCS box containing 8 |
| chr25_+_29161609 | 0.09 |
ENSDART00000180752
|
pkmb
|
pyruvate kinase M1/2b |
| chr23_-_4855122 | 0.09 |
ENSDART00000133701
|
slc6a1a
|
solute carrier family 6 (neurotransmitter transporter), member 1a |
| chr23_+_36087219 | 0.08 |
ENSDART00000154825
|
hoxc3a
|
homeobox C3a |
| chr2_-_36914281 | 0.08 |
ENSDART00000008322
|
si:dkey-193b15.5
|
si:dkey-193b15.5 |
| chr10_+_10636237 | 0.08 |
ENSDART00000136853
|
fam163b
|
family with sequence similarity 163, member B |
| chr16_-_47426482 | 0.08 |
ENSDART00000148631
ENSDART00000149723 |
sept7b
|
septin 7b |
| chr7_+_49681040 | 0.08 |
ENSDART00000176372
ENSDART00000192172 |
rassf7b
|
Ras association (RalGDS/AF-6) domain family (N-terminal) member 7b |
| chr19_+_3840955 | 0.08 |
ENSDART00000172305
|
lsm10
|
LSM10, U7 small nuclear RNA associated |
| chr11_-_13341483 | 0.08 |
ENSDART00000164978
|
mast3b
|
microtubule associated serine/threonine kinase 3b |
| chr17_-_31695217 | 0.08 |
ENSDART00000104332
ENSDART00000143090 |
lin52
|
lin-52 DREAM MuvB core complex component |
| chr14_-_10387377 | 0.08 |
ENSDART00000145118
|
commd5
|
COMM domain containing 5 |
| chr25_+_31122806 | 0.08 |
ENSDART00000067039
|
rassf8a
|
Ras association (RalGDS/AF-6) domain family (N-terminal) member 8a |
| chr7_-_43840418 | 0.08 |
ENSDART00000174592
|
cdh11
|
cadherin 11, type 2, OB-cadherin (osteoblast) |
| chr4_-_390431 | 0.08 |
ENSDART00000067482
ENSDART00000138500 |
dynlt1
|
dynein, light chain, Tctex-type 1 |
| chr6_+_3827751 | 0.08 |
ENSDART00000003008
ENSDART00000122348 |
gad1b
|
glutamate decarboxylase 1b |
| chr24_-_5713799 | 0.08 |
ENSDART00000137293
|
dia1b
|
deleted in autism 1b |
| chr18_+_31280984 | 0.08 |
ENSDART00000170285
ENSDART00000150608 ENSDART00000159720 |
def8
|
differentially expressed in FDCP 8 homolog (mouse) |
| chr11_+_24716837 | 0.07 |
ENSDART00000145217
|
zgc:153953
|
zgc:153953 |
| chr20_-_2355357 | 0.07 |
ENSDART00000085281
|
EPB41L2
|
si:ch73-18b11.1 |
| chr25_-_20024829 | 0.07 |
ENSDART00000140182
ENSDART00000174776 |
cnot4a
|
CCR4-NOT transcription complex, subunit 4a |
| chr7_-_42206720 | 0.07 |
ENSDART00000110907
|
itfg1
|
integrin alpha FG-GAP repeat containing 1 |
| chr25_+_32473277 | 0.07 |
ENSDART00000146451
|
sqor
|
sulfide quinone oxidoreductase |
| chr6_-_47953829 | 0.07 |
ENSDART00000157264
|
si:dkey-166d12.2
|
si:dkey-166d12.2 |
| chr22_+_20169352 | 0.07 |
ENSDART00000169055
ENSDART00000061617 |
hmg20b
|
high mobility group 20B |
| chr10_-_31805923 | 0.07 |
ENSDART00000077785
|
vps26bl
|
vacuolar protein sorting 26 homolog B, like |
| chr25_-_11057753 | 0.07 |
ENSDART00000186551
|
sv2bb
|
synaptic vesicle glycoprotein 2Bb |
| chr6_+_3828560 | 0.07 |
ENSDART00000185273
ENSDART00000179091 |
gad1b
|
glutamate decarboxylase 1b |
| chr23_-_27506161 | 0.07 |
ENSDART00000145007
|
asb8
|
ankyrin repeat and SOCS box containing 8 |
| chr2_+_47581997 | 0.07 |
ENSDART00000112579
|
scg2b
|
secretogranin II (chromogranin C), b |
| chr7_+_42206543 | 0.07 |
ENSDART00000112543
|
phkb
|
phosphorylase kinase, beta |
| chr18_+_41542542 | 0.07 |
ENSDART00000087445
|
tsen34
|
TSEN34 tRNA splicing endonuclease subunit |
| chr1_+_58282449 | 0.07 |
ENSDART00000131475
|
si:dkey-222h21.7
|
si:dkey-222h21.7 |
| chr19_+_42770041 | 0.06 |
ENSDART00000150930
|
clasp2
|
cytoplasmic linker associated protein 2 |
| chr12_+_46696867 | 0.06 |
ENSDART00000152928
ENSDART00000153445 ENSDART00000123357 ENSDART00000152880 ENSDART00000123834 ENSDART00000174765 ENSDART00000189923 |
exoc7
|
exocyst complex component 7 |
| chr12_+_13282797 | 0.06 |
ENSDART00000137757
ENSDART00000152397 |
irf9
|
interferon regulatory factor 9 |
| chr6_+_42338309 | 0.06 |
ENSDART00000015277
|
gpx1b
|
glutathione peroxidase 1b |
| chr25_+_18964782 | 0.06 |
ENSDART00000017299
|
tdg.1
|
thymine DNA glycosylase, tandem duplicate 1 |
| chr20_+_7398090 | 0.06 |
ENSDART00000108649
|
pcsk9
|
proprotein convertase subtilisin/kexin type 9 |
| chr9_-_2892250 | 0.06 |
ENSDART00000140695
|
cdca7a
|
cell division cycle associated 7a |
| chr22_-_15671525 | 0.06 |
ENSDART00000080286
|
si:ch1073-396h14.1
|
si:ch1073-396h14.1 |
| chr24_-_24796583 | 0.06 |
ENSDART00000144791
ENSDART00000146570 |
pde7a
|
phosphodiesterase 7A |
| chr22_-_36750589 | 0.06 |
ENSDART00000010824
|
acy1
|
aminoacylase 1 |
| chr15_-_20412286 | 0.06 |
ENSDART00000008589
|
chp2
|
calcineurin-like EF-hand protein 2 |
| chr2_+_15612755 | 0.06 |
ENSDART00000003035
|
amy2a
|
amylase, alpha 2A (pancreatic) |
| chr1_-_56213723 | 0.06 |
ENSDART00000142505
ENSDART00000137237 |
si:dkey-76b14.2
|
si:dkey-76b14.2 |
| chr7_+_42206847 | 0.06 |
ENSDART00000149250
|
phkb
|
phosphorylase kinase, beta |
| chr2_-_37458527 | 0.06 |
ENSDART00000146820
|
si:dkey-57k2.7
|
si:dkey-57k2.7 |
| chr5_-_31856681 | 0.06 |
ENSDART00000187817
|
pkn3
|
protein kinase N3 |
| chr11_+_20896122 | 0.06 |
ENSDART00000162339
ENSDART00000181111 |
CABZ01008730.1
|
|
| chr1_-_26131088 | 0.06 |
ENSDART00000193973
ENSDART00000054209 |
cdkn2a/b
|
cyclin-dependent kinase inhibitor 2A/B (p15, inhibits CDK4) |
| chr16_-_24561354 | 0.06 |
ENSDART00000193278
ENSDART00000126274 |
si:ch211-79k12.2
|
si:ch211-79k12.2 |
| chr8_+_20994433 | 0.06 |
ENSDART00000141736
|
si:dkeyp-82a1.8
|
si:dkeyp-82a1.8 |
| chr3_+_59851537 | 0.06 |
ENSDART00000180997
|
CU693479.1
|
|
| chr25_-_21066136 | 0.06 |
ENSDART00000109520
|
fbxl14a
|
F-box and leucine-rich repeat protein 14a |
| chr1_+_30103297 | 0.06 |
ENSDART00000146783
|
vps8
|
vacuolar protein sorting 8 homolog (S. cerevisiae) |
| chr5_-_26764880 | 0.06 |
ENSDART00000140392
ENSDART00000134728 |
rnf181
|
ring finger protein 181 |
| chr14_+_33507608 | 0.06 |
ENSDART00000038128
|
mcts1
|
malignant T cell amplified sequence 1 |
| chr6_-_51101834 | 0.06 |
ENSDART00000092493
|
ptprt
|
protein tyrosine phosphatase, receptor type, t |
| chr14_-_42997145 | 0.06 |
ENSDART00000172801
|
pcdh10b
|
protocadherin 10b |
| chr14_+_989733 | 0.06 |
ENSDART00000161487
ENSDART00000127317 |
si:ch73-308l14.2
|
si:ch73-308l14.2 |
| chr6_+_49742164 | 0.05 |
ENSDART00000024578
|
npepl1
|
aminopeptidase like 1 |
| chr25_-_21092222 | 0.05 |
ENSDART00000154765
|
prr5a
|
proline rich 5a (renal) |
| chr14_-_33936524 | 0.05 |
ENSDART00000112438
|
si:ch73-335m24.5
|
si:ch73-335m24.5 |
| chr14_+_30568961 | 0.05 |
ENSDART00000184303
|
mrpl11
|
mitochondrial ribosomal protein L11 |
| chr4_-_13156971 | 0.05 |
ENSDART00000182164
|
grip1
|
glutamate receptor interacting protein 1 |
| chr21_+_11865972 | 0.05 |
ENSDART00000081676
|
ubap1
|
ubiquitin associated protein 1 |
| chr18_-_17087138 | 0.05 |
ENSDART00000135597
|
zc3h18
|
zinc finger CCCH-type containing 18 |
| chr4_-_25836684 | 0.05 |
ENSDART00000142491
|
ndufa12
|
NADH dehydrogenase (ubiquinone) 1 alpha subcomplex, 12 |
| chr24_-_40668208 | 0.05 |
ENSDART00000171543
|
smyhc1
|
slow myosin heavy chain 1 |
| chr22_+_10215558 | 0.05 |
ENSDART00000063274
|
kctd6a
|
potassium channel tetramerization domain containing 6a |
| chr15_-_42736433 | 0.05 |
ENSDART00000154379
|
si:ch211-181d7.1
|
si:ch211-181d7.1 |
| chr17_+_6217704 | 0.05 |
ENSDART00000129100
|
BX000363.1
|
|
| chr22_-_11078988 | 0.05 |
ENSDART00000126664
ENSDART00000006927 |
use1
|
unconventional SNARE in the ER 1 homolog (S. cerevisiae) |
| chr21_+_37442458 | 0.05 |
ENSDART00000180161
ENSDART00000186950 |
cox7b
|
cytochrome c oxidase subunit VIIb |
| chr21_-_17296789 | 0.05 |
ENSDART00000192180
|
gfi1b
|
growth factor independent 1B transcription repressor |
| chr5_+_38744660 | 0.05 |
ENSDART00000180506
|
si:dkey-58f10.6
|
si:dkey-58f10.6 |
| chr12_-_11560794 | 0.05 |
ENSDART00000149098
ENSDART00000169975 |
plekha1b
|
pleckstrin homology domain containing, family A (phosphoinositide binding specific) member 1b |
| chr5_+_23598364 | 0.05 |
ENSDART00000132155
|
kat5b
|
K(lysine) acetyltransferase 5b |
| chr2_-_37874647 | 0.05 |
ENSDART00000039386
|
zgc:66427
|
zgc:66427 |
| chr15_-_14552101 | 0.05 |
ENSDART00000171169
|
numbl
|
numb homolog (Drosophila)-like |
| chr2_-_45510223 | 0.05 |
ENSDART00000113058
|
gpsm2
|
G protein signaling modulator 2 |
| chr20_+_38837238 | 0.05 |
ENSDART00000061334
|
ift172
|
intraflagellar transport 172 |
| chr24_-_26715174 | 0.05 |
ENSDART00000079726
|
pld1b
|
phospholipase D1b |
| chr19_+_29808699 | 0.04 |
ENSDART00000051799
ENSDART00000164205 |
hdac1
|
histone deacetylase 1 |
| chr5_-_10768258 | 0.04 |
ENSDART00000157043
|
rtn4r
|
reticulon 4 receptor |
| chr15_+_8767650 | 0.04 |
ENSDART00000033871
|
ap2s1
|
adaptor-related protein complex 2, sigma 1 subunit |
| chr16_+_5184402 | 0.04 |
ENSDART00000156685
|
soga3a
|
SOGA family member 3a |
| chr19_+_7595924 | 0.04 |
ENSDART00000140083
|
s100s
|
S100 calcium binding protein S |
| chr14_+_46410766 | 0.04 |
ENSDART00000032342
|
anxa5a
|
annexin A5a |
| chr18_-_2433011 | 0.04 |
ENSDART00000181922
ENSDART00000193276 |
CR769778.1
|
|
| chr8_-_19649617 | 0.04 |
ENSDART00000189033
|
fam78bb
|
family with sequence similarity 78, member B b |
| chr25_+_16689633 | 0.04 |
ENSDART00000073416
|
ada2a
|
adenosine deaminase 2a |
| chr21_-_22543611 | 0.04 |
ENSDART00000177084
|
myo5b
|
myosin VB |
| chr22_-_31517300 | 0.04 |
ENSDART00000164799
|
slc6a6b
|
solute carrier family 6 (neurotransmitter transporter), member 6b |
| chr19_+_29808471 | 0.04 |
ENSDART00000186428
|
hdac1
|
histone deacetylase 1 |
| chr8_+_23861461 | 0.04 |
ENSDART00000037109
|
srpk1a
|
SRSF protein kinase 1a |
| chr22_+_12366516 | 0.04 |
ENSDART00000157802
|
r3hdm1
|
R3H domain containing 1 |
| chr9_-_2892045 | 0.04 |
ENSDART00000137201
|
cdca7a
|
cell division cycle associated 7a |
| chr10_+_26896142 | 0.04 |
ENSDART00000188225
|
ehbp1l1b
|
EH domain binding protein 1-like 1b |
| chr15_-_5467477 | 0.04 |
ENSDART00000123839
|
arrb1
|
arrestin, beta 1 |
| chr2_+_371759 | 0.04 |
ENSDART00000153788
|
si:dkey-33c14.7
|
si:dkey-33c14.7 |
| chr16_-_43065164 | 0.04 |
ENSDART00000149431
|
sema4aa
|
sema domain, immunoglobulin domain (Ig), transmembrane domain (TM) and short cytoplasmic domain, (semaphorin) 4Aa |
| chr3_+_36284986 | 0.04 |
ENSDART00000059533
|
wipi1
|
WD repeat domain, phosphoinositide interacting 1 |
| chr4_+_76441200 | 0.03 |
ENSDART00000125751
|
si:dkey-16p6.1
|
si:dkey-16p6.1 |
| chr3_-_26191960 | 0.03 |
ENSDART00000113843
|
ypel3
|
yippee-like 3 |
| chr21_-_25618175 | 0.03 |
ENSDART00000133512
|
fosl1b
|
FOS-like antigen 1b |
| chr6_+_13207139 | 0.03 |
ENSDART00000185601
ENSDART00000182182 |
ino80db
|
INO80 complex subunit Db |
| chr25_-_34973211 | 0.03 |
ENSDART00000045177
|
cdk10
|
cyclin-dependent kinase 10 |
| chr13_-_36050303 | 0.03 |
ENSDART00000134955
ENSDART00000139087 |
lgmn
|
legumain |
| chr16_+_33143503 | 0.03 |
ENSDART00000058471
ENSDART00000179385 |
rhbdl2
|
rhomboid, veinlet-like 2 (Drosophila) |
| chr2_+_54696042 | 0.03 |
ENSDART00000074270
|
ankrd12
|
ankyrin repeat domain 12 |
| chr9_-_42730672 | 0.03 |
ENSDART00000136728
|
fkbp7
|
FK506 binding protein 7 |
| chr15_+_2857556 | 0.03 |
ENSDART00000157758
|
mre11a
|
MRE11 homolog A, double strand break repair nuclease |
| chr25_+_17920668 | 0.03 |
ENSDART00000093358
|
borcs5
|
BLOC-1 related complex subunit 5 |
| chr25_+_21176937 | 0.03 |
ENSDART00000163751
ENSDART00000188930 ENSDART00000187155 |
erc1a
|
ELKS/RAB6-interacting/CAST family member 1a |
| chr19_-_31007417 | 0.03 |
ENSDART00000048144
|
rbbp4
|
retinoblastoma binding protein 4 |
| chr20_-_20355577 | 0.03 |
ENSDART00000018500
|
hif1ab
|
hypoxia inducible factor 1, alpha subunit (basic helix-loop-helix transcription factor) b |
| chr23_+_33752275 | 0.03 |
ENSDART00000007260
|
si:ch211-210c8.6
|
si:ch211-210c8.6 |
| chr16_-_30655980 | 0.03 |
ENSDART00000146508
|
ldlrad4b
|
low density lipoprotein receptor class A domain containing 4b |
| chr11_-_26832685 | 0.03 |
ENSDART00000153519
|
iqsec1b
|
IQ motif and Sec7 domain 1b |
| chr10_-_41397663 | 0.03 |
ENSDART00000158038
|
gpat4
|
glycerol-3-phosphate acyltransferase 4 |
| chr13_+_39182099 | 0.03 |
ENSDART00000131434
|
fam135a
|
family with sequence similarity 135, member A |
| chr4_-_20108833 | 0.03 |
ENSDART00000100867
|
fam3c
|
family with sequence similarity 3, member C |
| chr25_+_7241084 | 0.03 |
ENSDART00000190501
ENSDART00000190588 |
hmg20a
|
high mobility group 20A |
| chr3_-_50147160 | 0.03 |
ENSDART00000191341
|
btr02
|
bloodthirsty-related gene family, member 2 |
| chr19_+_7424347 | 0.03 |
ENSDART00000004622
|
sf3b4
|
splicing factor 3b, subunit 4 |
| chr16_+_42481447 | 0.03 |
ENSDART00000037401
|
herpud2
|
HERPUD family member 2 |
| chr23_+_39611688 | 0.03 |
ENSDART00000034690
|
otud3
|
OTU deubiquitinase 3 |
| chr7_-_37622370 | 0.03 |
ENSDART00000173523
|
nkd1
|
naked cuticle homolog 1 (Drosophila) |
| chr24_+_39027481 | 0.03 |
ENSDART00000085565
|
capn15
|
calpain 15 |
| chr10_+_25947946 | 0.03 |
ENSDART00000064393
|
ufm1
|
ubiquitin-fold modifier 1 |
| chr14_+_8940326 | 0.03 |
ENSDART00000159920
|
rps6kal
|
ribosomal protein S6 kinase a, like |
| chr21_-_22357985 | 0.03 |
ENSDART00000101751
|
skp2
|
S-phase kinase-associated protein 2, E3 ubiquitin protein ligase |
| chr7_+_20966434 | 0.02 |
ENSDART00000185570
|
efnb3b
|
ephrin-B3b |
| chr10_+_19554604 | 0.02 |
ENSDART00000063806
|
atp6v1b2
|
ATPase H+ transporting V1 subunit B2 |
| chr19_-_7321221 | 0.02 |
ENSDART00000092375
|
oxr1b
|
oxidation resistance 1b |
| chr18_-_39188664 | 0.02 |
ENSDART00000162983
|
mapk6
|
mitogen-activated protein kinase 6 |
| chr9_-_29039506 | 0.02 |
ENSDART00000100744
|
tmem177
|
transmembrane protein 177 |
| chr17_+_38295847 | 0.02 |
ENSDART00000008532
|
mbip
|
MAP3K12 binding inhibitory protein 1 |
| chr18_+_18000887 | 0.02 |
ENSDART00000147797
|
si:ch211-212o1.2
|
si:ch211-212o1.2 |
| chr17_-_8886735 | 0.02 |
ENSDART00000121997
|
nkl.3
|
NK-lysin tandem duplicate 3 |
| chr16_-_10071886 | 0.02 |
ENSDART00000136168
ENSDART00000130382 |
ptpn2a
|
protein tyrosine phosphatase, non-receptor type 2, a |
| chr7_-_24994568 | 0.02 |
ENSDART00000002961
|
rcor2
|
REST corepressor 2 |
| chr25_+_17870799 | 0.02 |
ENSDART00000136797
|
btbd10a
|
BTB (POZ) domain containing 10a |
| chr18_+_3140682 | 0.02 |
ENSDART00000166382
|
clns1a
|
chloride channel, nucleotide-sensitive, 1A |
| chr2_-_43635777 | 0.02 |
ENSDART00000148633
|
itgb1b.1
|
integrin, beta 1b.1 |
| chr19_+_28291062 | 0.02 |
ENSDART00000163382
|
lpcat1
|
lysophosphatidylcholine acyltransferase 1 |
| chr4_-_72036230 | 0.02 |
ENSDART00000172108
|
pyroxd1
|
pyridine nucleotide-disulphide oxidoreductase domain 1 |
| chr21_+_20715020 | 0.02 |
ENSDART00000015224
|
gadd45gb.1
|
growth arrest and DNA-damage-inducible, gamma b, tandem duplicate 1 |
| chr16_+_38659475 | 0.02 |
ENSDART00000023238
|
emc2
|
ER membrane protein complex subunit 2 |
| chr7_-_55051692 | 0.02 |
ENSDART00000170637
|
tpcn2
|
two pore segment channel 2 |
| chr2_-_45510699 | 0.02 |
ENSDART00000024034
ENSDART00000145634 |
gpsm2
|
G protein signaling modulator 2 |
| chr7_-_23873173 | 0.02 |
ENSDART00000101408
|
zgc:162171
|
zgc:162171 |
| chr18_+_13275735 | 0.02 |
ENSDART00000148127
|
plcg2
|
phospholipase C, gamma 2 |
| chr7_-_23996133 | 0.02 |
ENSDART00000173761
|
si:dkey-183c6.8
|
si:dkey-183c6.8 |
| chr1_+_41465771 | 0.02 |
ENSDART00000010211
|
paip2b
|
poly(A) binding protein interacting protein 2B |
| chr8_-_36399884 | 0.02 |
ENSDART00000108538
|
si:zfos-2070c2.3
|
si:zfos-2070c2.3 |
| chr4_+_16777715 | 0.02 |
ENSDART00000148344
|
BX649476.1
|
|
| chr16_+_32014552 | 0.02 |
ENSDART00000047570
|
mboat7
|
membrane bound O-acyltransferase domain containing 7 |
Network of associatons between targets according to the STRING database.
First level regulatory network of hoxd12a
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | GO:0009449 | gamma-aminobutyric acid biosynthetic process(GO:0009449) |
| 0.0 | 0.1 | GO:0006564 | L-serine biosynthetic process(GO:0006564) |
| 0.0 | 0.1 | GO:1901890 | positive regulation of cell junction assembly(GO:1901890) |
| 0.0 | 0.1 | GO:0070221 | sulfide oxidation(GO:0019418) sulfide oxidation, using sulfide:quinone oxidoreductase(GO:0070221) |
| 0.0 | 0.1 | GO:0000379 | tRNA-type intron splice site recognition and cleavage(GO:0000379) |
| 0.0 | 0.1 | GO:0034080 | chromatin remodeling at centromere(GO:0031055) CENP-A containing nucleosome assembly(GO:0034080) CENP-A containing chromatin organization(GO:0061641) |
| 0.0 | 0.2 | GO:0006878 | cellular copper ion homeostasis(GO:0006878) |
| 0.0 | 0.2 | GO:0001964 | startle response(GO:0001964) |
| 0.0 | 0.1 | GO:0002188 | translation reinitiation(GO:0002188) |
| 0.0 | 0.2 | GO:0060142 | regulation of syncytium formation by plasma membrane fusion(GO:0060142) |
| 0.0 | 0.0 | GO:0001774 | microglial cell activation(GO:0001774) |
| 0.0 | 0.1 | GO:0033687 | osteoblast proliferation(GO:0033687) |
| 0.0 | 0.0 | GO:0032530 | regulation of microvillus organization(GO:0032530) regulation of microvillus assembly(GO:0032534) |
| 0.0 | 0.1 | GO:0006177 | GMP biosynthetic process(GO:0006177) GMP metabolic process(GO:0046037) |
| 0.0 | 0.0 | GO:0003245 | growth involved in heart morphogenesis(GO:0003241) cardiac chamber ballooning(GO:0003242) cardiac muscle tissue growth involved in heart morphogenesis(GO:0003245) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | GO:0016600 | flotillin complex(GO:0016600) |
| 0.0 | 0.1 | GO:0031371 | ubiquitin conjugating enzyme complex(GO:0031371) |
| 0.0 | 0.1 | GO:0000214 | tRNA-intron endonuclease complex(GO:0000214) |
| 0.0 | 0.1 | GO:0089701 | U2AF(GO:0089701) |
| 0.0 | 0.1 | GO:0005964 | phosphorylase kinase complex(GO:0005964) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | GO:0004351 | glutamate decarboxylase activity(GO:0004351) |
| 0.0 | 0.3 | GO:0001972 | retinoic acid binding(GO:0001972) |
| 0.0 | 0.2 | GO:0016531 | copper chaperone activity(GO:0016531) |
| 0.0 | 0.1 | GO:0070224 | sulfide:quinone oxidoreductase activity(GO:0070224) |
| 0.0 | 0.1 | GO:0003938 | IMP dehydrogenase activity(GO:0003938) |
| 0.0 | 0.2 | GO:0000340 | RNA 7-methylguanosine cap binding(GO:0000340) |
| 0.0 | 0.1 | GO:0004743 | pyruvate kinase activity(GO:0004743) |
| 0.0 | 0.1 | GO:0016892 | endoribonuclease activity, producing 3'-phosphomonoesters(GO:0016892) |
| 0.0 | 0.1 | GO:0005035 | death receptor activity(GO:0005035) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | ST TYPE I INTERFERON PATHWAY | Type I Interferon (alpha/beta IFN) Pathway |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | REACTOME PD1 SIGNALING | Genes involved in PD-1 signaling |
| 0.0 | 0.4 | REACTOME STRIATED MUSCLE CONTRACTION | Genes involved in Striated Muscle Contraction |
| 0.0 | 0.1 | REACTOME DIGESTION OF DIETARY CARBOHYDRATE | Genes involved in Digestion of dietary carbohydrate |
| 0.0 | 0.1 | REACTOME TRANSPORT OF ORGANIC ANIONS | Genes involved in Transport of organic anions |
| 0.0 | 0.1 | REACTOME AMINO ACID SYNTHESIS AND INTERCONVERSION TRANSAMINATION | Genes involved in Amino acid synthesis and interconversion (transamination) |