Project
PRJNA207719: Tissue specific transcriptome profiling
Navigation
Downloads
Results for hoxd4a
Z-value: 0.72
Transcription factors associated with hoxd4a
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
hoxd4a
|
ENSDARG00000059276 | homeobox D4a |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| hoxd4a | dr11_v1_chr9_-_1951144_1951144 | -0.22 | 7.2e-01 | Click! |
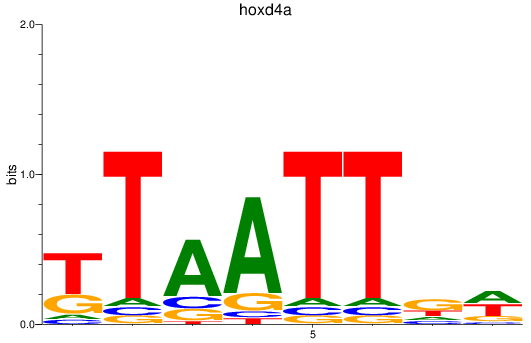
Activity profile of hoxd4a motif
Sorted Z-values of hoxd4a motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr22_+_7439186 | 0.73 |
ENSDART00000190667
|
zgc:92041
|
zgc:92041 |
| chr16_+_26017360 | 0.66 |
ENSDART00000149466
|
prss59.2
|
protease, serine, 59, tandem duplicate 2 |
| chr3_-_31258459 | 0.54 |
ENSDART00000177928
|
LO017965.1
|
|
| chr16_+_23403602 | 0.47 |
ENSDART00000159848
|
s100w
|
S100 calcium binding protein W |
| chr1_+_54655160 | 0.38 |
ENSDART00000190319
|
si:ch211-202h22.7
|
si:ch211-202h22.7 |
| chr2_+_16160906 | 0.38 |
ENSDART00000135783
|
selenoj
|
selenoprotein J |
| chr19_+_43297546 | 0.37 |
ENSDART00000168002
|
laptm5
|
lysosomal protein transmembrane 5 |
| chr23_+_4689626 | 0.36 |
ENSDART00000131532
|
gp9
|
glycoprotein IX (platelet) |
| chr3_-_32337653 | 0.35 |
ENSDART00000156918
ENSDART00000156551 |
si:dkey-16p21.8
|
si:dkey-16p21.8 |
| chr21_-_42831033 | 0.35 |
ENSDART00000160998
|
stk10
|
serine/threonine kinase 10 |
| chr5_+_27897504 | 0.35 |
ENSDART00000130936
|
adam28
|
ADAM metallopeptidase domain 28 |
| chr21_-_22689805 | 0.34 |
ENSDART00000157560
ENSDART00000110792 |
gig2e
|
grass carp reovirus (GCRV)-induced gene 2e |
| chr11_+_10975012 | 0.33 |
ENSDART00000192872
|
itgb6
|
integrin, beta 6 |
| chr10_-_41980797 | 0.33 |
ENSDART00000076575
|
rhof
|
ras homolog family member F |
| chr11_+_1608348 | 0.31 |
ENSDART00000162438
|
si:dkey-40c23.3
|
si:dkey-40c23.3 |
| chr20_+_15600167 | 0.31 |
ENSDART00000171991
|
faslg
|
Fas ligand (TNF superfamily, member 6) |
| chr4_-_14915268 | 0.31 |
ENSDART00000067040
|
si:dkey-180p18.9
|
si:dkey-180p18.9 |
| chr4_+_7822773 | 0.31 |
ENSDART00000171391
|
si:ch1073-67j19.2
|
si:ch1073-67j19.2 |
| chr12_+_20352400 | 0.30 |
ENSDART00000066383
|
hbae5
|
hemoglobin, alpha embryonic 5 |
| chr5_-_66768121 | 0.29 |
ENSDART00000141095
|
FERMT3 (1 of many)
|
im:7154036 |
| chr2_+_36121373 | 0.29 |
ENSDART00000187002
|
CT867973.2
|
|
| chr22_+_38037530 | 0.28 |
ENSDART00000012212
|
commd2
|
COMM domain containing 2 |
| chr23_-_1017605 | 0.28 |
ENSDART00000138290
|
cdh26.1
|
cadherin 26, tandem duplicate 1 |
| chr23_+_19813677 | 0.27 |
ENSDART00000139192
ENSDART00000142308 |
emd
|
emerin (Emery-Dreifuss muscular dystrophy) |
| chr14_+_15620640 | 0.27 |
ENSDART00000188867
|
si:dkey-203a12.9
|
si:dkey-203a12.9 |
| chr6_+_36839509 | 0.26 |
ENSDART00000190605
ENSDART00000104160 |
zgc:110788
|
zgc:110788 |
| chr23_-_16485190 | 0.26 |
ENSDART00000155038
|
si:dkeyp-100a5.4
|
si:dkeyp-100a5.4 |
| chr6_+_36381709 | 0.26 |
ENSDART00000004727
|
rhcgl1
|
Rh family, C glycoprotein, like 1 |
| chr13_+_30692669 | 0.25 |
ENSDART00000187818
|
CR762483.1
|
|
| chr17_+_25481466 | 0.25 |
ENSDART00000139451
|
aim1a
|
crystallin beta-gamma domain containing 1a |
| chr6_+_39098397 | 0.25 |
ENSDART00000003716
ENSDART00000188655 |
prss60.2
|
protease, serine, 60.2 |
| chr9_+_29616854 | 0.25 |
ENSDART00000033902
ENSDART00000143493 |
phf11
|
PHD finger protein 11 |
| chr14_-_5678457 | 0.24 |
ENSDART00000012116
|
tlx2
|
T cell leukemia homeobox 2 |
| chr12_-_990149 | 0.24 |
ENSDART00000054367
|
kdelr2b
|
KDEL (Lys-Asp-Glu-Leu) endoplasmic reticulum protein retention receptor 2b |
| chr2_+_33326522 | 0.23 |
ENSDART00000056655
|
klf17
|
Kruppel-like factor 17 |
| chr7_-_3429874 | 0.23 |
ENSDART00000132330
|
si:ch211-285c6.1
|
si:ch211-285c6.1 |
| chr14_+_22022441 | 0.23 |
ENSDART00000149121
|
clcf1
|
cardiotrophin-like cytokine factor 1 |
| chr6_-_27891961 | 0.23 |
ENSDART00000155116
|
im:7152348
|
im:7152348 |
| chr13_-_37631092 | 0.22 |
ENSDART00000108855
|
si:dkey-188i13.7
|
si:dkey-188i13.7 |
| chr11_+_1602916 | 0.22 |
ENSDART00000184434
ENSDART00000112597 ENSDART00000192165 |
si:dkey-40c23.2
si:dkey-40c23.3
|
si:dkey-40c23.2 si:dkey-40c23.3 |
| chr7_+_12927944 | 0.22 |
ENSDART00000027329
|
fth1a
|
ferritin, heavy polypeptide 1a |
| chr12_-_34435604 | 0.22 |
ENSDART00000115088
|
birc5a
|
baculoviral IAP repeat containing 5a |
| chr10_-_24759616 | 0.21 |
ENSDART00000079528
|
ilk
|
integrin-linked kinase |
| chr10_+_17776981 | 0.20 |
ENSDART00000141693
|
ccl19b
|
chemokine (C-C motif) ligand 19b |
| chr18_+_13309862 | 0.20 |
ENSDART00000131555
|
si:ch211-260p9.3
|
si:ch211-260p9.3 |
| chr9_-_14084743 | 0.20 |
ENSDART00000056105
|
fer1l6
|
fer-1-like family member 6 |
| chr25_+_18563476 | 0.20 |
ENSDART00000170841
|
cav1
|
caveolin 1 |
| chr22_+_8753365 | 0.19 |
ENSDART00000106086
|
si:dkey-182g1.2
|
si:dkey-182g1.2 |
| chr5_+_9246458 | 0.19 |
ENSDART00000081772
|
susd1
|
sushi domain containing 1 |
| chr8_+_22377770 | 0.19 |
ENSDART00000132841
|
si:dkey-23c22.5
|
si:dkey-23c22.5 |
| chr4_+_15944245 | 0.19 |
ENSDART00000134594
|
si:dkey-117n7.3
|
si:dkey-117n7.3 |
| chr11_-_22303678 | 0.19 |
ENSDART00000159527
ENSDART00000159681 |
tfeb
|
transcription factor EB |
| chr6_-_8480815 | 0.19 |
ENSDART00000162300
|
rasal3
|
RAS protein activator like 3 |
| chr23_-_6865946 | 0.18 |
ENSDART00000056426
|
ftr58
|
finTRIM family, member 58 |
| chr24_+_2495197 | 0.18 |
ENSDART00000146887
|
f13a1a.1
|
coagulation factor XIII, A1 polypeptide a, tandem duplicate 1 |
| chr18_-_48296793 | 0.18 |
ENSDART00000032184
ENSDART00000193076 |
CABZ01069595.1
|
|
| chr15_-_1822548 | 0.17 |
ENSDART00000082026
ENSDART00000180230 |
mmp28
|
matrix metallopeptidase 28 |
| chr25_+_31277415 | 0.17 |
ENSDART00000036275
|
tnni2a.4
|
troponin I type 2a (skeletal, fast), tandem duplicate 4 |
| chr1_-_999556 | 0.17 |
ENSDART00000170884
ENSDART00000172235 |
gart
|
phosphoribosylglycinamide formyltransferase |
| chr15_-_17813680 | 0.17 |
ENSDART00000158556
|
CT573342.2
|
|
| chr11_-_40101246 | 0.17 |
ENSDART00000161083
|
tnfrsf9b
|
tumor necrosis factor receptor superfamily, member 9b |
| chr24_-_37640705 | 0.16 |
ENSDART00000066583
|
zgc:112496
|
zgc:112496 |
| chr5_+_66433287 | 0.16 |
ENSDART00000170757
|
kntc1
|
kinetochore associated 1 |
| chr1_+_58375871 | 0.16 |
ENSDART00000166736
|
CU914631.1
|
|
| chr1_-_44157273 | 0.16 |
ENSDART00000166618
|
slc43a3a
|
solute carrier family 43, member 3a |
| chr23_-_31913231 | 0.16 |
ENSDART00000146852
ENSDART00000085054 |
mtfr2
|
mitochondrial fission regulator 2 |
| chr16_+_29509133 | 0.15 |
ENSDART00000112116
|
ctss2.1
|
cathepsin S, ortholog2, tandem duplicate 1 |
| chr10_-_26202766 | 0.15 |
ENSDART00000136393
|
fhdc3
|
FH2 domain containing 3 |
| chr16_+_7242610 | 0.15 |
ENSDART00000081477
|
sri
|
sorcin |
| chr19_-_43757568 | 0.15 |
ENSDART00000058491
|
ppt1
|
palmitoyl-protein thioesterase 1 (ceroid-lipofuscinosis, neuronal 1, infantile) |
| chr23_-_31913069 | 0.15 |
ENSDART00000135526
|
mtfr2
|
mitochondrial fission regulator 2 |
| chr15_-_20400423 | 0.15 |
ENSDART00000081290
ENSDART00000156813 ENSDART00000192427 |
rnaset2l
|
ribonuclease T2, like |
| chr5_+_28770273 | 0.15 |
ENSDART00000114473
|
trafd1
|
TRAF-type zinc finger domain containing 1 |
| chr25_+_31276842 | 0.15 |
ENSDART00000187238
|
tnni2a.4
|
troponin I type 2a (skeletal, fast), tandem duplicate 4 |
| chr14_-_899170 | 0.15 |
ENSDART00000165211
ENSDART00000031992 |
rgs14a
|
regulator of G protein signaling 14a |
| chr22_+_8753092 | 0.14 |
ENSDART00000140720
|
si:dkey-182g1.2
|
si:dkey-182g1.2 |
| chr11_+_37096520 | 0.14 |
ENSDART00000181503
|
card19
|
caspase recruitment domain family, member 19 |
| chr7_-_2129076 | 0.14 |
ENSDART00000182385
|
si:cabz01007807.1
|
si:cabz01007807.1 |
| chr25_+_29474982 | 0.14 |
ENSDART00000130410
|
il17rel
|
interleukin 17 receptor E-like |
| chr14_-_35462148 | 0.13 |
ENSDART00000045809
|
srpx2
|
sushi-repeat containing protein, X-linked 2 |
| chr18_+_3572314 | 0.13 |
ENSDART00000169814
ENSDART00000157819 |
serp1
|
stress-associated endoplasmic reticulum protein 1 |
| chr1_-_35695614 | 0.13 |
ENSDART00000133813
ENSDART00000145291 ENSDART00000176451 |
si:dkey-27h10.2
|
si:dkey-27h10.2 |
| chr14_+_26247319 | 0.13 |
ENSDART00000192793
|
CCDC69
|
coiled-coil domain containing 69 |
| chr3_+_26123736 | 0.13 |
ENSDART00000010477
|
hsp70.3
|
heat shock cognate 70-kd protein, tandem duplicate 3 |
| chr2_+_3201345 | 0.12 |
ENSDART00000130349
|
wnt9a
|
wingless-type MMTV integration site family, member 9A |
| chr1_-_45213565 | 0.12 |
ENSDART00000145757
|
ddx39aa
|
DEAD (Asp-Glu-Ala-Asp) box polypeptide 39Aa |
| chr1_-_18811517 | 0.12 |
ENSDART00000142026
|
si:dkey-167i21.2
|
si:dkey-167i21.2 |
| chr1_-_45662774 | 0.12 |
ENSDART00000042158
|
serhl
|
serine hydrolase-like |
| chr14_-_3381303 | 0.12 |
ENSDART00000171601
|
im:7150988
|
im:7150988 |
| chr16_-_47427016 | 0.11 |
ENSDART00000074575
|
sept7b
|
septin 7b |
| chr22_+_37631234 | 0.11 |
ENSDART00000007346
|
psmd1
|
proteasome 26S subunit, non-ATPase 1 |
| chr24_-_2450597 | 0.11 |
ENSDART00000188080
ENSDART00000093331 |
rreb1a
|
ras responsive element binding protein 1a |
| chr17_-_2578026 | 0.11 |
ENSDART00000065821
|
zp3.2
|
zona pellucida glycoprotein 3, tandem duplicate 2 |
| chr22_+_18166660 | 0.11 |
ENSDART00000105432
|
borcs8
|
BLOC-1 related complex subunit 8 |
| chr25_+_29474583 | 0.11 |
ENSDART00000191189
|
il17rel
|
interleukin 17 receptor E-like |
| chr12_+_25223843 | 0.11 |
ENSDART00000077180
ENSDART00000127454 ENSDART00000122665 |
mta3
|
metastasis associated 1 family, member 3 |
| chr17_+_30843881 | 0.11 |
ENSDART00000149600
ENSDART00000148547 |
tpp1
|
tripeptidyl peptidase I |
| chr23_+_24085531 | 0.10 |
ENSDART00000139710
|
ttll10
|
tubulin tyrosine ligase-like family, member 10 |
| chr19_-_1855368 | 0.10 |
ENSDART00000029646
|
rplp1
|
ribosomal protein, large, P1 |
| chr17_-_25649079 | 0.10 |
ENSDART00000130955
|
ppp1cb
|
protein phosphatase 1, catalytic subunit, beta isozyme |
| chr2_-_45135591 | 0.10 |
ENSDART00000014691
ENSDART00000123916 |
eif4e2
|
eukaryotic translation initiation factor 4E family member 2 |
| chr11_+_11303458 | 0.10 |
ENSDART00000162486
ENSDART00000160703 |
si:dkey-23f9.4
|
si:dkey-23f9.4 |
| chr7_-_26532089 | 0.10 |
ENSDART00000121698
|
senp3b
|
SUMO1/sentrin/SMT3 specific peptidase 3b |
| chr3_+_21225750 | 0.09 |
ENSDART00000139213
|
zgc:153968
|
zgc:153968 |
| chr11_+_36665359 | 0.09 |
ENSDART00000166144
|
si:ch211-11c3.9
|
si:ch211-11c3.9 |
| chr23_+_39695827 | 0.09 |
ENSDART00000113893
ENSDART00000186679 |
tmco4
|
transmembrane and coiled-coil domains 4 |
| chr18_+_7073130 | 0.09 |
ENSDART00000101216
ENSDART00000148947 |
si:dkey-88e18.2
|
si:dkey-88e18.2 |
| chr15_+_45640906 | 0.09 |
ENSDART00000149361
ENSDART00000149079 |
sagb
|
S-antigen; retina and pineal gland (arrestin) b |
| chr24_+_39638555 | 0.09 |
ENSDART00000078313
|
luc7l
|
LUC7-like (S. cerevisiae) |
| chr25_-_2723682 | 0.09 |
ENSDART00000113382
|
adpgk
|
ADP-dependent glucokinase |
| chr16_-_17541890 | 0.08 |
ENSDART00000131328
|
clcn1b
|
chloride channel, voltage-sensitive 1b |
| chr24_-_3426620 | 0.08 |
ENSDART00000184346
|
nck1b
|
NCK adaptor protein 1b |
| chr8_-_50888806 | 0.08 |
ENSDART00000053750
|
acsl2
|
acyl-CoA synthetase long chain family member 2 |
| chr22_+_37631034 | 0.08 |
ENSDART00000159016
ENSDART00000193346 |
psmd1
|
proteasome 26S subunit, non-ATPase 1 |
| chr9_+_48219111 | 0.08 |
ENSDART00000111225
ENSDART00000145972 |
ccdc173
|
coiled-coil domain containing 173 |
| chr24_+_21621654 | 0.08 |
ENSDART00000002595
|
rpl21
|
ribosomal protein L21 |
| chr10_+_39893439 | 0.08 |
ENSDART00000003435
|
smfn
|
small fragment nuclease |
| chr7_-_30280934 | 0.08 |
ENSDART00000126741
|
serf2
|
small EDRK-rich factor 2 |
| chr11_+_18183220 | 0.08 |
ENSDART00000113468
|
LO018315.10
|
|
| chr25_+_8407892 | 0.08 |
ENSDART00000153536
|
muc5.2
|
mucin 5.2 |
| chr14_-_7137808 | 0.08 |
ENSDART00000054803
|
trpt1
|
tRNA phosphotransferase 1 |
| chr23_-_35491587 | 0.08 |
ENSDART00000153794
|
fam110c
|
family with sequence similarity 110, member C |
| chr24_+_38301080 | 0.07 |
ENSDART00000105672
|
mybpc2b
|
myosin binding protein C, fast type b |
| chr6_-_15641686 | 0.07 |
ENSDART00000135583
|
mlpha
|
melanophilin a |
| chr4_+_14981854 | 0.07 |
ENSDART00000067046
|
cax1
|
cation/H+ exchanger protein 1 |
| chr11_-_41220794 | 0.07 |
ENSDART00000192895
|
mrps16
|
mitochondrial ribosomal protein S16 |
| chr22_-_19552796 | 0.07 |
ENSDART00000148088
ENSDART00000105485 |
si:dkey-78l4.14
|
si:dkey-78l4.14 |
| chr7_-_73851280 | 0.07 |
ENSDART00000190053
|
FP236812.3
|
|
| chr25_-_2723902 | 0.07 |
ENSDART00000143721
ENSDART00000175224 |
adpgk
|
ADP-dependent glucokinase |
| chr2_-_47923803 | 0.07 |
ENSDART00000056306
|
ftr24
|
finTRIM family, member 24 |
| chr8_+_23783066 | 0.07 |
ENSDART00000062968
|
si:ch211-163l21.8
|
si:ch211-163l21.8 |
| chr13_+_48359573 | 0.07 |
ENSDART00000161959
ENSDART00000165311 |
msh6
|
mutS homolog 6 (E. coli) |
| chr9_-_22918413 | 0.07 |
ENSDART00000007392
|
arl5a
|
ADP-ribosylation factor-like 5A |
| chr4_-_19921410 | 0.07 |
ENSDART00000184359
ENSDART00000179965 |
BX323820.1
|
|
| chr25_-_1348370 | 0.07 |
ENSDART00000154865
|
itga11b
|
integrin, alpha 11b |
| chr22_-_9618867 | 0.07 |
ENSDART00000131246
|
si:ch73-286h23.4
|
si:ch73-286h23.4 |
| chr18_-_17147803 | 0.07 |
ENSDART00000187986
ENSDART00000169080 |
zc3h18
|
zinc finger CCCH-type containing 18 |
| chr14_-_3070613 | 0.07 |
ENSDART00000193729
ENSDART00000090196 |
slc35a4
|
solute carrier family 35, member A4 |
| chr18_+_29156827 | 0.06 |
ENSDART00000137587
ENSDART00000135633 |
ppfibp2a
|
PTPRF interacting protein, binding protein 2a (liprin beta 2) |
| chr14_-_30905288 | 0.06 |
ENSDART00000173449
ENSDART00000173451 |
si:ch211-126c2.4
|
si:ch211-126c2.4 |
| chr4_-_49582108 | 0.06 |
ENSDART00000154999
|
si:dkey-159n16.2
|
si:dkey-159n16.2 |
| chr19_+_45962016 | 0.06 |
ENSDART00000169710
|
utp23
|
UTP23, small subunit (SSU) processome component, homolog (yeast) |
| chr10_-_43771447 | 0.06 |
ENSDART00000052307
|
arrdc3b
|
arrestin domain containing 3b |
| chr3_-_39259919 | 0.06 |
ENSDART00000124845
|
nmt1a
|
N-myristoyltransferase 1a |
| chr6_-_39270851 | 0.06 |
ENSDART00000148839
|
arhgef25b
|
Rho guanine nucleotide exchange factor (GEF) 25b |
| chr3_+_27770110 | 0.06 |
ENSDART00000017962
|
eci1
|
enoyl-CoA delta isomerase 1 |
| chr22_-_20950448 | 0.06 |
ENSDART00000002029
|
fkbp8
|
FK506 binding protein 8 |
| chr8_-_4972916 | 0.06 |
ENSDART00000101576
|
tmem230b
|
transmembrane protein 230b |
| chr14_-_31618243 | 0.06 |
ENSDART00000016592
|
mmgt1
|
membrane magnesium transporter 1 |
| chr23_-_29553430 | 0.06 |
ENSDART00000157773
ENSDART00000126384 |
ube4b
|
ubiquitination factor E4B, UFD2 homolog (S. cerevisiae) |
| chr13_+_7049823 | 0.06 |
ENSDART00000178997
ENSDART00000161443 |
rnaset2
|
ribonuclease T2 |
| chr25_+_31868268 | 0.06 |
ENSDART00000022325
|
parp16
|
poly (ADP-ribose) polymerase family, member 16 |
| chr6_+_4387150 | 0.06 |
ENSDART00000181283
|
rbm26
|
RNA binding motif protein 26 |
| chr1_+_43686251 | 0.06 |
ENSDART00000074604
ENSDART00000137791 |
cisd2
|
CDGSH iron sulfur domain 2 |
| chr7_+_56676838 | 0.06 |
ENSDART00000112242
|
zgc:194679
|
zgc:194679 |
| chr12_-_2869565 | 0.06 |
ENSDART00000152772
|
zdhhc16b
|
zinc finger, DHHC-type containing 16b |
| chr3_+_25825043 | 0.06 |
ENSDART00000153749
|
pik3r6b
|
phosphoinositide-3-kinase, regulatory subunit 6b |
| chr4_+_16885854 | 0.06 |
ENSDART00000017726
|
etnk1
|
ethanolamine kinase 1 |
| chr10_+_40629616 | 0.05 |
ENSDART00000147476
|
CR396590.9
|
|
| chr13_+_36585399 | 0.05 |
ENSDART00000030211
|
gmfb
|
glia maturation factor, beta |
| chr7_-_20582842 | 0.05 |
ENSDART00000169750
ENSDART00000111719 |
si:dkey-19b23.11
|
si:dkey-19b23.11 |
| chr2_+_16696052 | 0.05 |
ENSDART00000022356
ENSDART00000164329 |
ppp1r7
|
protein phosphatase 1, regulatory (inhibitor) subunit 7 |
| chr14_-_31489410 | 0.05 |
ENSDART00000018347
|
cab39l1
|
calcium binding protein 39, like 1 |
| chr20_-_29633507 | 0.05 |
ENSDART00000040292
|
cpsf3
|
cleavage and polyadenylation specific factor 3 |
| chr12_+_18899396 | 0.05 |
ENSDART00000105858
|
xrcc6
|
X-ray repair complementing defective repair in Chinese hamster cells 6 |
| chr3_-_32873641 | 0.05 |
ENSDART00000075277
|
zgc:113090
|
zgc:113090 |
| chr6_+_41191482 | 0.05 |
ENSDART00000000877
|
opn1mw3
|
opsin 1 (cone pigments), medium-wave-sensitive, 3 |
| chr23_-_16980213 | 0.05 |
ENSDART00000046889
|
dnmt3bb.3
|
DNA (cytosine-5-)-methyltransferase beta, duplicate b.3 |
| chr7_+_69905307 | 0.05 |
ENSDART00000035907
|
sec24d
|
SEC24 homolog D, COPII coat complex component |
| chr7_+_22657566 | 0.04 |
ENSDART00000141048
|
ponzr5
|
plac8 onzin related protein 5 |
| chr20_+_30939178 | 0.04 |
ENSDART00000062556
|
sod2
|
superoxide dismutase 2, mitochondrial |
| chr6_-_2627488 | 0.04 |
ENSDART00000044089
ENSDART00000158333 ENSDART00000155109 |
hyi
|
hydroxypyruvate isomerase |
| chr11_+_42641404 | 0.04 |
ENSDART00000172641
ENSDART00000169938 |
il17rd
|
interleukin 17 receptor D |
| chr25_+_18965430 | 0.04 |
ENSDART00000169742
|
tdg.1
|
thymine DNA glycosylase, tandem duplicate 1 |
| chr6_-_14292307 | 0.04 |
ENSDART00000177852
ENSDART00000061745 |
inpp4ab
|
inositol polyphosphate-4-phosphatase type I Ab |
| chr22_-_2922053 | 0.04 |
ENSDART00000178290
|
pak2b
|
p21 protein (Cdc42/Rac)-activated kinase 2b |
| chr6_+_3693441 | 0.04 |
ENSDART00000065256
|
ppig
|
peptidylprolyl isomerase G (cyclophilin G) |
| chr1_+_50987535 | 0.04 |
ENSDART00000140657
|
mdh1aa
|
malate dehydrogenase 1Aa, NAD (soluble) |
| chr4_-_55592720 | 0.04 |
ENSDART00000164747
|
znf1029
|
zinc finger protein 1029 |
| chr2_-_1364678 | 0.04 |
ENSDART00000011919
ENSDART00000164674 |
ss18
|
synovial sarcoma translocation, chromosome 18 (H. sapiens) |
| chr4_-_11098539 | 0.04 |
ENSDART00000135511
|
si:dkey-21h14.12
|
si:dkey-21h14.12 |
| chr20_-_51186524 | 0.04 |
ENSDART00000027836
ENSDART00000114407 |
rbm25b
|
RNA binding motif protein 25b |
| chr4_+_20536120 | 0.04 |
ENSDART00000125685
|
zmp:0000000650
|
zmp:0000000650 |
| chr4_-_56898328 | 0.04 |
ENSDART00000169189
|
si:dkey-269o24.6
|
si:dkey-269o24.6 |
| chr16_+_8695595 | 0.04 |
ENSDART00000173249
|
si:cabz01093077.1
|
si:cabz01093077.1 |
| chr3_-_34136368 | 0.04 |
ENSDART00000136900
ENSDART00000186125 |
clpp
|
caseinolytic mitochondrial matrix peptidase proteolytic subunit |
| chr4_+_20566371 | 0.04 |
ENSDART00000127576
|
BX248410.1
|
|
| chr24_-_11506054 | 0.04 |
ENSDART00000140217
ENSDART00000106310 |
prpf4bb
|
pre-mRNA processing factor 4Bb |
| chr7_+_5910467 | 0.04 |
ENSDART00000173232
|
si:ch211-113a14.22
|
si:ch211-113a14.22 |
| chr17_-_28097760 | 0.04 |
ENSDART00000149861
|
kdm1a
|
lysine (K)-specific demethylase 1a |
| chr9_+_38457806 | 0.04 |
ENSDART00000142512
|
mcm3ap
|
minichromosome maintenance complex component 3 associated protein |
| chr13_+_9468535 | 0.04 |
ENSDART00000135088
ENSDART00000164270 ENSDART00000099619 ENSDART00000164656 |
HTRA2 (1 of many)
|
si:dkey-265c15.6 |
| chr17_+_21477892 | 0.04 |
ENSDART00000155309
|
pla2g4f.2
|
phospholipase A2, group IVF, tandem duplicate 2 |
| chr11_-_35171768 | 0.03 |
ENSDART00000192896
|
traip
|
TRAF-interacting protein |
| chr3_+_55031685 | 0.03 |
ENSDART00000132587
|
mpg
|
N-methylpurine DNA glycosylase |
| chr7_-_59311165 | 0.03 |
ENSDART00000171105
|
m1ap
|
meiosis 1 associated protein |
| chr18_-_49318823 | 0.03 |
ENSDART00000098419
|
sb:cb81
|
sb:cb81 |
| chr22_-_12337781 | 0.03 |
ENSDART00000188357
ENSDART00000123574 |
zranb3
|
zinc finger, RAN-binding domain containing 3 |
| chr2_-_1468258 | 0.03 |
ENSDART00000114431
|
gpaa1
|
glycosylphosphatidylinositol anchor attachment 1 |
| chr4_+_20576857 | 0.03 |
ENSDART00000125340
|
BX248410.2
|
|
Network of associatons between targets according to the STRING database.
First level regulatory network of hoxd4a
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.2 | GO:0048785 | hatching gland development(GO:0048785) |
| 0.0 | 0.2 | GO:0030299 | intestinal cholesterol absorption(GO:0030299) |
| 0.0 | 0.2 | GO:0035989 | tendon development(GO:0035989) |
| 0.0 | 0.2 | GO:0006621 | protein retention in ER lumen(GO:0006621) |
| 0.0 | 0.2 | GO:0046084 | adenine metabolic process(GO:0046083) adenine biosynthetic process(GO:0046084) |
| 0.0 | 0.2 | GO:0048680 | positive regulation of axon regeneration(GO:0048680) positive regulation of neuron projection regeneration(GO:0070572) |
| 0.0 | 0.3 | GO:0072488 | ammonium transmembrane transport(GO:0072488) |
| 0.0 | 0.2 | GO:0006880 | intracellular sequestering of iron ion(GO:0006880) sequestering of iron ion(GO:0097577) |
| 0.0 | 0.5 | GO:2000401 | regulation of lymphocyte migration(GO:2000401) |
| 0.0 | 0.1 | GO:0006290 | somatic diversification of immune receptors via somatic mutation(GO:0002566) pyrimidine dimer repair(GO:0006290) somatic hypermutation of immunoglobulin genes(GO:0016446) |
| 0.0 | 0.1 | GO:0018008 | N-terminal protein myristoylation(GO:0006499) N-terminal peptidyl-glycine N-myristoylation(GO:0018008) protein myristoylation(GO:0018377) |
| 0.0 | 0.1 | GO:0051876 | pigment granule dispersal(GO:0051876) |
| 0.0 | 0.1 | GO:1903292 | protein localization to Golgi membrane(GO:1903292) |
| 0.0 | 0.4 | GO:0006825 | copper ion transport(GO:0006825) |
| 0.0 | 0.3 | GO:0015671 | oxygen transport(GO:0015671) |
| 0.0 | 0.0 | GO:0070278 | extracellular matrix constituent secretion(GO:0070278) |
| 0.0 | 0.3 | GO:0008625 | extrinsic apoptotic signaling pathway via death domain receptors(GO:0008625) |
| 0.0 | 0.1 | GO:0010259 | multicellular organism aging(GO:0010259) |
| 0.0 | 0.1 | GO:0036075 | endochondral ossification(GO:0001958) replacement ossification(GO:0036075) |
| 0.0 | 0.2 | GO:0002084 | protein depalmitoylation(GO:0002084) macromolecule depalmitoylation(GO:0098734) |
| 0.0 | 0.2 | GO:0072378 | blood coagulation, fibrin clot formation(GO:0072378) |
| 0.0 | 0.3 | GO:0033627 | cell adhesion mediated by integrin(GO:0033627) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.3 | GO:0034685 | integrin alphav-beta6 complex(GO:0034685) |
| 0.1 | 0.2 | GO:0005775 | vacuolar lumen(GO:0005775) lysosomal lumen(GO:0043202) |
| 0.1 | 0.2 | GO:0034515 | proteasome storage granule(GO:0034515) |
| 0.0 | 0.3 | GO:0031838 | haptoglobin-hemoglobin complex(GO:0031838) |
| 0.0 | 0.1 | GO:0032301 | MutSalpha complex(GO:0032301) |
| 0.0 | 0.2 | GO:0030663 | COPI-coated vesicle membrane(GO:0030663) |
| 0.0 | 0.1 | GO:0008180 | COP9 signalosome(GO:0008180) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.2 | GO:0046923 | ER retention sequence binding(GO:0046923) |
| 0.1 | 0.4 | GO:0016531 | copper chaperone activity(GO:0016531) |
| 0.1 | 0.2 | GO:0033897 | ribonuclease T2 activity(GO:0033897) |
| 0.0 | 0.1 | GO:0004377 | GDP-Man:Man3GlcNAc2-PP-Dol alpha-1,2-mannosyltransferase activity(GO:0004377) |
| 0.0 | 0.3 | GO:0031720 | haptoglobin binding(GO:0031720) |
| 0.0 | 0.3 | GO:0030368 | interleukin-17 receptor activity(GO:0030368) |
| 0.0 | 0.1 | GO:0032357 | guanine/thymine mispair binding(GO:0032137) single base insertion or deletion binding(GO:0032138) single thymine insertion binding(GO:0032143) oxidized DNA binding(GO:0032356) oxidized purine DNA binding(GO:0032357) mismatch repair complex binding(GO:0032404) MutLalpha complex binding(GO:0032405) |
| 0.0 | 0.1 | GO:0004379 | glycylpeptide N-tetradecanoyltransferase activity(GO:0004379) |
| 0.0 | 0.1 | GO:0015369 | calcium:proton antiporter activity(GO:0015369) metal ion:proton antiporter activity(GO:0051139) |
| 0.0 | 0.2 | GO:0008199 | ferric iron binding(GO:0008199) |
| 0.0 | 0.5 | GO:0048306 | calcium-dependent protein binding(GO:0048306) |
| 0.0 | 0.2 | GO:0043028 | cysteine-type endopeptidase inhibitor activity involved in apoptotic process(GO:0043027) cysteine-type endopeptidase regulator activity involved in apoptotic process(GO:0043028) |
| 0.0 | 0.1 | GO:0016742 | hydroxymethyl-, formyl- and related transferase activity(GO:0016742) |
| 0.0 | 0.1 | GO:0001965 | G-protein alpha-subunit binding(GO:0001965) |
| 0.0 | 0.3 | GO:0008519 | ammonium transmembrane transporter activity(GO:0008519) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.3 | SA FAS SIGNALING | The TNF-type receptor Fas induces apoptosis on ligand binding. |
| 0.0 | 0.3 | PID INTEGRIN5 PATHWAY | Beta5 beta6 beta7 and beta8 integrin cell surface interactions |
| 0.0 | 0.3 | PID INTEGRIN A4B1 PATHWAY | Alpha4 beta1 integrin signaling events |
| 0.0 | 0.4 | PID ALK1 PATHWAY | ALK1 signaling events |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.4 | REACTOME PLATELET ADHESION TO EXPOSED COLLAGEN | Genes involved in Platelet Adhesion to exposed collagen |
| 0.0 | 0.3 | REACTOME HORMONE SENSITIVE LIPASE HSL MEDIATED TRIACYLGLYCEROL HYDROLYSIS | Genes involved in Hormone-sensitive lipase (HSL)-mediated triacylglycerol hydrolysis |
| 0.0 | 0.3 | REACTOME EXTRINSIC PATHWAY FOR APOPTOSIS | Genes involved in Extrinsic Pathway for Apoptosis |
| 0.0 | 0.2 | REACTOME CELL EXTRACELLULAR MATRIX INTERACTIONS | Genes involved in Cell-extracellular matrix interactions |
| 0.0 | 0.2 | REACTOME PURINE RIBONUCLEOSIDE MONOPHOSPHATE BIOSYNTHESIS | Genes involved in Purine ribonucleoside monophosphate biosynthesis |